Introduction
Acute pancreatitis (AP) is a common acute digestive
tract disease, associated with high rates of morbidity and
mortality. The pathogenesis of AP remains unclear; however,
tryp-sinogen activation is considered to be an important cause and
early event in the development of AP. Its activation in pancreatic
acinar cells has been suggested to be the primary mechanism
underlying the development of AP (1). A previous study suggested that
advanced activation of intracellular trypsinogen may be associated
with an abnormal calcium signaling pathway, low extracellular pH,
lysosome activation and plasminogen aggregation (2). A previous study demonstrated that
autophagy and changes in lysosomal cathepsin are associated with
the occurrence of AP (3), which
has resulted in this area becoming a focus of research.
Autophagy, a form of programmed cell death, involves
the formation of autophagosomes by the engulfing of cellular
degradation products with single- or double-layered membranes. The
products are then transported to lysosomes to form autolysosomes,
and are digested and degraded by intralysosomal hydrolases to
complete the cell metabolism and renewal of cell organelles
(4). A number of studies have
shown that autophagy is associated with the pathogenesis of AP.
Sherwood et al (5)
demonstrated that during early AP, trypsinogen activation occurred
in pancreatic acinar vacuoles, suggesting that autophagy may be
involved in the pathogenesis of AP. During autophagy, trypsinogen
is transported to the lysosomes, which have an acidic environment,
resulting in zymogen activation (5). Mareninova et al (6) detected abnormal autophagy in an in
vitro AP rat model, in which an increased number of zymogens
accumulated in autolysosomes, resulting in trypsinogen activation.
These studies suggested that autophagy participates in the
formation of acinar cell vacuoles in AP and in the process of
trypsinogen activation, therefore, autophagy may be an important
cause of AP.
MicroRNAs are non-coding endogenous RNAs expressed
in eukaryotes and are typically 18–25 nucleotides in length. Mature
microRNAs bind to the 3′-untranslated region (UTR) of target genes
through complementary base pairing to degrade the target mRNA or
inhibit the translation of the mRNAs to regulate gene expression.
Thus, miRNAs post-transcriptionally regulate gene expression
(7). It has been demonstrated that
miRNAs have regulatory functions in numerous pathological and
physiological processes, including cell proliferation,
differentiation and cell death. Recent studies have confirmed that
miRNAs maintain the autophagy process by regulating the expression
of autophagy-related genes, which may result in autophagy
inhibition (8,9). By contrast, selected miRNAs activate
autophagy (10,11). Tekirdag et al (12) demonstrated that miR-181a may
downregulate the mRNA level and protein expression of the
autophagy-related gene ATG5 through binding to the 3′-UTR of ATG5.
miR-181a thus blocks the formation of the ATG12-ATG5-ATG16L1
complex and inhibits the occurrence of autophagy (12). Korkmaz et al (13) demonstrated that autophagy induced
by starvation may be inhibited upon transfection of miR-376b into
the MCF-7 breast cancer cell line and the Huh-7 human liver cancer
cell line. In addition, miR-101, the miR-30 family and miR-130a
were demonstrated to be associated with autophagy (14–16).
However, it is unknown which miRNAs are capable of regulating
autophagy in the pancreatic acinar cells. As autophagy is
associated with trypsinogen activation in the pancreatic acinar
cells in AP and a variety of miRNAs may regulate autophagy,
identifying differential expression of miRNAs and their functions
in pancreatic acinar cell autophagy may be important in the
understanding of AP pathogenesis and in the development of novel
treatments.
The present study established an in vitro
pancreatic acinar cell autophagy model using the AR42J
starvation-induced pancreatic acinar cell line. The differential
expression of miRNAs in AR42J cells during autophagy was detected
using miRNA chips. Bioinformatics methods were used to analyze the
functions of differentially expressed miRNAs with the aim of
identifying novel targets for AP treatment and a greater
understanding of autophagy-promoted AP.
Materials and methods
Cell lines and cell culture
conditions
The AR42J rat pancreatic acinar cell line (China
Center for Type Culture Collection, Wuhan, China) was cultured in
Ham's F12K medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented
with 10% Gibco fetal bovine serum and Gibco antibiotics (100 U/ml
of penicillin and 100 mg/ml of streptomycin) from Thermo Fisher
Scientific, Inc. (Waltham, MA, USA), and maintained in a 5%
CO2 and 37°C incubator.
