Introduction
The primary treatment of sinoatrial node (SAN)
dysfunction due to congenital defects, acquired diseases, gene
mutations and aging is electronic pacemaker implantation. However,
such devices are not optimal choices due to deficiencies in
biological responsiveness and other shortages, such as lack of
autonomic modulation, limited battery life, the need for permanent
catheter implantation into the heart, unstable electrode position,
and electronic and magnetic interference (1). A cell therapy approach to produce a
biological pacemaker focuses on the overexpression of
hyperpolarization-activated cyclic nucleotide-gated cation channel
(HCN4) which can induce 'funny' current If. The
If current is known to be essential for the spontaneous
diastolic depolarization of SAN cells (2–4). In
our previous study, it was demonstrated that HCN4-transfected cMSCs
in vivo can induce spontaneous activity; however, the
spontaneous rates were lower than that of normal SAN cells
(3). In addition, it was indicated
that HCN4 loss was a cause of the dysfunction of SAN cells and
resulted in abnormally slow heart rates (3–5). To
overcome this obstacle, identification of an upstream gene is
required in order to maintain the high expression of HCN4.
SAN development is a strictly regulated process, and
a number of signaling molecules are involved. Shox2, a member of
the short stature homeobox family, is an early cardiac
transcription factor which has been identified to be uniquely
expressed in the SAN region (6).
Studies in Shox2 knockout mouse models demonstrated that Shox2 is
crucial in the formation and differentiation of SAN by regulating
the genetic cascade. A Shox2 null mutation may lead to heart
defects, including cardiac edema and hypoplasia of the SAN due to a
reduced level of cell proliferation, which results in a decrease in
the heart rate (6). Shox2 mutation
has also been shown to impact the SAN genetic network resulting in
the downregulation of HCN4 in this region (6). Conversely, overexpression of Shox2 in
mice and Xenopus embryos showed a decreased expression of working
myocardium markers (6,7). However, whether overexpression of
Shox2 in MSCs is able to establish a phenotype similar to native
pacemaker cells and improve pacemaker function remains unknown.
The aim of the present study was to identify an
upstream gene to maintain high expression of HCN4. A previous study
demonstrated that Shox2 is important in the differentiation of SAN
and is an upstream gene of HCN4 (6). A number of studies have indicated
that co-culture of MSCs with neonatal cardiomyocytes (CMs) can
induce MSC differentiation into CMs in vitro, providing a
model for heart tissue engineering research (8–10).
Based on these data, canine MSCs (cMSCs) effectively transfected
with a lentiviral vector encoding a mouse Shox2 (mShox2) gene was
employed in this study, and a model of direct co-culture of cMSCs
with rat neonatal CMs (RNCMs) was established. Then levels of
functional markers characterizing mature SAN cells, such as T box 3
(Tbx3), HCN4 and Connexin 45 (Cx45) were evaluated. It was
investigated whether the regulation of the differentiation of cMSCs
into pacemaker-like cells through the overexpression of Shox2 is
feasible.
Materials and methods
cMSCs culture and identification
cMSCs were isolated from the bone marrow of three
adult dogs (two males and one female; Third Military Medical
University, Chongqing, China), weighing 10–14 kg, as previously
described (3). In brief, bone
marrow aspiration from the femurs and tibias was performed on dogs
anesthetized with 30 mg/kg intravenous sodium pentobarbital
(Sigma-Aldrich, St. Louis, MO, USA). The cells obtained were grown
in α-mimium essential medium (Hyclone Laboratories, Inc., Logan,
UT, USA), supplemented with 0.22% HEPES, 0.22%
Na2CO3, 10% fetal calf serum (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and
100 µg/ml streptomycin (Hyclone Laboratories, Inc.), and
incubated in a 37°C, 5% CO2 humidified atmosphere. These
cMSCs were identified via flow cytometry with
CD34−/CD45−/CD29+/CD44+
and for the ability to differentiate into adipogenic, osteogenic
and chondrogenic lineages as previously described (3). The study was approved by the ethics
committee of the Third Military Medical University.
Construction of mShox2 lentiviral vector
and mShox2 infection
The lentiviral vector expressing mShox2
(pLentis-mShox2-RFP) was constructed by inserting the mShox2 gene
into a pLentis-RFP vector using BamHI (FD0054) and
EcoRI (N41890) restriction sites, all obtained from
Invitrogen (Thermo Fisher Scientific, Inc.). The lentiviral
particles were prepared using a calcium phosphate method, as
previously described (2–4). Third generation cMSCs were
transfected with pLentis-mShox2-RFP or pLentis-RFP in the presence
of 2 µg/ml polybrene (Sigma-Aldrich) at a multiplicity of
infection (MOI) of 20 for 24 h. The expression of RFP after 48 h
was >90% of the infected cells.
