Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer in Eastern Asia (1).
However, it remains to be a type of cancer with a low 5-year
survival rate worldwide (<10%) when in the advanced stages, due
to difficulties in detection and the high frequency of tumor
recurrence (2,3). Hepatocarcinogenesis involves complex
molecular mechanisms, involving alterations in the structure or
expression of several tumor suppressor genes and oncogenes
(4,5). However, the detailed molecular
mechanisms underlying HCC metastasis remain unclear. Thus, the
identification of novel biomarkers to increase specificity or
sensitivity for early diagnosis and to improve the prognosis of HCC
is required.
Forkhead box J2 (FoxJ2), a member of the forkhead
transcription factor family (6),
has been identified in several mammals and vertebrates (7–9) and
is widely distributed in different organs and tissues, in adults
and in the fetus. FOXJ2 has been suggested to be involved in
positively regulating the progression of the cell cycle or aiding
in tumorigenesis (10). Wang et
al (11) reported that
overexpression of FOXJ2 was able to reduce the migration of breast
cancer cells, and inhibit the metastasis of human breast cancer by
regulating the epithelial-mesenchymal transition (EMT) key markers
E-cadherin and vimentin. In human glioma cells, overexpression of
FOXJ2 has been reported to increase E-cadherin expression and
reduce vimentin expression (12).
Overexpression of FOXJ2 has been observed to significantly inhibit
cell migration, and knockdown of FOXJ2 to promote cellular
motility, thus it was suggested that FOXJ2 suppresses cell
migration and invasion in glioma (12).
The current study aimed to investigate the potential
involvement of FOXJ2 in HCC pathology and to evaluate the
prognostic value of FOXJ2 expression in HCC. It was identified that
FOXJ2 was significantly downregulated in HCC specimens, compared
with adjacent nontumorous tissues. Furthermore, it was observed
that the expression of FOXJ2 was correlated with histological
differentiation, the size of the largest tumor and metastasis, and
Ki-67 expression levels. In vitro, knockdown of FOXJ2
expression was able to promote HepG2 cell proliferation. These
observations may provide novel insight into the mechanisms
underlying HCC development.
Materials and methods
Patients and tissue samples
All the HCC samples used in the current study were
from 110 patients who underwent hepatic resection without
preoperative systemic chemotherapy at The Third People's Hospital
of Liaocheng (Liaocheng, China). The main clinical and pathological
characteristics are presented in Table
I.
 | Table IClinicopathological features of
hepatocellular carcinoma in relation to the FOXJ2/Ki67 expression
pattern. |
Table I
Clinicopathological features of
hepatocellular carcinoma in relation to the FOXJ2/Ki67 expression
pattern.
| Clinicopathological
feature | Total number per
group | FOXJ2 expression
| P-value | χ2
value |
|---|
| Low | High |
|---|
| All cases | 110 | 63 | 47 | | |
| Age (years) | | | | | |
| ≤50 | 61 | 36 | 25 | 0.680 | 0.170 |
| >50 | 49 | 27 | 22 | | |
| Gender | | | | | |
| Male | 89 | 52 | 37 | 0.614 | 0.254 |
| Female | 21 | 11 | 10 | | |
| Histological
differentiation | | | | | |
| Well | 11 | 2 | 9 | 0.005a | 10.638 |
| Moderate | 83 | 48 | 35 | | |
| Poor | 16 | 13 | 3 | | |
| Size of largest tumor
(cm) | | | | | |
| ≤5 | 63 | 28 | 35 | 0.002a | 9.916 |
| >5 | 47 | 35 | 12 | | |
| Tumor number | | | | | |
| Solitary | 74 | 41 | 33 | 0.570 | 0.322 |
| Multiple | 36 | 22 | 14 | | |
| HBV | | | | | |
| Positive | 88 | 54 | 34 | 0.083 | 3.009 |