Establishment of autophagy model in AR42J
cells
In the current study, AR42J cells were treated with
Earle's Balanced Salt Solution (EBSS; Leagene Biotech Co., Ltd.,
Beijing, China) to establish an in vitro autophagy model of
pancreatic acinar cells. EBSS is a balanced salt solution that
maintains cell activity for a short time in a CO2
environment. EBSS maintains the osmotic pressure and activity of
cells, without providing nutrients. Therefore, EBSS is widely used
in the preparation of cell starvation models. For the autophagy
group in the present study, AR42J pancreatic acinar cells growing
at the logarithmic phase were collected. The original medium was
discarded, the cells were rinsed gently three times with
phosphate-buffered saline solution, and EBSS (5 ml/25
cm2 cell culture flask) was added to the AR42J cells for
2, 4 and 6 h at 37°C with an atmosphere of 5% humidified
CO2. The cells were then harvested using Gibco trypsin
(Thermo Fisher Scientific, Inc.), and RNA and protein were
extracted using TRIzol and protein lysis buffer, respectively
(Invitrogen; Thermo Fisher Scientific, Inc.). For the control
group, AR42J cells growing at the logarithmic phase were collected,
and Gibco complete nutrient medium (Thermo Fisher Scientific, Inc.)
was added for 2, 4 and 6 h.
Western blot analysis
AR42J cells were homogenized in the protein lysis
buffer, and the debris was removed by centrifugation at 12,000 × g
for 10 min at 4°C. The bicinchoninic acid assay (Thermo Fisher
Scientific, Inc.) was used to detect protein concentration
according to the protocol of Walker (17). Proteins (50 µg/lane) were
separated by 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred onto Invitrogen polyvinylidene
fluoride membranes (Thermo Fisher Scientific, Inc.). The membranes
were then washed with Tris-buffered saline (10 mM Tris and 150 mM
NaCl) containing 0.05% Tween-20 (TBST; Sigma-Aldrich) and blocked
with 3% bovine serum albumin (BSA; Sigma-Aldrich) for 1 h on an
orbital shaker at room temperature. Rabbit poly-clonal beclin-1
(1:1,000; sc-11427), LC3-II (1:1,000; sc-28266) and GAPDH (1:1,000;
sc-25778) antibodies were purchased from Santa Cruz Biotechnology,
Inc. (Dallas, TX, USA). Goat anti-rabbit IgG horseradish
peroxidase-conjugated secondary antibodies (1:2,000; ZDR-5306) were
purchased from ZSGB Biotechnology (Beijing, China). TBST with 3%
BSA was used to dilute the antibodies. Membranes were incubated
with primary antibodies overnight at 4°C then washed in TBST for 30
min 3 times. Following washes, membranes were incubated with the
secondary antibodies for 2 h at room temperature, and washed again
in TBST for 30 min 3 times. Proteins were visualized by enhanced
chemiluminescence (ECL), with an Amersham ECL Plus kit (GE
Healthcare Life Sciences, Chalfont, UK). After mixing 1 ml ECL
reagents A and B, the membrane was incubated with the mixing ECL
for 2 min, according to the manufacturer's instructions. GAPDH
protein levels were used as a loading control. X-ray film (Thermo
Fisher Scientific, Inc.) and a Medical Film Processor (AFP Imaging
Corporation, Mount Kisco, NY, USA) were used to develop protein
bands in the dark. After using an Epson Perfection 3490 Photo
scanner (Epson America, Inc, Plainfield, IN, USA) to obtain the TIF
images of the protein bands, Image J software, version 1.49
(imagej.nih.gov/ij/) was used to quantify
the expression. The time point of maximum autophagy was
investigated for the following miRNA experiments.
RNA extraction
Total RNA of the AR42J cells treated with EBSS (6 h)
was extracted and isolated using TRIzol reagent according to the
manufacturer's instructions. RNA quantity and quality were measured
using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions with
the following parameters: OD A260/A280 ratio, ~2.0 for pure RNA
(ratios between 1.8 and 2.1 were acceptable); and OD A260/A230
ratio, >1.8. RNA integrity was assessed by standard denaturing
agarose gel electrophoresis.
miRNA microarray
Following RNA extraction, the miRCURY LNA microRNA,
Hi-Power Labeling kit, Hy3/Hy5 (Exiqon A/S, Vedbaek, Denmark) was
used for miRNA labeling according to the manufacturer's
instructions. Briefly, 1 µg of each sample was
3′-end-labeled with the Hy3TM fluorescent label, using the T4 RNA
ligase as follows: RNA (diluted in 2.0 µl water) was mixed
with 1.0 µl CIP buffer and CIP (Exiqon A/S). The mixture was
incubated for 30 min at 37°C, and the interaction of the mixture
was terminated by incubation for 5 min at 95°C. Following
incubation, 3.0 µl labeling buffer, 1.5 µl
fluorescent label (Hy3TM), 2.0 µl dimethyl sulfoxide and 2.0
µl labeling enzyme were added to the mixture. The labeling
reaction was incubated for 1 h at 16°C, and terminated by
incubation for 15 min at 65°C. Upon termination of the labeling
procedure, the Hy3TM-labeled samples were hybridized on the miRCURY
LNA microRNA Array system (version 18.0; Exiqon A/S) according to
the manufacturer's instructions. The total 25 µl mixture
from the Hy3TM-labeled samples with 25 µl hybridization
buffer were first denatured for 2 min at 95°C, incubated on ice for
2 min and then hybridized on the miRCURY LNA microRNA Array system
for 16–20 h at 56°C in a NimbleGen 12-Bay Hybridization system
(Roche Diagnostics, Basel, Switzerland). Following hybridization,
the slides were created, washed numerous times with the wash buffer
(Exiqon A/S) and dried using centrifugation for 5 min at 35.7 × g.