Co-culture conditions
Primary RNCMs were isolated from newborn Sprague
Dawley rats in 2 days following by the steps used in our lab
(11). Rats were obtained from the
Third Military Medical University. Animal experiments were
performed in accordance with the Guide for the Care and Use of
Laboratory Animals published by the Third Military Medical
University and approved by the Committee on the Ethics of Animal
Experiments of the Third Military Medical University (permit
number: SYXK201012). Experiments were conducted at least in
triplicate and >30 rats were used. Briefly, RNCMs were isolated
by digestion with 1 mg/ml type I collagenase (Sigma-Aldrich) with
0.08% trypsin (Ameresco, Solon, OH, USA) diluted with
Ca+ Mg+-free D-Hanks (Wuhan Boster Biological
Technology, Ltd., Wuhan, China). The harvested RNCMs were purified
by pre-plating (30 min, 37°C). For co-culture experiments, cMSCs
and RNCMs were mixed and plated at a ratio of 1:4 (20% cMSCs) onto
the 60-cm2 culture dishes (Corning, Inc., Corning, NY,
USA). The cell mixtures were co-cultured for 5–7 days as isotropic
monolayers, with a medium change after 24 h. Fluorescence mapping
was performed after 5 days in culture, and cMSCs were identified
with red fluorescence observed using a BX41 microscope (Olympus
Corporation, Tokyo, Japan). RNCMs mature to form rod-shaped
striated cells after 48 h in culture. The co-culture experiment was
performed three times to validate the results. For reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blotting, the cMSCs were selected by treatment with
puromycin (3 µg/ml; Sigma-Aldrich) for 3 days in addition to
fluorescence-activated cell sorting for RFP.
Immunofluorescence
Immunofluorescence was conducted as previously
described (11). Transfected cMSCs
were fixed with 4% paraformaldehyde (Wuhan Boster Biological
Technology, Ltd., Wuhan, China) for 15 min at room temperature,
washed in phosphate-buffered saline, then treated with 0.2% Triton
(Sigma-Aldrich) for 15 min. Cells were incubated with goat
polyclonal anti-Shox2 (cat. no. sc-21898, Santa Cruz Biotechnology
Inc., Santa Cruz Biotechnology Inc.; 1:50) and rabbit polyclonal
anti-Cx45 (cat. no. sc-25716, Santa Cruz Biotechnology Inc.; 1:50)
primary antibodies overnight at 4°C. They were then incubated with
donkey anti-goat IgG antibodies conjugated to Alexa Fluor 488 (cat.
no. A-11055, Invitrogen, Thermo Fisher Scientific, Inc.; 1:100) for
Shox2 and donkey anti-rabbit IgG antibody conjugated to Alexa Fluor
488 (A21206, Invitrogen, Thermo Fisher Scientific, Inc.; 1:100) for
Cx45 for 60 min. After further washing, the cells were mounted with
antifade mounting medium (Beyotime Institute of Biotechnology,
Shanghai, China). The nuclei stained with
4′,6-diamidino-2-phenylindole (Sigma-Aldrich) was used as a
location control. The fluorescent images were obtained with a Zeiss
LSM710 laser confocal microscope (Carl Zeiss Microscopy GmbH, Jena,
Germany). The results were analyzed with ZEN lite 2011 software
(Carl Zeiss Microscopy GmbH).
RT-qPCR analyses
RT-qPCR was performed according a previous study
(12). All primers (Table I) were synthesized by Invitrogen,
Thermo Fisher Scientific, Inc. (Shanghai, China). The total
cellular mRNA was extracted with TRIzol (Invitrogen Thermo Fisher
Scientific, Inc.). Subsequently, cDNA synthesis was performed
according to the manufacturer's instructions. RT-qPCR was performed
with SYBR Green Realtime PCR Master mix (QPK-201, Toyobo, Co.,
Ltd., Osaka, Japan) on a Stratagene Mx3000P (Agilent Technologies,
Inc., Santa Clara, CA, USA) instrument. Quantitative measurements
were determined using the comparative Cq (2−ΔΔCq) method
(13). All samples were normalized
by endogenous level of glyceraldehyde 3-phosphate dehydrogenase
(GAPDH). All results were repeated three times.
 | Table IPrimers used in reverse
transcription-quantitative polymerase chain reaction analyses. |
Table I
Primers used in reverse
transcription-quantitative polymerase chain reaction analyses.