| Negative | 22 | 9 | 13 | | |
| Liver cirrosis | | | | | |
| Yes | 89 | 11 | 10 | 0.614 | 0.254 |
| No | 21 | 52 | 37 | | |
| Microvascular
invasion | | | | | |
| Yes | 31 | 19 | 12 | 0.594 | 0.285 |
| No | 79 | 44 | 35 | | |
| Metastasis | | | | | |
| Yes | 16 | 50 | 44 | 0.036a | 4.399 |
| No | 94 | 13 | 3 | | |
| Serum AFP value
(ng/ml) | | | | | |
| ≤50 | 42 | 20 | 22 | 0.108 | 2.587 |
| >50 | 68 | 43 | 25 | | |
| Ki67 | | | | | |
| Low | 59 | 17 | 42 | <0.001a | 22.848 |
| High | 51 | 38 | 13 | | |
Immunohistochemistry
Tissues were formalin-fixed and paraffin-embedded
for immunohistochemical study. The sections were dewaxed in xylene
and rehydrated in graded ethanol. Thereafter, the sections were
processed in 10 mmol/l citrate buffer (pH 6.0) and heated to 105°C
in an autoclave for three cycles of 5 min each for antigen
retrieval. Subsequently, endogenous peroxidase activity was blocked
by soaking in 0.3% hydrogen peroxide for 15 min subsequent to
resting of the sections room temperature for 1 h. Goat serum (Wuhan
Huamei Biotech Co., Ltd., Wuhan, China) was applied to block any
nonspecific reactions for 15 min. The sections were then incubated
overnight at 4°C with goat anti-human FOXJ2 poly-clonal antibody
(1:150; sc-54374; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
and anti-Ki67 mouse monoclonal antibody (1:100; sc-23900; Santa
Cruz Biotechnology, Inc.). Negative control slides were processed
in parallel using a nonspecific immunoglobulin IgG (Sigma-Aldrich,
St. Louis, MO, USA) at the same concentration as the primary
antibody. All slides were processed using the
peroxidase-antiperoxidase method (Dako, Hamburg, Germany).
Subsequent to rinsing in water, the sections were counterstained
with hematoxylin, dehydrated and coverslipped. All of the
immunostained sections were evaluated in a blinded manner without
knowledge of the clinical and pathological features of the
patients. For assessment of FOXJ2, five high-power fields in each
specimen were selected randomly, and nuclear (cytoplasmic) staining
was examined. Greater than 500 cells were counted to determine the
labeling index, which represented the percentage of immunostained
cells relative to the total number of cells. In half of the
samples, staining was repeated twice to avoid possible technical
errors, however similar results were obtained in these samples. A
second investigator using a multihead microscope assessed the
obtained results, and a consensus was achieved.
Cell culture
One normal hepatocyte cell lines (L02) and 3 HCC
cell lines (HepG2, Huh7 and Hep1) were obtained from the Institute
of Cell Biology of the Chinese Academy of Sciences (Shanghai).
Cells were cultured in Roswell Park Memorial Institute 1640 and
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS), and 100 U/ml penicillin-streptomycin
mixture (all mediums from Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) in 5% CO2 at 37°C.
The sequences of the primers for FOXJ2 were as
follows: 5′-AAG CTT GGA AGT GCC TCC CAG-3′, and the control 5′-TGA
TGA CAT CAA GAA GGT GGT GAAG-3′. In addition, a mock group with no
transfection was also used as a control. Cells were seeded the day
prior to transfection using DMEM with 10% FBS however without
antibiotics. Transfection was performed using Lipofectamine 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The cells were incubated
with the control vectors and Lipofectamine plus reagent complexes
for 4–6 h at 37°C, and FBS was added to DMEM at a final
concentration of 10%. The cells were used for subsequent
experiments at 48 h subsequent to transfection.
