Introduction
Colorectal cancer (CRC) is one of the major causes
of cancer-associated mortality worldwide (1). Although the treatment strategy for
CRC has improved substantially over the last few decades, targeted
therapy for CRC remains in its infancy (2). In addition, the pathogenesis of CRC
remains to be fully elucidated, and understanding the molecular
mechanisms, which drives the development of CRC is essential for
identifying novel molecular targets for CRC.
MicroRNAs (miRs) are highly conserved, small
non-coding RNA strands, which act as negative regulators of gene
expression by incompletely complementary pairing with the
3′-untranslated region (UTR) of target mRNAs, which, in the
majority of cases, inhibits the translation process of target genes
(3–5). miRs are expressed in a restricted
spatial and temporal manner, which confers their ability to
specifically regulate different important biological processes in a
variety of settings (6,7). Previous investigations have
elucidated the pivotal role of miRs in CRC and suggested several
novel miR-based therapeutic strategies (8). However, due to the extensive miR-mRNA
interaction network in every facet of tumor biology, it is possible
that a several additional miRs may be involved in critical
processes, including proliferation, apoptosis and
trans-differentiation in CRC cells.
B cell lymphoma (Bcl)-2 homologous antagonist/killer
(Bak1) is an essential cell death regulator, which initiates
mitochondria-mediated apoptosis by interacting with proteins,
including Bcl-extra large (xL) (9)
and voltage-dependent anion channels (VDACs) (10). Previous studies have shown that
Bcl-2 family proteins, including Bcl-2-associated X protein (Bax),
are capable of predicting CRC prognosis, and that Bax agonists
efficiently induce apoptosis in tumors (11–14).
However, how their expression is intrinsically controlled in CRC
remains to be fully elucidated. In the present study, the miRanda
database (www.microrna.org) was used to search for
potential microRNAs that target Bak1. It was suggested that Bak1
may be targeted by miR-410; the data in the present study may show
a differential expression of miR-410 in CRC tumor tissues and
characterize the role of miR-410 in the apoptosis of CRC cells for
the first time. These data will help to under-stand the implication
of miR-410 in CRC tumorigenesis and its potential therapeutic value
in CRC treatment.
Materials and methods
Tissue collection
A total of 20 samples of human colorectal
adenocarcinoma tumor tissues and their paired adjacent normal
tissues were collected from Binzhou Medical University Hospital
(Binzhou, China); the tissues were immediately transferred into
liquid nitrogen following laparoscopic resection. All patients were
diagnosed with adenocarcinoma and were aged between 35 and 60
years. Among them, 12 were male and 8 were female. Tumor samples
were collected between February and May 2013. The cancer tissues
and adjacent tumor tissues were collected, and the negative margin
of the adjacent tissue was confirmed by histological examination.
All diagnoses were histologically confirmed. Informed consent was
obtained from the patients, and the experimental protocol was
approved by the institutional Ethics Committee of Binzhou Medical
University Hospital, Binzhou, China.
Cell culture and transfection
Cell lines of human colorectal cancer (SW480, HT29
and HCT116) and normal colon epithelial cells (FHC) were obtained
from American Type Culture Collection (Vanassas, MA, USA). The
cells were cultured in Dulbecco's modified Eagle's medium
supplemented with 10% fetal bovine serum at a temperature of 37°C
with 5% CO2. The culture media were replaced every other
day. The pan-caspase inhibitor, ZVAD-fmk (Sigma-Aldrich, St. Louis,
MO, USA) was dissolved in dimethyl sulfoxide (DMSO) and used at the
final concentration of 20 µM. Small interfering (si)RNA for
Bak1, miR-410 mimics, miR-410-inhibitor and their negative control
sequences were transfected into cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the protocols provided by the manufacturer. Cells at
70–80% confluence were transfected in the absence of antibiotics.
After 24 h of transfection the medium was replaced. miR-410 and the
miR-410-inhibitor were purchased from Guangzhou RiboBio Co., Ltd,
(Guangzhou, China), and Bak1-siRNA was purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Cells were scraped from the culture plate, and total
RNA from the tissues and treated cells was isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA
samples were then reverse transcribed using a specific stem-loop
primer (Guangzhou RiboBio Co., Ltd.) for miR-410, followed by
amplification with SYBR Green master mix on the ABI7000 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The cDNA templates were diluted to 1:10 and 5 µl per
reaction was analyzed. The thermal cycling conditions were as
follows: 95°C for 10 min, 95°C for 15 sec, 50–60°C for 5 sec and
72°C for 15 sec for 40 cycles. The Bulge-Loop™ hsa-miR-410 qRT-PCR
primer set was purchased from Guangzhou RiboBio Co., Ltd. U6 small
nuclear RNA was used as the internal control to normalize the
expression of miR-410. The relative expression of miR-410 was
determined using the 2−ΔΔCq method (15).