| Gene | Forward | Reverse |
|---|
| Shox2 |
5′-ACTATCCAGACGCTTTCATGCG-3′ |
5′-TTCGATTTTGAAACCAAACCTGTAC-3′ |
| Tbx3 |
5′-GTAAGATGTTCTGGGCTGGATAAA-3′ | 5′-GTAGCAGGGCTGTCTGGG
TG-3′ |
| HCN4 |
5′-AGGGCACCATCGGCAAGA-3′ |
5′-CCACGCTCAGCGAATACAGG-3′ |
| Cx45 |
5′-CAGCAGACTTCCTTGCCCTCATA-3′ |
5′-CTTAGCATTGGACAGTTCGGTGT-3′ |
| Nkx2.5 |
5′-CCGAGCCTGGTAGGAAAGGG-3′ |
5′-AAATCCAAGGGACGTGGAGACA-3′ |
| Cx43 |
5′-TGCTATGACAAATCCTTCCCAATC-3′ |
5′-GCCGTGCTCTTCAATTCCATACTT-3′ |
| GAPDH |
5′-GAGATCCCGCCAACATCAAA-3′ |
5′-GGCATCAGCAGAAGGAGCAG-3′ |
Western blotting
Western blotting was conducted as previously
described (11). Briefly, cMSCs,
washed in PBS, were lysed in radioimmunoprecipitation assay buffer
containing phenylmethylsulfonyl fluoride (RIPA/PMSF), then the
total protein in the sample was quantified using a bicinchoninic
acid protein assay kit (Beyotime Institute of Biotechnology),
according to the manufacturer's instructions. Briefly, the
membranes were blocked with 5% bovine serum albumin (Gibco; Thermo
Fisher Scientific, Inc.) in TBS solution for 3 h at room
temperature with shaking. The membranes were then incubated with
primary goat polyclonal antibody against Shox2 (cat. no. sc-21898,
Santa Cruz Biotechnology Inc.; 1:200) and GAPDH (cat. no. sc-48166,
Santa Cruz Biotechnology Inc.; 1:200) and rabbit polyclonal
antibodies against Tbx3 (cat. no. sc-48781, Santa Cruz
Biotechnology Inc.; 1:200), Cx45 (cat. no. sc-25716, Santa Cruz
Biotechnology Inc.; 1:200), Cx43 (cat. no. sc-9059, Santa Cruz
Biotechnology Inc.; 1:200), HCN4 (cat. no. ab69054, Abcam,
Cambridge, UK; 1:100) and Nkx2.5 (cat. no. ab97355, Abcam, UK;
1:500) were incubated separately overnight at 4°C with gentle
agitation. After >3 washes in Tris-buffered saline with
Tween-20, the membranes were incubated for 2 h at room temperature
with corresponding secondary antibodies: Donkey anti-goat
IgG-horseradish peroxidase (HRP) (cat no. sc-2020, Santa Cruz
Biotechnology Inc.; 1:5,000) and mouse anti-rabbit IgG-HRP (cat no.
sc-2357; Santa Cruz Biotechnology Inc.; 1:5,000). Then specific
bands of target proteins were visualized using an enhanced
chemiluminescence detection kit (Beyotime Institute of
Biotechnology) according to the manufacturer's recommendations.
Finally, the target signals were normalized to the GAPDH signal.
Experiments were performed several times to verify results.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Statistical comparisons among multiple groups were
analyzed by one-way analysis of variance with Dunnett's T3 test
with SPSS 19.0 software (IBM, Armonk, NY, USA). P≤0.05 was
considered to indicate a statistically significant difference.