Western blot analysis
Tissue and cell proteins were homogenized in a
homogenization buffer containing 1 M Tris HCl (pH 7.5), 1% Triton
X-100, 1% Nonidet P-40, 10% sodium dodecyl sulfate (SDS), 0.5%
sodium deoxycholate, 0.5 M ethylenediaminetetraacetic acid,
leupeptin 10 µg/ml, aprotinin 10 µg/ml and 1 mM
phenylmethylsulfonyl fluoride, then were centrifuged at 10,000 × g
for 30 min at 4°C to collect the supernatant liquid. Protein
concentrations were determined with a Bio-Rad protein assay
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The total cellular
protein extracts were separated by 4–15% SDS-polyacrylamide gel
(Bio-Rad Laboratories, Inc.) electrophoresis and transferred to a
nitrocellulose membrane. Membranes were blocked with 5% non-fat dry
milk in phosphate-buffered saline (PBS) for 2 h at room
temperature, then were incubated with antibodies against anti-FOXJ2
(1:1,000; cat. no. sc-54374; Santa Cruz Biotechnology, Inc.) and
GAPDH (1:1,000; cat. no. G5262; Sigma-Aldrich) in PBS with the milk
about 1 h at the room temperature. Blots were washed three times in
PBS buffer, followed by incubation with the horseradish
peroxidase-linked secondary antibodies (1:500; cat. no. sc-51625;
Santa Cruz Biotechnology, Inc.). The specific proteins in the blots
were visualized using the enhanced chemiluminescence reagent (NEN
Life Science Products, Inc., Boston, MA, USA). The experiments were
conducted on three separate occasions.
Cell proliferation assay
Cell proliferation was determined using the Cell
Counting Kit-8 (CCK-8) assay following the manufacturer's
instructions (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). In brief, cells that were transfected with small
interfering RNA (siRNA) were seeded at a density of
2×104/well into a 96-well cell culture cluster (Corning
Incorporated, Corning, NY, USA) in 100 µl culture medium and
incubated overnight. CCK-8 reagents were added to a subset of the
wells, and they were incubated for 2 h at 37°C. The absorbance was
recorded with an automated plate reader. Each experiment was
performed in triplicate and repeated a minimum of three times.
Statistical analysis
Statistical analysis was performed using the SPSS
17.0 software package (SPSS, Inc, Chicago, IL, USA). The
association between FOXJ2 and Ki-67 expression and
clinicopathological features was analyzed using the χ2
test. FOXJ2 and Ki67 expression was studied using the Spearman rank
correlation test due to the nonparametric nature of the data. For
analysis of survival data, Kaplan-Meier curves were constructed,
and the log-rank test was performed. Multivariate analysis was
performed using Cox's proportional hazards model, with P<0.05
considered to indicate a statistically significant difference. The
results were expressed as the mean ± standard deviation.
Results
FOXJ2 expression in HCC and HCC