Methylthiazolyldiphenyl-tetrazolium
bromide (MTT) assay
The SW480 cells were seeded into 96-well
plates at 2.5×104/ml, and cell viability was determined
by the MTT method using a kit (cat. no. C0009) obtained from
Beyotime Institute of Biotechnology (Shanghai, China). The formazan
produced by the living cells in the presence of MTT were visualized
by DMSO dissolution. The absorbance value at 570 nm in each group
was measured using a microplate reader and used to compare the
growth activity of the cells in different groups.
Cytochrome c apoptosis assay
Cytochrome c release was used to determine
apoptosis. The present study assessed the release of cytochrome
c in the cytosol using an ELISA kit (cat. no. H190; Nanjing
Jiancheng Bioengineering Institute, Nanjing, China), according to
the manufacturer's protocol. The experiments were performed in
triplicate.
Western blot analysis
Protein extraction was performed by homogenizing the
cells with radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology), followed by mixing with an appropriate
quantity (1/5 of total sample volume) of SDS-sample buffer
(Beyotime Institute of Biotechnology) and heating at 100°C for 3
min. Protein was quantified using a BCA Protein Assay kit (cat. no.
P0011; Beyotime Institute of Biotechnology). The proteins (30
µg) were then subjected to 10% SDS-PAGE and transferred onto
nitrocellulose membranes. The membranes were blocked with 5% milk
for 1 h at room temperature. The membranes were incubated with the
following primary rabbit antibodies at 4°C for 16 h: Anti-Bak1
(1:1,000; cat. no. 12105), anti-Bcl-2 (1:1,000; cat. no. 2876),
anti-c-caspase-3 (1:500; cat. no. 9664), anti-VDAC (1:1,000; cat.
no. 4866) and anti-β-actin (1:1,000; cat. no. 4976). The membranes
were then incubated with horseradish peroxidase-conjugated rabbit
IgG secondary antibody (1:3,000; cat. no. 7074) at room temperature
for 50 min, and the bands were detected using enhanced
chemiluminescence methods with a kit (cat. no. P0018; Beyotime
Institute of Biotechnology). The membranes were washed with
phosphate-buffered saline with 0.5% Tween 20 three times (10 min
each) after primary antibody and secondary antibody incubation. The
bands were quantified using ImageJ software (version 1.45; National
Institutes of Health, Bethesda, MA, USA). The relative expression
levels of the proteins of interest were obtained by normalizing
their band densities to that of β-actin. All antibodies were
purchased from Cell Signaling Technology, Inc.
Luciferase assay
A 415bp-fragment of the Bak1 3′-UTR, which contained
a putative binding site of miR-410, was subcloned into the 3′-UTR
region of the pmirGLO firefly luciferase construct (Promega,
Madison, WI, USA). The following primers were used for PCR
amplification: Sense 5′-CGAGCTCGTCCTCTCAGTTCTCTCCCT-3′ and
antisense 5′-GCTCTAGAAGGCTGTGCCCAATAGAGAA-3′. The product was then
inserted into the site between SacI and XbaI. The
empty vector and the PCR products were digested with SacI and XbaI,
and subjected to T4 DNA ligation (Promega) at 16°C overnight. The
ligation product was then transformed. Single colonies were picked
and the construct was confirmed by sequencing. The constructs were
then transfected into SW480 cells, accompanied with the internal
control vector, pRL-TK, and miR-410 mimics or miR-410-inhibitor
(Guangzhou RiboBio Co., Ltd.). At 48 h post-transfection, the
luciferase activity was measured using a Dual-Luciferase Reporter
assay kit (cat. no. E1901; Promega).