The slides were then scanned using the Axon GenePix 4000B
microarray scanner (Molecular Devices, LLC, Sunnyvale, CA, USA).
Scanned images were imported into the GenePix Pro 6.0 software
(Molecular Devices, LLC) for grid alignment and data extraction.
Replicated miRNAs were averaged and miRNAs with intensities ≥30 in
all samples were selected for calculation of normalization factor.
Expressed data were normalized using the median normalization.
Following normalization, differentially expressed miRNAs were
identified through P-value filtering in the R package siggenes
(18). The Tree-view software,
version 1.6 (GubuSoft, LLC, Belmont, MA, USA) was used to conduct
the cluster analysis of differentially expressed miRNAs.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miRNA expression levels were detected in the
autophagy and control groups by RT-qPCR. Samples were selected from
the original microarray experiments for further RT-qPCR testing
based on sufficient RNA remaining. cDNA synthesis for RT-qPCR
quantification of miRNAs was performed using the TaqMan MicroRNA
Reverse Transcription kit (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the protocol of the manufacturer. RT was
performed using 3µl custom-designed miRNA-specific primers
(Invitrogen; Thermo Fisher Scientific, Inc.). An RT master mix was
prepared as follows: 3 µl RT primer, 5 ng total RNA and 50
units of MultiScribe Reverse Transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc.), 1.5 µl RT Buffer (10X), 0.15
µl dNTPs (100 mM) and 4 units of RNase inhibitor (all from
TaqMan kit) were incubated in PCR a tube for 16°C for 30 min, 42°C
for 30 min and 85°C for 5 min. The cDNA product was then stored at
4°C. The sequencing information of selected miRNAs was obtained
using a TaqMan miRNA assay (Invitrogen; Thermo Fisher Scientific,
Inc.).
The primers were designed as follows:
rno-miR-148b-3p, 5′-ucagugcaucacagaacuuugu-3′; rno-miR-6215,
5′-uuuaggguugcagagccagg-3′; rno-miR-200a-3p,
5′-uaacacugucugguaacgaugu-3′ and rno-miR-425-5p,
5′-aaugacacgaucacucccguuga-3′. The RT-qPCR was performed using the
LightCycler system (Roche Diagnostics). The 20 µl reaction
mixture contained 1.0 µl TaqMan Small RNA Assay (20X), 1.33
µl product from the RT reaction, 10.0 µl TaqMan
Universal PCR Master mix II (2X) and 7.67 µl nuclease-free
water. The reactions were incubated in 96-well optical plates. PCR
conditions were as follows: 10 min hold at 95°C, followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. GAPDH expression was
used as an internal control and the relative expression of each
miRNA was determined by the ΔΔCq method, comparing expression of
the test miRNA to average GAPDH expression, followed by comparison
with the autophagy group to the control group (19).
Bioinformatics analysis
Two databases, miRecords (c1.accurascience.com/miRecords) and miRTarBase
(mirtarbase.mbc.nctu.edu.tw/) were
utilized to screen the known significant target genes of miRNAs.
The miRecords is a database of animal miRNA-mRNA interactions,
including the manual collection of experimental verification of
interactions. Thus far, the miRecords database has collected 548
miRNAs from 9 species and their verified target genes (20). The miRTarBase database
comprehensively collects experimental verification of interactions.
The latest data demonstrated that the database has collected 773
microRNAs and their 2,632 target genes from 14 species (21). Only differentially expressed genes
and miRNAs may be screened as miRNA-mRNA interactions (P-value
cut-off was 0.01). Following integration of the data from the two
databases, Cytoscape 2.8.2 software (www.cytoscape.org) was used to generate the miRNA-mRNA
network. The Human Protein Reference Database (www.hprd.org/) was used to obtain a protein-protein
interaction (PPI) network that included 36,874 edges and 9,453
nodes. The target genes were mapped to the PPI network, and pairs
of interacting genes in which both genes were differentially
expressed were noted, creating sub-networks using the String
software, version 10.0 (www.string-db.org). The Gene Ontology (GO) categories
(geneontology.org) comprise three structured
networks of defined terms to describe gene product attributes
(P-value cut-off was 0.05). Based on the latest data from the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/kegg), a pathway analysis was conducted
using the differentially expressed genes (P-value and FDR cut-offs
were 0.05). Literature mining was conducted to determine which of
the target genes were autophagy-associated. Briefly, the Perl
language was utilized to write a literature review program using
the ActivePerl software, version 5.16.2 (ActiveState Software,
Inc., Vancouver, BC, Canada). The software was then used to
retrieve literature data from the PubMed database (www.ncbi.nlm.nih.gov/pubmed). The search
locations were the titles and abstracts of publications, and the
search keywords were the names of the differentially expressed
genes, and the word 'autophagy'. Finally, manual screening was used
to screen for autophagy-associated genes accurately.