tissues
To investigate whether FOXJ2 served a role in the
development and progression of HCC, its expression pattern was
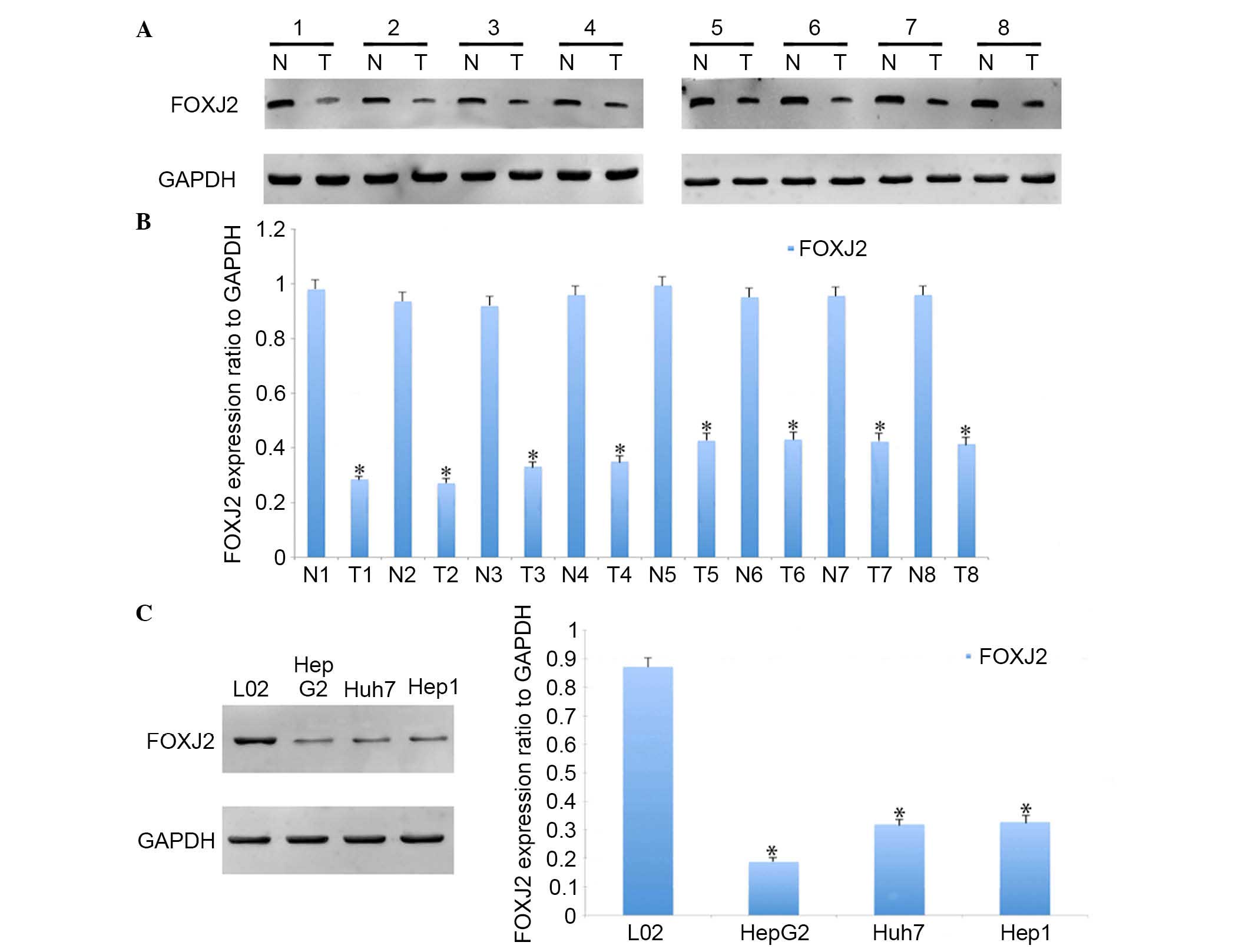
assessed in HCC and adjacent normal tissues. Using western blot
analysis, significantly lower expression levels of FOXJ2 were
identified in HCC tissues compared with the matched normal tissues
(Fig. 1A and B). Furthermore,
samples collected from 110 patients with HCC who underwent hepatic
resection without preoperative systemic chemotherapy at the Third
People's Hospital of Liaocheng. These samples were used to prepare
a tissue microarray in order to examine the expression of FOXJ2 and
the proliferation marker Ki-67. Immunohistochemistry was conducted
to determine the expression of FOXJ2 and Ki-67 in HCC tissues and
adjacent normal tissues. The expression rate of FOXJ2 and Ki-67 in
each tumorous specimen were counted and summarized. Coinciding with
the western blot results, immunohistochemical analysis indicated
that the expression of FOXJ2 was significantly lower in HCC
specimens compared with the nontumorous samples, with the inverse
observed for Ki67 expression (Fig.
2). Notably, the expression rate of FOXJ2 was significantly
correlated with Ki67 expression in HCC specimens with a
co-efficient of −0.679 (Fig. 3).
These observations indicated that FOXJ2 was evidently downregulated
in HCC specimens.
The correlation between FOXJ2 expression
and clinicopathologic variables in HCC specimens
In order to further tackle the involvement of FOXJ2
expression in HCC progression, the correlation between FOXJ2
expression and clinicopathologic factors was analyzed. According to
the expression rate of FOXJ2, the HCC specimens were separated into
two groups, the high and low FOXJ2 expression groups. It was
identified that the expression of FOXJ2 was significantly
correlated with histological differentiation, size of largest
tumor, metastasis and Ki67 expression (Table I). However, there is no correlation
between FOXJ2 expression and other clinicopathologic factors,
including age, gender, tumor number, hepatitis B virus status,
liver cirrosis, microvascular invasion and serum α-fetoprotein
value. Furthermore, the survival status of the patients was
measured and the association between different clinicopathological
variables and survival was analyzed. It was identified that the
survival status of the patients was significantly correlated with
various clinicopathological parameters, including histological
differentiation, size of largest tumor, metastasis, FOXJ2
expression and Ki67 expression (Table
II). In addition, multivariate analysis using Cox's
proportional hazards model demonstrated that the histological
differentiation, size of largest tumor, metastasis and FOXJ2
expression were independent prognostic indicators for patient
overall survival (Table III).