Statistical analysis
All the data are reported as the mean ± standard
deviation. SPSS version 19.0 (IBM SPSS, Amronk, NY, USA) was used
to perform statistical analyses. Differences in the expression
levels of miR-410 and Bak1 in the paired tissue samples were
determined using a paired t-test. Comparisons of three or
more groups were performed using one way analysis of variance
followed by a Newman-Keuls post-hoc test. Correlations between the
expression levels of Bak1 and miR-410 were analyzed using
Spearman's correlation. Two-tailed P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-410 is upregulated in CRC tumor and
cell lines
To evaluate the potential role of miR-410 in CRC,
the present study firstly compared the expression levels of miR-410
in CRC tissues and the matched adjacent normal tissues. As shown in
Fig. 1A, a significant increase in
the expression of miR-410 was observed in the tumor tissues. The
expression of miR-410 was also examined in several CRC cell lines,
and it was found that miR-410 was expressed at a higher levels in
the SW480, HCT-116 and HT-29 CRC cell lines, compared with the FHC
normal colon epithelial cell line (Fig. 1B).
miR-410 suppresses apoptosis and promotes
growth of CRC cells
To further understand the role of miR-410 in CRC,
the present study transfected the SW480 cell line with miR-410
mimics or miR-410 antisense inhibitor to promote the expression of
miR-410 or inhibit the endogenous expression of miR-410,
respectively. RT-qPCR analysis confirmed the transfection
efficiency (Fig. 2A). The
transfected SW480 cells were allowed to grow for 4 days
consecutively, and the viability of the cells in each group were
determined at indicated time points using an MTT assay. As shown in
Fig. 2B, the forced expression
miR-410 promoted the growth activity of CRC cells, whereas the
inhibition of miR-410 decreased the growth activity. Of note, the
ZVAD-fmk apoptosis inhibitor markedly attenuated the growth
inhibitory effect of the miR-410-inhibitor, suggesting that miR-410
may have altered the growth activity of CRC cells by affecting the
apoptotic activity. The results of the western blot analysis of the
apoptosis marker, cleaved-caspase-3, are shown in Fig. 2. Consistent with expectations, the
overexpression of miR-410 decreased the expression of
cleaved-caspase-3, and the inhibition of miR-410 enhanced its
expression. The present study also observed alteration in the
release of cytochrome c following the gain or loss of
function of miR-410, and the results were consistent with the
results obtained from the western blot analysis of
cleaved-caspase-3 (Fig. 2D).
miR-410 suppresses apoptosis by targeting
Bak1 in CRC cells
The present study then aimed to determine the
mechanism by which miR-410 regulates apoptosis in CRC cells.
Bioinformatics analysis predicted a putative binding site of
miR-410 in the human Bak1 3′-UTR (Fig.
3A), and the present study assessed whether Bak1 was targeted
by miR-410 in the CRC cells. The results of the RT-qPCR analysis
showed that the loss or gain of function of miR-410 had no
significant affect on the mRNA level of Bak1 (Fig. 3B). The protein level of Bak1 was
significantly inhibited when miR-410 was overexpressed, whereas the
inhibition of miR-410 had the opposite effect (Fig. 3C). Bcl-2, an anti-apoptotic
protein, which is important in the mitochondrial apoptotic pathway,
was positively correlated with the expression of miR-410. Previous
studies have shown that VDAC is controlled by Bak1 and mediates the
release of cytochrome c to facilitate apoptosis (10,16),
and the present study observed that the expression of VDAC was in
agreement with Bak1, which suggested that the Bak1/VDAC interaction
was involved in miR-410-associated apoptotic signaling. Further
investigation showed that miR-410 decreased the luciferase activity
in the SW480 cells, whereas the inhibition of miR-410 increased the
luciferase activity (Fig. 3D).
Additionally, as shown in Fig. 3E,
the silencing of Bak1 using siRNA significantly attenuated the
pro-apoptotic effect of the miR-410 inhibitor on the CRC cells.
Taken together, these results showed that Bak1 was a direct target
of miR-410 and was required for the apoptosis induced by miR-410
inhibition.