Statistical analysis
Data are expressed as the mean ± standard deviation
and comparisons were made using Student's t-test. P<0.01 was
considered to indicate a statistically significant difference in
gene expression according to the microarray manufacturer's analysis
(Exiqon A/S). Categorical variables were given as numbers and
percentages, and Fisher's exact test was utilized to test their
associations. Statistical analyses of western blotting and RT-qPCR
data were performed using the SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA), and P<0.05 was considered to indicate a
statistically significant difference.
Results
Detection of autophagy in the AR42J
cells
The results of the western blot analysis (Fig. 1) demonstrated that the expression
intensity values of microtubule-associated protein 1 light chain 3
(LC3-II) were 0.81±0.13 and 0.82±0.12 (2 h); 0.76±0.06 and
1.03±0.08 (4 h); and 0.76±0.10 and 1.28±0.10 (6 h) for the control
and autophagy groups, respectively. At the 4 and 6 h points, the
expression of LC3-II in autophagy group was significantly increased
compared with that in control group (4 h, P<0.05; 6 h,
P<0.01). The expression of LC3-II gradually increased in the
autophagy groups in a time-dependent manner (P<0.05).
Furthermore, the expression intensity values of Beclin-1 were
0.42±0.11 and 0.52±0.11 (2 h); 0.51±0.10 and 0.91±0.08 (4 h); and
1.08±0.07 (6 h), for the control and autophagy groups,
respectively. At the 4 and 6 h points, the expression level of
beclin-1 in the autophagy group was significantly increased
compared with that in control group (P<0.01). The expression of
LC3-II gradually increased in the autophagy groups in a
time-dependent manner. The expression of beclin-1 at 4 h was
significantly increased compared with that at 2 h (P<0.01) and
at 6 h was higher than that at 4 h (P<0.05).
Differentially expressed miRNAs in AR42J
autophagy-induced cells
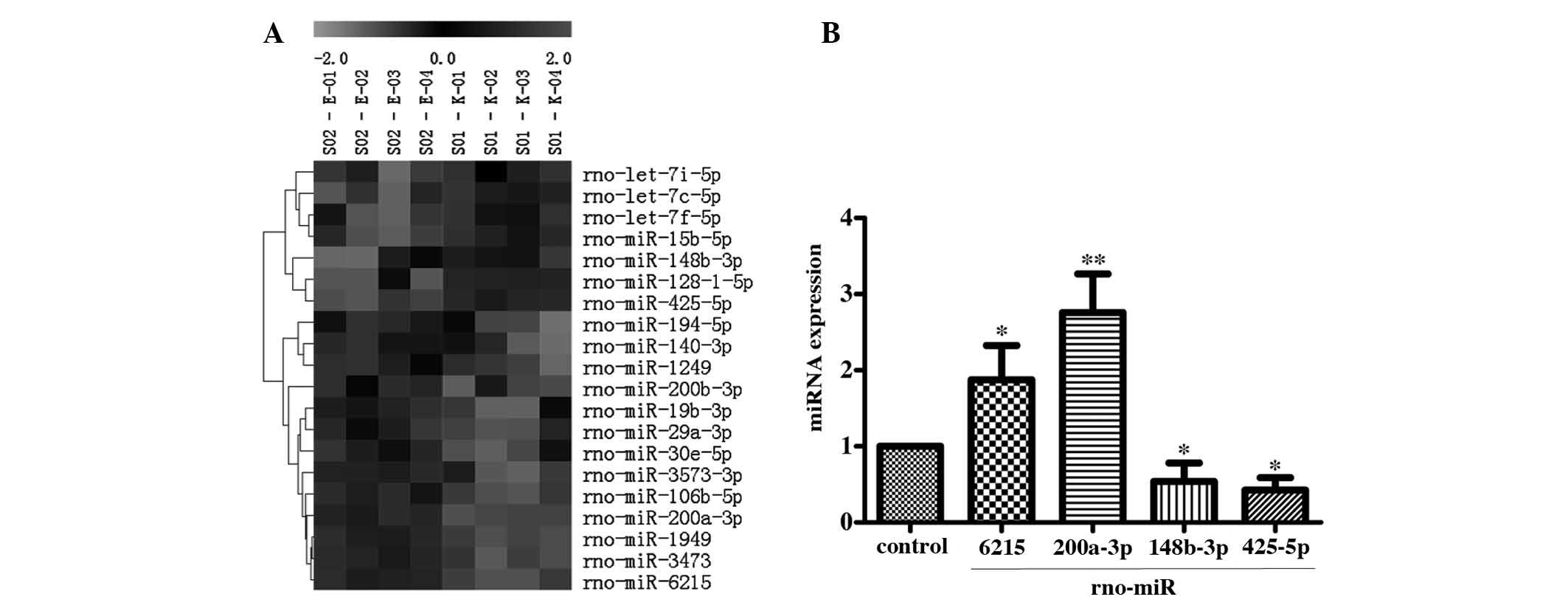
The results of miRNA microarray analysis
demonstrated that 20 miRNAs were differentially expressed (13
upregulated; 7 downregulated). The results of cluster analysis
showed that there was expression correlation among these
differentially expressed miRNAs (Fig.