Due to the fact that our observations indicated that FOXJ2
expression was associated with the survival of patients with HCC,
the survival curves of patients with HCC with low and high levels
of FOXJ2 expression were assessed. Patients with low expression
levels of FOXJ2 had significantly poorer survival, compared with
those in the high FOXJ2 expression group (Fig. 4).
 | Table IISurvival status and
clinicopathological parameters in hepatocellular carcinoma
specimens. |
Table II
Survival status and
clinicopathological parameters in hepatocellular carcinoma
specimens.
| Parameter | Total | Survival status
| P-value | χ2
value |
|---|
| Dead (n=64) | Alive (n=46) |
|---|
| Age (years) | | | | | |
| ≤50 | 61 | 39 | 22 | 0.770 | 0.085 |
| >50 | 49 | 30 | 19 | | |
| Gender | | | | | |
| Male | 89 | 59 | 30 | 0.111 | 2.534 |
| Female | 21 | 10 | 11 | | |
| Histological
differentiation | | | | | |
| Well | 11 | 3 | 8 | 0.016a | 8.307 |
| Moderate | 83 | 53 | 30 | | |
| Poor | 16 | 13 | 3 | | |
| Size of largest
tumor (cm) | | | | | |
| ≤5 | 63 | 33 | 30 | 0.009a | 6.751 |
| >5 | 47 | 36 | 11 | | |
| Tumor number | | | | | |
| Solitary | 74 | 43 | 31 | 0.151 | 2.063 |
| Multiple | 36 | 26 | 10 | | |
| HBV | | | | | |
| Positive | 88 | 58 | 30 | 0.167 | 1.905 |
| Negative | 22 | 11 | 11 | | |
| Liver cirrosis | | | | | |
| Yes | 89 | 59 | 30 | 0.111 | 2.534 |
| No | 21 | 10 | 11 | | |
| Microvascular
invasion | | | | | |
| Yes | 31 | 22 | 9 | 0.263 | 1.254 |
| No | 79 | 47 | 32 | | |
| Metastasis | | | | | |
| Yes | 16 | 14 | 2 | 0.027a | 4.915 |
| No | 94 | 55 | 39 | | |
| Serum AFP value
(ng/ml) | | | | | |
| ≤50 | 42 | 25 | 17 | 0.585 | 0.298 |
| >50 | 68 | 44 | 24 | | |
| FOXJ2 | | | | | |
| Low | 63 | 55 | 8 | <0.001a | 38.085 |
| High | 47 | 14 | 33 | | |
| Ki67 | | | | | |
| Low | 59 | 23 | 36 | <0.001a | 19.278 |
| High | 51 | 41 | 10 | | |
 | Table IIIUnivariate and multivariate analysis
of prognostic factors using Cox proportional hazards model. |
Table III
Univariate and multivariate analysis
of prognostic factors using Cox proportional hazards model.
| Factor | Univariate analysis
| Multivariate
analysis
|
|---|
| RR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age
(≤50/>50) | 0.895 | 0.556–1.440 | 0.674 | – | – | – |
| Gender (M/F) | 1.670 | 0.854–3.267 | 0.134 | – | – | – |
| Differentiation
(Well/Moderate/Poor) | 3.557 | 1.116–11.337 | 0.032a | 1.707 | 0.508–5.735 | 0.387 |
| Tumor size (≤5
cm/>5 cm) | 2.263 | 1.404–3.647 | 0.001a | 1.429 | 0.862–2.369 | 0.167 |
| Tumor number
(Solitary/Multiple) | 1.466 | 0.898–2.392 | 0.126 | – | – | – |
| HBV (+/−) | 1.514 | 0.795–2.886 | 0.207 | – | – | – |
| Liver cirrosis
(+/−) | 1.740 | 0.889–3.405 | 0.106 | – | – | – |
| Microvascular
invasion (+/−) | 1.572 | 0.946–2.613 | 0.081 | – | – | – |
| Metastasis
(+/−) | 3.077 | 1.687–5.612 | <0.001a | 2.151 | 1.146–4.036 | 0.017a |
| AFP value (≤50
ng/ml/>50 ng/ml) | 1.222 | 0.748–1.997 | 0.424 | – | – | – |
| FOXJ2
(low/high) | 0.171 | 0.094–0.311 | <0.001a | 4.692 | 2.490–8.844 | <0.001a |
FOXJ2 was downregulated in HCC cell
lines
HCC cell lines and a L02 hepatocyte cell line were
used to analyze the role of FOXJ2 in HCC cell physiology. The
expression levels of FOXJ2 in these cell lines were first
determined using western blot analysis. It was identified that L02
cells had the highest expression of FOXJ2 of all the cell lines
examined (Fig. 1C).