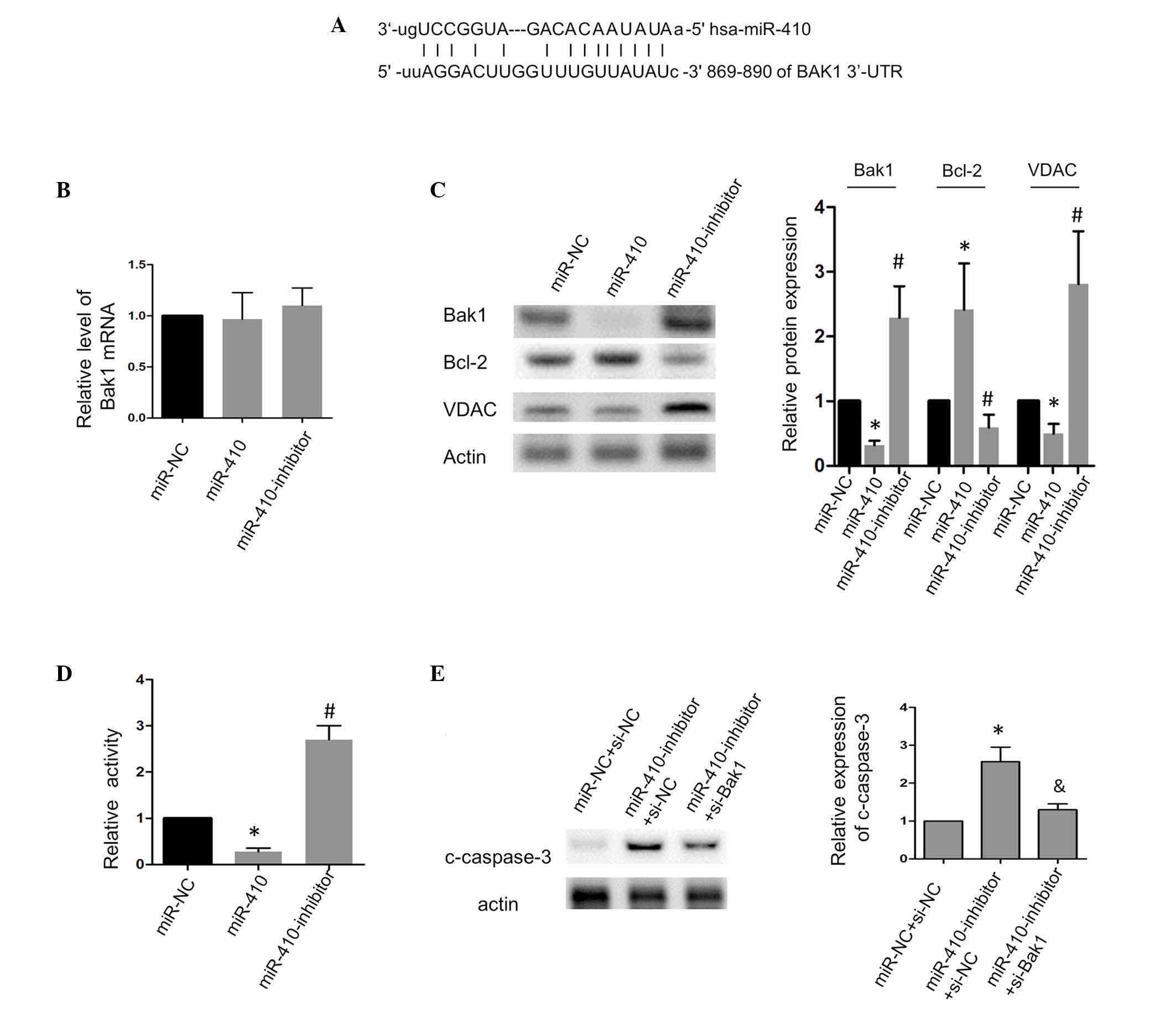 | Figure 3miR-410 suppresses apoptosis by
targeting Bak1 in SW480 cells. (A) Schematic showing the putative
base-pair binding between miR-410 and the BAK1 3′-UTR. (B) Effects
of miR-410 and the miR-410 inhibitor on the mRNA levels of Bak1.
(C) Effects of miR-410 and the miR-410 inhibitor on the protein
levels of Bak1, Bcl-2 and VDAC. (D) Effects of miR-410 and the
miR-410 inhibitor on the luciferase activity of the
pmirGLO-BAK1-3′-UTR in SW480 cells. (E) Silencing Bak1 attenuated
the caspase-3 cleavage induced by the miR-410-inhibitor.
*P<0.05 and #P<0.05, vs. miR-NC or
miR-NC+si-NC. &P<0.05, vs.
miR-410-inhibitor+si-NC. miR, microRNA; Bak1, B cell
lymphoma-2-agonist/killer 1; VADC, voltage-dependent anion channel;
c-caspase-3, cleaved-caspase-3; NC, negative control; UTR,
untranslated region; si-, small interfering-RNA. |
Expression of Bak1 is inversely
correlated with the expression of miR-410 in CRC
As the Bak1 was confirmed as a direct target of
miR-410 in vitro, the present study next investigated
whether the expression of Bak1 was also altered in tumor tissues.