2A).
Validation of the expression level of
miRNA in AR42J autophagy-induced cells
The result of RT-qRCR detection of the selected
miRNAs showed that the expression levels of two miRNAs,
rno-miR-6215 (1.88±0.45) and rno-miR-200a-3p (2.76±0.50), were
significantly upregulated. In addition, the expression levels of
rno-miR-148b-3p (0.54±0.24) and rno-miR-425-5p (0.42±0.17) were
significantly downregulated in the AR42J autophagy-induced cells
(Fig. 2B) compared with control.
The result was consistent with the results of the microarray.
According to the principle of targeted gene suppression by miRNAs,
the inhibitory effect of the down-regulated miRNA on the target
gene was reduced, resulting in high expression of the target gene.
Therefore, the downregulated miRNAs in autophagy group, such as
rno-miR-148b-3p and rno miR 425 5p, may increase target gene
expression and thus contribute to the progression of autophagy.
Prediction of differentially expressed
miRNA target genes and screening of autophagy-associated genes
Following the combination of data from two miRNA
databases, miRanda and miRecords, the target genes predicted in the
databases as affected genes of differentially expressed miRNAs were
selected. As demonstrated in Fig.
3, only the downregulation of miRNA rno-miR-148b-3p and its 593
target genes were shown to be significant (P<0.05) compared with
the control group.
Through literature mining and manual screening, 10
autophagy-related genes from the 593 target genes were identified;
Atg12, Hspa5, Hspb6, Sqstm1, Cdc37, Tlr4, Pik3c3, Atf3, Irf1 and
Bag3. These genes, directly or indirectly, participated in the
autophagy process (Fig. 4).
 | Figure 4Screening of autophagy-related genes
from the target genes and their affect on the autophagy pathway.
Through literature mining and manual screening, 10
autophagy-related genes were screened from the 593 target genes:
Atg12, Hspa5, Hspb6, Sqstm1, Cdc37, Tlr4, Pik3c3, Atf3, Irf1 and
Bag3. All of these genes, directly or indirectly, participated in
the autophagy pathway. |
Results of the target gene GO analysis
and pathway enrichment
Using DAVID software (david.ncifcrf.gov), GO function analysis was performed
on the target genes. The results demonstrated that the target genes
of miRNAs were enriched in the regulation of phosphorylation,
regulation of transferase activity, regulation of kinase activity,
regulation of protein modification process, response to insulin
stimulus, regulation of programmed cell death and regulation of
cell death pathways. The results analysis of the pathway enrichment
of the miRNA target genes using the KEGG database showed that only
the insulin signaling pathway simultaneously satisfied the
requirements of P<0.05 and FDR<0.05 (Table I).
 | Table IGO analysis. |
Table I
GO analysis.
| Term | Name | P-value | FDR |
|---|
| GO:0042325 | Regulation of
phosphorylation | 8.56E-10 | 1.49E-06 |
| GO:0051338 | Regulation of
transferase activity | 3.86E-08 | 6.72E-05 |
| GO:0043549 | Regulation of
kinase activity | 8.44E-08 | 1.47E-04 |
| GO:0031399 | Regulation of
protein modification process | 1.31E-06 | 2.28E-03 |
| GO:0032868 | Response to insulin
stimulus | 2.74E-06 | 4.75E-03 |
| GO:0043067 | Regulation of
programmed cell death | 1.47E-05 | 2.55E-02 |
| GO:0010941 | Regulation of cell
death | 1.59E-05 | 2.77E-02 |
| Pathway
analysis | | | |
| rno04910 | Insulin signaling
pathway | 7.10E-05 | 8.23E-03 |
PPI network of target genes
The ultimate goal of the post-genomic era is to
recognize the expression patterns and biological functions of all
proteins that implement the spectrum of biological activities. One
of the key challenges is the analysis of the PPI network, which may
increase understanding of cell structures and functions from a
systems point of view. At present, the investigation of the PPI
network using bioinformatics methods demonstrates significant
advantages. In the current study, the String software was utilized
to create the PPI network of the 593 target genes (Fig. 5).
Discussion
Autophagy is a mechanism of self-digestion that
represents the only way to degrade organelles on the cellular
level. Thus, it has been suggested to be involved in pancreatic
self-digestion. Zymogen activation may be the result of lysosomal
degradation of zymogen granules during the autophagy process.