Depletion of FOXJ2 led to enhanced cell
proliferation
Due to the fact that FOXJ2 expression was observed
to be inversely correlated with histological differentiation and
Ki-67 expression in HCC specimens, it was hypothesized that FOXJ2
may influence the proliferation of HCC cells. Transfection of
FOXJ2-siRNA oligos was performed to deplete FOXJ2 expression in
three HCC cell lines. Subsequent to transfection of FOXJ2-targeting
siRNAs, three HCC cell lines were subjected to western blot
analysis to determine the interference efficiencies of different
siRNA oligos. As presented in Fig. 5A
and B, si-FOXJ2 of the HepG2 cell line was significantly
knocked down. Subsequently, the impact of FOXJ2 depletion on HepG2
cell proliferation was determined using the CCK-8 assay. As
presented in Fig. 5C,
FOXJ2-depleted cells exhibited enhanced cell growth, compared with
the mock and control siRNA-transfected cells. The results obtained
demonstrated that FOXJ2 depletion could promote the proliferation
of hepatocytes.
Discussion
HCC prognosis remains unsatisfactory due to its high
rates of recurrence and metastasis. Despite significant
improvements in surveillance and clinical treatment strategies
(13), traditional therapeutic
methods, including chemotherapy and surgical operation, remain
inadequate. Therefore, it is critical to identify patients with
poor prognosis for timely intervention, and to develop novel
targeted therapeutic strategies. Therefore, previous studies have
focused on the development of therapeutic strategies based on
molecular targeting in human cancer, including in HCC (14,15).
The current study demonstrated that reduced expression levels of
FOXJ2 were significantly associated with the poor prognosis of
patients with HCC, which is partially attributed to the cell
proliferation of HCC cells. Thus, it is suggested that the
observations of the current study may benefit the development of
novel therapeutic strategies for patients with HCC based on
FOXJ2.
Members of the Fox gene family share an
evolutionarily conserved DNA-binding domain, termed the 'fork-head'
or 'winged-helix' domain (16,17).
The Fox family is a large and diverse group of transcription
factors that serve various important roles in a wide variety of
biological processes including development, metabolism, immunology
and senescence (16,18,19).
In addition, it has been reported that several members of this
family are involved in the etiology of cancer (20,21).
The FOXA1 gene has been observed to be amplified and overexpressed
in esophageal and lung cancer (22), and the FOXP1 transcription factor
is suggested to function as either an oncogene or as a tumor
suppressor that is cell type-dependent (23–26).
Furthermore, FOXJ1 was observed to be markedly upregulated in human
HCC specimens (27), and increased
expression of FOXM1 protein was identified in variety of human
tumors, including in HCC (28) and
intrahepatic cholangiocarcinoma (29).
FOXJ2 (FHX) is a recently characterized human
forkhead transcriptional activator that binds DNA with a dual
sequence specicity (30) and can
be expressed as two isoforms, FHX.L and FHX.S, which differ in
their C-terminal ends (6). In
humans and mice, FOXJ2 expression can be detected in almost all
tissues of the adult or the fetus. It has been identified to be
localized constitutively at the nucleus of the cell and may be
involved in the nuclear translocation mechanism of all forkhead
factors. A previous study demonstrated that FOXJ2 served an
important role in lipopolysaccharide-induced inflammatory
responses, and that FOXJ2 may be involved in the activation of
astrocytes (31). In addition,
studies have demonstrated the association between FOXJ2 and cancer.