As shown in Fig. 4A, the
expression of Bak1 was found to be downregulated in the CRC
tissues, compared with the adjacent tumor tissues. Of note,
Spearman's correlation coefficient showed a significant inverse
correlation between the expression of Bak1 and miR-410 (Fig. 4B). This result further confirmed
that Bak1 was targeted by miR-410 in the clinical setting of
CRC.
Discussion
In the present study, it was demonstrated that
miR-410 was crucial in regulating the apoptosis of CRC cells. It
was found that miR-410 was upregulated in CRC tissues and cells.
Mechanistically, the present study revealed that the pro-apoptotic
factor, Bak1, was targeted by miR-410, which in turn decreased the
basal level of apoptosis and thus enhanced the growth activity of
CRC cells. The results of the present study, which reported the
tumorgenicity of miR-410, may not only expand on current
understanding of the regulatory mechanism of cell death by
non-coding RNAs, but also provided insights into the targeted
therapy for CRC.
Accumulating evidence has confirmed that miRs are
important regulators either in the early development or in the
progress of CRC (8), and previous
reports have identified that several miRs are aberrantly expressed
(17,18). For example miR-143 and miR-145,
which have been confirmed as tumor suppressors, are markedly
downregulated in CRC (17,19). By contrast, miR-21 has been
identified as an oncogenic miR, which is important in several
aspects of CRC, including apoptosis, proliferation and metastasis
(20–22). Previous studies have shown that
miR-410 acts in a diverse manner to control multiple cellular
processes, including angiogenesis, differentiation and
proliferation (23–25). However, the roles of miR-410 in
regulating cell behaviors remain to be fully elucidated, and
conflicting results have been reported with regards to its role in
cancer cell growth. Chien et al reported that miR-410 is
involved in cell cycle progression by regulating the
cyclin-dependent kinase 1-dependent pRb/E2F pathway in breast
cancer cells (26). Chen et
al reconfirmed the antitumor activity of miR-410 in glioma
cells, showing that MET, the receptor for hepatocyte growth factor
receptor, is targeted by miR-410 (27). Notably, a study by Wang et
al demonstrated the increased expression and oncogenic role of
miR-410 in hepatic and colorectal tumors; they suggested that
miR-410 binds to the 3′-UTR of four and a half LIM domains 1, which
promoted its methylation and thus inhibited its transcription
(28). The divergent conclusions
of these studies suggest that the role of miR-410 in cancer may be
temporally- or spatially-specific. Despite reports of its role in
CRC, whether miR-410 functions to regulate the apoptotic machinery
remain to be elucidated. In agreement with Wang et al
(28), the present study
demonstrated that miR-410 enhanced the growth activity of CRC
cells. However, these results are the first, to the best of our
knowledge, to suggest that miR-410 may be a component of the
apoptotic regulatory network, as the apoptosis inhibitor, ZVAD-fmk,
efficiently inhibited the growth inhibitory activity of the miR-410
inhibitor, and altered the activation of caspase-3 by miR-410 gain
or loss of function.
miRs are important negative regulators of gene
expression. Using bioinformatics analysis, the present study found
a segment of complementary sequence of miR-410 within the 3′-UTR of
the Bak1-encoding gene. Therefore, it was suggested that Bak1 was a
putative functional target in this cellular model. Previous studies
have shown that Bak1 is a critical regulator of
mitochondria-mediated apoptosis (9,10).
Bak1 and Bax, two pro-apoptotic proteins, form a complex upon
apoptotic stimuli, which mediates the release of cytochrome
c (10). It has also been
shown that several Bcl-2 family proteins, including Bak1 and Bax,
act downstream of the transcriptional factor, p53 (9). As p53 mutations are often observed in
CRC (29–31), it is possible that the function of
Bak1 may have important implications in CRC. Although several
studies have preliminarily characterized the role of Bak1 in
vitro and in vivo (32–34),
how the expression of Bak1 is regulated under the circumstance
remains to be fully elucidated. miRs constitute important
regulators in apoptosis, and a previous study showed that Bak1 is
targeted by miR-125b and mediates its anti-apoptotic effect in
neural crest cells (35).