Hashimoto et al (22)
demonstrated that in an ATG5 knockout mouse model of AP, pancreatic
acinar cell autophagy was decreased, trypsinogen activity was
significantly reduced and the severity of AP was reduced. Thus, it
was proposed that the autophagy in the mouse pancreatic acinar
cells had a role in damage at the early stages of AP (22). Previous studies demonstrated that a
large number of abnormal autophagic vacuoles accumulated within
pancreatic acinar cells during AP. A decrease in the lysosomal
proteases led to an increase in autolysosomes, and the deficiency
and imbalance of lysosomal proteases resulted in a decrease of the
degradation capacity of autolysosomes. The accumulation of large
amounts of zymogens in autolysosomes resulted in zymogen activation
and cell injury (6). These
findings support the view of Hashimoto et al (22) that excessive autophagy in
pancreatic acinar cells in the early stages of AP exaggerated the
degree of the disease. Identification of the changes in gene
expression and function of pancreatic acinar cells during autophagy
is of great significance for studies investigating the mechanisms
and treatments of AP.
A variety of proteins are involved in the process of
autophagy, including LC3. LC3 is divided into type I and type II
proteins. LC3-II is located on the membrane of autophagosomes
within cells. The content of this protein is proportional to the
number of autophagic vacuoles (23). Beclin-1, another protein in the
autophagic process, serves a role in the formation of the
proautophagic complex and mediates the localization of other
autophagy proteins to the autophagosome membrane (24). In the present study, an autophagy
model was established using the EBSS starvation method to induce
autophagy in the AR42J cells. Western blot analysis demonstrated
that the increase in the relative protein expression of LC3-II and
Beclin-1 was correlated with the duration of starvation.
Starvation-induced autophagy occurred in a time-dependent manner,
confirming the successful establishment of an AR42J model of
autophagy. It was also observed in order to determine the best time
point for subsequent miRNA chip experiments.
The regulation mechanisms of autophagy are
complicated. Multiple signaling pathways are known to be involved
autophagy, such as the mTOR, insulin and AMPK pathways. miRNAs
regulate autophagy through regulating autophagy-related target
genes in the above mentioned pathways. It is known that in the
process of regulating autophagy, miRNAs mostly inhibit the
occurrence of autophagy, while only a few miRNAs are known to
activate autophagy. Furthermore, the inhibition of autophagy by
miRNAs is mediated by downregulating key molecules of the
autophagosome formation process, such as LC3 and Beclin-1, to
inhibit the processes of autophagosome formation and membrane
extension (10,11,25).
The present study identified 20 differentially expressed miRNAs in
the AR42J autophagy-induced model and predicted their target genes.
Only the downregulated miRNA rno-miR-148b-3p predicted 593 target
genes with statistical significance (P<0.05). Consistent with
the fact that miRNAs mediate their effects through the inhibition
of their target genes, the expression of rno-miR-148b-3p in the
autophagic AR42J cells decreased, leading to a decrease in its
ability to inhibit autophagy-associated target genes, thus
promoting autophagy. The pathway analysis showed that the target
genes of rno-miR-148b-3p were primarily enriched in the insulin
signaling pathway. Yamamoto et al (26) demonstrated that the activation of
the insulin signaling pathway lead to the removal of accumulated
proteins mediated by intracellular macro-autophagy and that this
process required the participation of macroautophagy-associated
proteins, such as Beclin-1 and hvps34 (26). This result suggests that the low
expression of rno-miR-148b-3p during AR42J autophagy may be
involved in the induction of autophagy (Fig. 3). The GO analysis results showed
that the target genes of rno-miR-148b-3p were primarily involved in
the regulation of phosphorylation and other functions, including
'regulation of kinase activity', 'regulation of transferase
activity' and 'regulation of cell death'. Autophagy is a programmed
cell death pathway, and the process of autophagy depends on the
participation of numerous kinases, such as Akt, MAPK and AMPK.
These kinases regulate autophagy by phosphorylating downstream
autophagy-associated genes (27,28).
Therefore, the functions of rno-miR-148b-3p target genes cover a
number of processes of autophagy, and the under-expressed
rno-miR-148b-3p may be an miRNA regulating the autophagy
process.
To further understand the functions of
rno-miR-148b-3p in autophagy in the present study, 10 target genes
associated with autophagy were screened and the pathways of their
participation in the process of autophagy were demonstrated
(Fig. 3). Among these target
genes, ATG12 and Sqstm1/p62 are most closely associated with
autophagy. Previous studies demonstrated that ATG12 and Sqstm1/p62
are necessary factors in the process of autophagy. Atg12, an
autophagy-associated gene, is activated by the E1-like enzyme,
Atg7, and then transported to the E2-like enzyme, Atg10, to combine
with Atg5 and form an autophagosome precursor (29). Sqstm1/p62 was demonstrated to
recognize and engulf ubiquitinated proteins flagged for degradation
through binding to LC3-II in the process of autophagy (30). Thus, rno-miR-148b-3p may directly
participate in the regulation of autophagy through inhibition of
Atg12 and Sqstm1/p62. Furthermore, interferon regulatory factor-1
expression was demonstrated to be correlated with that of Atg7 and
Beclin-1, and affects the survival of breast cancer cells by
regulating autophagy (31). Joo
et al (32) demonstrated
that Cdc37 formed a complex with Hsp90 to regulate Atg13-mediated
autophagy. Merabova et al (33) indicated that over-expressed Bag3
may increase the interaction between Bcl2 and Beclin-1 and then
induce Beclin-1-dependent autophagy. A previous study demonstrated
that TLR4 and p62 mediate lipopolysaccharide-induced autophagy
(34). As demonstrated in Fig. 3, the rno-miR-148b-3p target genes
indirectly affected autophagy by regulating key genes of the
autophagic signaling pathway, further suggesting that
rno-miR-148b-3p has the function of regulating autophagy.