For example, Wang et al (11) identified that the expression of
FOXJ2 was higher in primary breast cancer tissues without lymph
nodes metastases compared with those with, demonstrating that FOXJ2
can inhibit the metastasis of human breast cancer by regulating EMT
key markers (E-cadherin and vimentin). Qiu et al (12) identified that FOXJ2 suppressed cell
migration and invasion in glioma, and that overexpression of FOXJ2
increased E-cadherin expression and reduced vimentin expression,
and significantly inhibited migration in U87 cells. Knockdown of
FOXJ2 promoted cellular motility.
The current study identified that FOXJ2 may be an
important prognotic factor in HCC. Western blotting and
immunohistochemistry analysis identified that FOXJ2 was
downregulated in HCC tissues and HCC cells, and observed that there
was a significant negative correlation between FOXJ2 expression
levels and HCC. FOXJ2 and Ki-67 were identified to be present
predominantly in the nucleus, and FOXJ2 expression was negatively
correlated with Ki67 expression. Accordingly, Kaplan-Meier survival
analysis indicated that low expression of FOXJ2 was associated with
poor prognosis of patients with HCC. Furthermore, it was
demonstrated that FOXJ2 inhibited the proliferation of HCC using a
CCK-8 assay. Knockdown of FOXJ2 expression was suggested to promote
cell proliferation.
In summary, the results of the present study suggest
that FOXJ2 is a novel and promising prognostic biomarker for HCC
progression and prognosis. To the best of our knowledge, the
current study is the first to investigate the clinical significance
of FOXJ2 in HCC. With technological development, and using
microarray analysis, numerous novel treatments may be developed
based on the gene expression of tumors. The results of the current
study may be useful in aiding in the prediction of prognosis, and
may as a result be beneficial in the future treatment of patients
with HCC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Altekruse SF, McGlynn KA and Reichman ME:
Hepatocellular carcinoma incidence, mortality, and survival trends
in the United States from 1975 to 2005. J Clin Oncol. 27:1485–1491.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruix J, Boix L, Sala M and Llovet JM:
Focus on hepatocellular carcinoma. Cancer Cell. 5:215–219. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lau WY and Lai EC: Hepatocellular
carcinoma: Current management and recent advances. Hepatobiliary
Pancreat Dis Int. 7:237–257. 2008.PubMed/NCBI
|
|
5
|
Blum HE and Moradpour D: Viral
pathogenesis of hepatocellular carcinoma. J Gastroenterol Hepatol.
17(Suppl 3): S413–S420. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pérez-Sánchez C, Gómez-Ferrería MA, de La
Fuente CA, Granadino B, Velasco G, Esteban-Gamboa A and Rey-Campos
J: FHX, a novel fork head factor with a dual DNA binding
specificity. J Biol Chem. 275:12909–12916. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pérez-Sánchez C, Arias-de-la-Fuente C,
Gómez-Ferrería MA, Granadino B and Rey-Campos J: FHX.L and FHX.S,
two isoforms of the human fork-head factor FHX (FOXJ2) with
differential activity. J Mol Biol. 301:795–806. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Choi VM, Harland RM and Khokha MK:
Developmental expression of FOXJ1.2, FOXJ2 and FOXQ1 in xenopus
tropicalis. Gene Expr Patterns. 6:443–447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wijchers PJ, Hoekman MF, Burbach JP and
Smidt MP: Identification of forkhead transcription factors in
cortical and dopaminergic areas of the adult murine brain. Brain
Res. 1068:23–33. 2006. View Article : Google Scholar
|
|
10
|
Kehn K, Berro R, Alhaj A, Bottazzi ME, Yeh
WI, Klase Z, Van Duyne R, Fu S and Kashanchi F: Functional
consequences of cyclin D1/BRCA1 interaction in breast cancer cells.