Consistent with the previous reports, the present study provided
compelling evidence that Bak1 is an important regulator of
apoptosis, which is under the control of miR-410. miRs can have
multiple targets and, in the present study, it was shown that
miR-410 not only altered the expression of Bak1, but also affected
the expression of Bcl-2 and VDAC. Whether these changes were the
result of alterations in the expression of Bak1 or the functional
readout by other possible uncharacterized target genes remains to
be elucidated. However, the overall anti-apoptotic effect of
miR-410 suggests its potential application in cancer treatment.
Of note, the present study demonstrated that miR-410
is upregulated in CRC tissues, compared with normal adjacent
tissues, and was well correlated with the expression of Bak1.
However, how miR-410 is constantly regulated under oncogenic
stimuli remains to be elucidated. Previous studies have
demonstrated that the expression of miRs can be regulated at the
transcriptional level. For example, miR-34a is transactivated by
p53, which exerts multiple antitumor effects in CRC (36-38),
and the expression of miR-21 expression is induced by AP-1 during
tumorgenesis (39). As miR-410 is
aberrantly regulated in CRC, identifying the nodal points, which
regulate its expression may be a suitable topic for future
investigation.
In conclusion, the present study identified miR-410
as an miR with tumorigenic potential, and provided compelling
evidence that miR-410 is integrated into the apoptotic program of
CRC. It was shown that the upregulated expression of miR-410 may
contribute to decreased levels of basal apoptosis, which may be the
possible mechanism driving the development of CRC. These results
may assist in the development of targeted molecular therapy for
CRC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hagan S, Orr MC and Doyle B: Targeted
therapies in colorectal cancer-an integrative view by PPPM. EPMA J.
4:32013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schetter AJ, Okayama H and Harris CC: The
role of microRNAs in colorectal cancer. Cancer J. 18:244–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nieminen AI, Eskelinen VM, Haikala HM,
Tervonen TA, Yan Y, Partanen JI and Klefström J: Myc-induced
AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial
apoptosis. Proc Natl Acad Sci USA. 110:E1839–E1848. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimizu S, Narita M and Tsujimoto Y: Bcl-2
family proteins regulate the release of apoptogenic cytochrome c by
the mitochondrial channel VDAC. Nature. 399:483–487. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pryczynicz A, Gryko M, Niewiarowska K,
Cepowicz D, Ustymowicz M, Kemona A and Guzińska-Ustymowicz K: Bax
protein may influence the invasion of colorectal cancer. World J
Gastroenterol. 20:1305–1310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
García-Flórez LJ, Gómez-Álvarez G, Frunza
AM, Barneo-Serra L, Martínez-Alonso C and Fresno-Forcelledo MF:
Predictive markers of response to neoadjuvant therapy in rectal
cancer. J Surg Res. 194:120–126. 2015. View Article : Google Scholar
|
|
13
|
Xin M, Li R, Xie M, Park D, Owonikoko TK,
Sica GL, Corsino PE, Zhou J, Ding C, White MA, et al:
Small-molecule Bax agonists for cancer therapy. Nat Commun.
5:49352014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ogura E, Senzaki H, Yamamoto D, Yoshida R,
Takada H, Hioki K and Tsubura A: Prognostic significance of Bcl-2,
Bcl-xL/S, Bax and Bak expressions in colorectal carcinomas. Oncol
Rep. 6:365–369. 1999.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
16
|
Naghdi S, Varnai P and Hajnoczky G: Motifs
of VDAC2 required for mitochondrial Bak import and tBid-induced
apoptosis. Proc Natl Acad Sci USA. 112:E5590–E5599. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
18
|
Luo X, Burwinkel B, Tao S and Brenner H:
MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer
Epidemiol Biomarkers Prev. 20:1272–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh
T, Kojima K, Nakashima R, Kitade Y and Naoe T: Role of
anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer
Gene Ther. 17:398–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
21
|
Peacock O, Lee AC, Cameron F, Tarbox R,
Vafadar-Isfahani N, Tufarelli C and Lund JN: Inflammation and
MiR-21 pathways functionally interact to downregulate PDCD4 in
colorectal cancer. PloS One. 9:e1102672014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin PL, Wu DW, Huang CC, He TY, Chou MC,
Sheu GT and Lee H: MicroRNA-21 promotes tumour malignancy via
increased nuclear translocation of β-catenin and predicts poor
outcome in APC-mutated but not in APC-wild-type colorectal cancer.