Various biological activities are realized by the
association and dissociation of protein molecules. The interaction
between proteins can form a signal transduction network system that
regulates a variety of cellular physiological activities and
cellular reactions to the change of microenvironment (35). Therefore, PPI networks can serve an
important role in regulating cell activities in different diseases.
An in-depth analysis of PPI network would aid the understanding of
the progress of disease and its molecular mechanisms (36). The current study demonstrated the
interactions among rno-miR-148b-3p target genes through a PPI
network analysis. In addition, numerous interactions were indicated
among the rno-miR-148b-3p target genes and the autophagy pathway
genes. These results suggest that the regulation of autophagy by
rno-miR-148b-3p may involve more complicated mechanisms than a
single gene or pathway.
In conclusion, the current study established an
in vitro model of rat pancreatic acinar cell autophagy in
AR42J cells, identified 20 differentially expressed miRNAs using
the miRNA chip and demonstrated that rno-miR-148b-3p may be a
regulatory miRNA in the autophagic process, using bioinformatics.
The results of the present study provide a starting point for
studies on the pathogenesis and treatment of AP. However, further
research is required to verify the regulatory effects and mechanism
of rno-miR-148b-3p and its target genes on autophagy.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81570579, 81570581,
81370566 and 81170397) and the Harbin Medical University
Postgraduate Innovative Research Projects (grant no.
YJSCX2015-19HYD).
References
|
1
|
Bhatia M, Wong FL, Cao Y, Lau HY, Huang J,
Puneet P and Chevali L: Pathophysiology of acute pancreatitis.
Pancreatology. 5:132–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sah RP and Saluja A: Molecular mechanisms
of pancreatic injury. Curr Opin Gastroenterol. 27:444–451. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohmuraya M and Yamamura K: Autophagy and
acute pancreatitis: A novel autophagy theory for trypsinogen
activation. Autophagy. 4:1060–1062. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimizu S, Yoshida T, Tsujioka M and
Arakawa S: Autophagic cell death and cancer. Int J Mol Sci.
15:3145–3153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherwood MW, Prior IA, Voronina SG, Barrow
SL, Woodsmith JD, Gerasimenko OV, Petersen OH and Tepikin AV:
Activation of trypsinogen in large endocytic vacuoles of pancreatic
acinar cells. Proc Natl Acad Sci USA. 104:5674–5679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mareninova OA, Hermann K, French SW,
O'Konski MS, Pandol SJ, Webster P, Erickson AH, Katunuma N,
Gorelick FS, Gukovsky I and Gukovskaya AS: Impaired autophagic flux
mediates acinar cell vacuole formation and trypsinogen activation
in rodent models of acute pancreatitis. J Clin Invest.
119:3340–3355. 2009.PubMed/NCBI
|
|
7
|
Ambros V: MicroRNAs and developmental
timing. Curr Opin Genet Dev. 21:511–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JK, Yuk JM, Kim SY, Jin HS, Yang CS
and Jo EK: MicroRNA-125a inhibits autophagy activation and
antimicrobial responses during mycobacterial infection. J Immunol.
194:5355–5365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su M, Wang J, Wang C, Wang X, Dong W, Qiu
W, Wang Y, Zhao X, Zou Y, Song L, et al: MicroRNA-221 inhibits
autophagy and promotes heart failure by modulating the
p27/CDK2/mTOR axis. Cell Death Differ. 22:986–999. 2015. View Article : Google Scholar :
|
|
10
|
Jing Z, Han W, Sui X, Xie J and Pan H:
Interaction of autophagy with microRNAs and their potential
therapeutic implications in human cancers. Cancer Lett.
356:332–338. 2015. View Article : Google Scholar
|
|
11
|
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang
JH and Wei YQ: Oncogenic and tumor suppressive roles of microRNAs
in apoptosis and autophagy. Apoptosis. 19:1177–1189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tekirdag KA, Korkmaz G, Ozturk DG, Agami R
and Gozuacik D: MIR181A regulates starvation- and rapamycin-induced
autophagy through targeting of ATG5. Autophagy. 9:374–385. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Korkmaz G, le Sage C, Tekirdag KA, Agami R
and Gozuacik D: miR-376b controls starvation and mTOR
inhibition-related autophagy by targeting ATG4C and BECN1.