Oncogene. 26:5060–5069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Yang S, Ni Q, He S, Zhao Y, Yuan
Q, Li C, Chen H, Zhang L, Zou L, et al: Overexpression of forkhead
box J2 can decrease the migration of breast cancer cells. J Cell
Biochem. 113:2729–2737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiu X, Ji B, Yang L, Huang Q, Shi W, Ding
Z, He X, Ban N, Fan S, Zhang J and Tian Y: The role of FOXJ2 in the
migration of human glioma cells. Pathol Res Pract. 211:389–397.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kudo M: Hepatocellular carcinoma 2009 and
beyond: From the surveillance to molecular targeted therapy.
Oncology. 75(Suppl 1): S1–S12. 2008. View Article : Google Scholar
|
|
14
|
Patel A and Sun W: Molecular targeted
therapy in hepatocellular carcinoma: From biology to clinical
practice and future. Curr Treat Options Oncol. 15:380–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsuchiya N, Sawada Y, Endo I, Saito K,
Uemura Y and Nakatsura T: Biomarkers for the early diagnosis of
hepatocellular carcinoma. World J Gastroenterol. 21:10573–10583.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaufmann E and Knöchel W: Five years on
the wings of fork head. Mech Dev. 57:3–20. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hannenhalli S and Kaestner KH: The
evolution of fox genes and their role in development and disease.
Nat Rev Genet. 10:233–240. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jonsson H and Peng SL: Forkhead
transcription factors in immunology. Cell Mol Life Sci. 62:397–409.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benayoun BA, Caburet S and Veitia RA:
Forkhead transcription factors: Key players in health and disease.
Trends Genet. 27:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin L, Miller CT, Contreras JI, Prescott
MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et
al: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1),
on chromosome band 14q13 is amplified and overexpressed in
esophageal and lung adenocarcinomas. Cancer Res. 62:5273–5279.
2002.PubMed/NCBI
|
|
23
|
Banham AH, Connors JM, Brown PJ, Cordell
JL, Ott G, Sreenivasan G, Farinha P, Horsman DE and Gascoyne RD:
Expression of the FOXP1 transcription factor is strongly associated
with inferior survival in patients with diffuse large B-cell
lymphoma. Clin Cancer Res. 11:1065–1072. 2005.PubMed/NCBI
|
|
24
|
Schuster MB and Porse BT: C/EBPalpha: A
tumour suppressor in multiple tissues? Biochim Biophys Acta.
1766:88–103. 2006.PubMed/NCBI
|
|
25
|
Barrans SL, Fenton JA, Banham A, Owen RG
and Jack AS: Strong expression of FOXP1 identifies a distinct
subset of diffuse large B-cell lymphoma (DLBCL) patients with poor
outcome. Blood. 104:2933–2935. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Banham AH, Beasley N, Campo E, Fernandez
PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P,
et al: The FOXP1 winged helix transcription factor is a novel
candidate tumor suppressor gene on chromosome 3p. Cancer Res.
61:8820–8829. 2001.PubMed/NCBI
|
|
27
|
Chen HW, Huang XD, Li HC, He S, Ni RZ,
Chen CH, Peng C, Wu G, Wang GH, Wang YY, et al: Expression of FOXJ1
in hepatocellular carcinoma: Correlation with patients' prognosis
and tumor cell proliferation. Mol Carcinog. 52:647–659. 2013.
View Article : Google Scholar
|
|
28
|
Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z,
Roskams T, Durnez A, Demetris AJ and Thorgeirsson SS:
Classification and prediction of survival in hepatocellular
carcinoma by gene expression profiling. Hepatology. 40:667–676.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Obama K, Ura K, Li M, Katagiri T, Tsunoda
T, Nomura A, Satoh S, Nakamura Y and Furukawa Y: Genome-wide
analysis of gene expression in human intrahepatic
cholangiocarcinoma. Hepatology. 41:1339–1348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Granadino B, Arias-de-la-Fuente C,
Pérez-Sánchez C, Párraga M, López-Fernández LA, del Mazo J and
Rey-Campos J: FHX (FOXJ2) expression is activated during
spermatogenesis and very early in embryonic development. Mech Dev.
97:157–160. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Cao X, Tao G, Cao Z, Wang S, Zhou
F, Xie W, Zhao P, Zhang Z and Cui Z: FOXJ2 expression in rat spinal
cord after injury and its role in inflammation. J Mol Neurosci.
47:158–165. 2012. View Article : Google Scholar : PubMed/NCBI
|