Carcinogenesis. 35:2175–2182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen J, Niu W, Zhou M and Zhang H, Ma J,
Wang L and Zhang H: MicroRNA-410 suppresses migration and invasion
by targeting MDM2 in gastric cancer. PloS One. 9:e1045102014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen N, Wang J, Hu Y, Cui B, Li W, Xu G,
Liu L and Liu S: MicroRNA-410 reduces the expression of vascular
endothelial growth factor and inhibits oxygen-induced retinal
neovascularization. PloS One. 9:e956652014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi SW, Kim JJ, Seo MS, Park SB, Kang TW,
Lee JY, Lee BC, Kang I, Shin TH, Kim HS, et al: miR-410 inhibition
induces RPE differentiation of amniotic epithelial stem cells via
overexpression of OTX2 and RPE65. Stem Cell Rev. 11:376–386. 2015.
View Article : Google Scholar
|
|
26
|
Chien WW, Domenech C, Catallo R, Kaddar T,
Magaud JP, Salles G and Ffrench M: Cyclin-dependent kinase 1
expression is inhibited by p16(INK4a) at the post-transcriptional
level through the microRNA pathway. Oncogene. 30:1880–1891. 2011.
View Article : Google Scholar
|
|
27
|
Chen L, Zhang J, Feng Y, Li R, Sun X, Du
W, Piao X, Wang H, Yang D, Sun Y, et al: MiR-410 regulates MET to
influence the proliferation and invasion of glioma. Int J Biochem
Cell Biol. 44:1711–1717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Fu J, Jiang M, Zhang X, Cheng L,
Xu X, Fan Z, Zhang J, Ye Q and Song H: MiR-410 is overexpressed in
liver and colorectal tumors and enhances tumor cell growth by
silencing FHL1 via a direct/indirect mechanism. PloS One.
9:e1087082014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Y and Bodmer WF: Analysis of P53
mutations and their expression in 56 colorectal cancer cell lines.
Proc Natl Acad Sci USA. 103:976–981. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oden-Gangloff A, Di Fiore F, Bibeau F,
Lamy A, Bougeard G, Charbonnier F, Blanchard F, Tougeron D, Ychou
M, Boissière F, et al: TP53 mutations predict disease control in
metastatic colorectal cancer treated with cetuximab-based
chemotherapy. Br J Cancer. 100:1330–1335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo HY and Xu RH: Predictive and
prognostic biomarkers with therapeutic targets in advanced
colorectal cancer. World J Gastroenterol. 20:3858–3874. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baltaziak M, Koda M, Wincewicz A,
Sulkowska M, Kanczuga-Koda L and Sulkowski S: Relationships of P53
and Bak with EPO and EPOR in human colorectal cancer. Anticancer
Res. 29:4151–4156. 2009.PubMed/NCBI
|
|
33
|
Kondo S, Shinomura Y, Miyazaki Y, Kiyohara
T, Tsutsui S, Kitamura S, Nagasawa Y, Nakahara M, Kanayama S and
Matsuzawa Y: Mutations of the bak gene in human gastric and
colorectal cancers. Cancer research. 60:4328–4330. 2000.PubMed/NCBI
|
|
34
|
Krajewska M, Moss SF, Krajewski S, Song K,
Holt PR and Reed JC: Elevated expression of Bcl-X and reduced Bak
in primary colorectal adenocarcinomas. Cancer Res. 56:2422–2427.
1996.PubMed/NCBI
|
|
35
|
Chen X, Liu J, Feng WK, Wu X and Chen SY:
MiR-125b protects against ethanol-induced apoptosis in neural crest
cells and mouse embryos by targeting Bak 1 and PUMA. Exp Neurol.
271:104–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li
Y, Li Z, Ng SS, Sung JJ, et al: miR-34a-5p suppresses colorectal
cancer metastasis and predicts recurrence in patients with stage
II/III colorectal cancer. Oncogene. 34:4142–4152. 2015. View Article : Google Scholar
|
|
38
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Talotta F, Cimmino A, Matarazzo MR,
Casalino L, De Vita G, D'Esposito M, Di Lauro R and Verde P: An
autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1
activity in RAS transformation. Oncogene. 28:73–84. 2009.
View Article : Google Scholar
|


