Autophagy. 8:165–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Frankel LB, Wen J, Lees M, Høyer-Hansen M,
Farkas T, Krogh A, Jäättelä M and Lund AH: microRNA-101 is a potent
inhibitor of autophagy. EMBO J. 30:4628–4641. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kovaleva V, Mora R, Park YJ, Plass C,
Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer
A, Lichter P and Seiffert M: miRNA-130a targets ATG2B and DICER1 to
inhibit autophagy and trigger killing of chronic lymphocytic
leukemia cells. Cancer Res. 72:1763–1772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang X, Zhong X, Tanyi JL, Shen J, Xu C,
Gao P, Zheng TM, DeMichele A and Zhang L: mir-30d Regulates
multiple genes in the autophagy pathway and impairs autophagy
process in human cancer cells. Biochem Biophys Res Commun.
431:617–622. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Walker JM: The bicinchoninic acid (BCA)
assay for protein quantitation. Methods Mol Biol. 32:5–8.
1994.PubMed/NCBI
|
|
18
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37(Database): D105–D110. 2009.
View Article : Google Scholar
|
|
21
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42(D1):
D78–D85. 2014. View Article : Google Scholar :
|
|
22
|
Hashimoto D, Ohmuraya M, Hirota M,
Yamamoto A, Suyama K, Ida S, Okumura Y, Takahashi E, Kido H, Araki
K, et al: Involvement of autophagy in trypsinogen activation within
the pancreatic acinar cells. J Cell Biol. 181:1065–1072. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meijer AJ and Codogno P: Regulation and
role of autophagy in mammalian cells. Int J Biochem Cell Biol.
36:2445–2462. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mizushima N: Methods for monitoring
autophagy. Int J Biochem Cell Biol. 36:2491–2502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang
Z and Ni X: MiR-204 regulates cardiomyocyte autophagy induced by
ischemia-reperfusion through LC3-II. J Biomed Sci. 18:352011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto A, Cremona ML and Rothman JE:
Autophagy-mediated clearance of huntingtin aggregates triggered by
the insulin-signaling pathway. J Cell Biol. 172:719–731. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan ST, Qin Y, Zhou ZW, He ZX, Zhang X,
Yang T, Yang YX, Wang D, Qiu JX and Zhou SF: Plumbagin induces G2/M
arrest, apoptosis, and autophagy via p38 MAPK- and
PI3K/Akt/mTOR-mediated pathways in human tongue squamous cell
carcinoma cells. Drug Des Devel Ther. 9:1601–1626. 2015.PubMed/NCBI
|
|
28
|
Heras-Sandoval D, Pérez-Rojas JM,
Hernández-Damián J and Pedraza-Chaverri J: The role of
PI3K/AKT/mTOR pathway in the modulation of autophagy and the
clearance of protein aggregates in neurodegeneration. Cell Signal.
26:2694–2701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakatogawa H: Two ubiquitin-like
conjugation systems that mediate membrane formation during
autophagy. Essays Biochem. 55:39–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park S, Choi SG, Yoo SM, Son JH and Jung
YK: Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3
and stimulate mitophagy. Autophagy. 10:1906–1920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schwartz-Roberts JL, Cook KL, Chen C,
Shajahan-Haq AN, Axelrod M, Wärri A, Riggins RB, Jin L, Haddad BR,
Kallakury BV, et al: Interferon regulatory factor-1 signaling
regulates the switch between autophagy and apoptosis to determine
breast cancer cell fate. Cancer Res. 75:1046–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Joo JH, Dorsey FC, Joshi A,
Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R,
Jung CH, Suen DF, et al: Hsp90-Cdc37 chaperone complex regulates
Ulk1- and Atg13-mediated mitophagy. Mol Cell. 43:572–585. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Merabova N, Sariyer IK, Saribas AS,
Knezevic T, Gordon J, Turco MC, Rosati A, Weaver M, Landry J and
Khalili K: WW domain of BAG3 is required for the induction of
autophagy in glioma cells. J Cell Physiol. 230:831–841. 2015.
View Article : Google Scholar :
|
|
34
|
Neal MD, Sodhi CP, Dyer M, Craig BT, Good
M, Jia H, Yazji I, Afrazi A, Richardson WM, Beer-Stolz D, et al: A
critical role for TLR4 induction of autophagy in the regulation of
enterocyte migration and the pathogenesis of necrotizing
enterocolitis. J Immunol. 190:3541–3551. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Giot L, Bader JS, Brouwer C, Chaudhuri A,
Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al: A protein
interaction map of Drosophila melanogaster. Science. 302:1727–1736.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li S, Armstrong CM, Bertin N, Ge H,
Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al:
A map of the interactome network of the metazoan C elegans.
Science. 303:540–543. 2004. View Article : Google Scholar : PubMed/NCBI
|