Results
Infection, expression, co-culture of
cMSCs and morphological changes
At 2 days after transfection, the infected cMSCs
exhibited red fluorescence. The transfection rates of cMSCs were
94±2.5%, which was analyzed by confocal laser microscope images in
at least four different random fields. The Shox2-RFP transfected
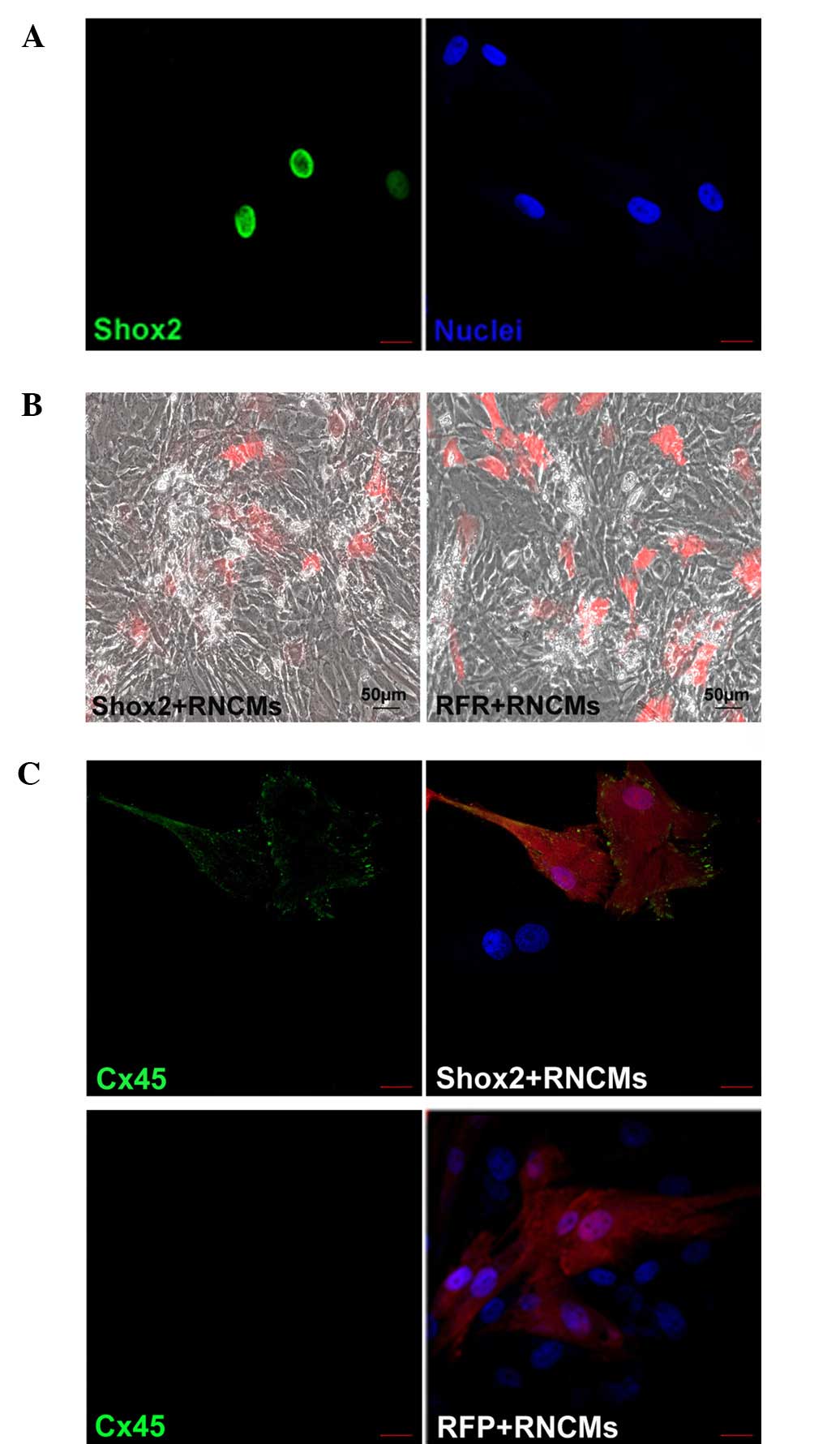
cMSCs expressed Shox2 protein (Fig.
1A), in concordance with the result obtained by Liu et
al (14). The growth rate of
these cMSCs decreased. In addition, their morphology was observed
to change and some cells were long-rod or furcation shaped. After
co-culture with RNCMs, the cMSCs were larger with spindle and
spider-like morphologies.
Fluorescence microscopy revealed that cMSCs were
randomly distributed in culture and that a large number existed in
a plane below the RNCMs with some interspersed between the RNCMs
(Fig. 1B). This is perhaps due to
the fact that cMSCs adhere to the coverslips within 3 to 4 h,
whereas RNCMs take up to 24 h to fully adhere.
An important characteristic of cMSCs is their
ability to assemble gap junctions between themselves and with
neighboring CMs (15). Connexin
molecule Cx45 is a marker of the SAN, Fig. 1C shows Cx45 expression in cMSC
cultures by immunofluorescence. It was demonstrated that the
expression of Cx45 in Shox2-transfected cMSCs co-cultured with
RNCMs was higher than that of the control cells.
Co-culturing Shox2-transfected cMSCs with
RNCMs upregulates SAN-marker expression
To investigate the role of Shox2 in the
differentiation of SAN, the expression status of several genes was
investigated. These genes included those that have been used to
identify SAN differentiation or are known to be important for SAN
formation and function, including Tbx3, a transcription factor
expressed in SAN with a role in SAN development; HCN4, a molecular
marker of pacemaker cells; and Cx45, which prevents the areas of
conductivity inside the SAN. The results demonstrated that
overexpression of Shox2 significantly increased the expression of
Tbx3, HCN4 and Cx45 at the mRNA (P<0.05) and protein levels, and
the difference increased markedly when co-cultured with RNCMs
(Figs. 2 and 3).
 | Figure 2Shox2, Tbx3, HCN4, Cx45, Nkx2.5 and
Cx43 gene expression was examined using reverse
transcription-quantitative polymerase chain reaction. Similar
results were obtained in three independent experiments. Data are
presented as the mean ± standard error of the mean.
*P<0.05 vs. control. Shox2, Short stature homeobox 2;
Tbx3, T box 3; HCN4, hyperpolarization-activated cyclic
nucleotide-gated cation channel; Cx45, connexin 45; Cx43, Connexin
43; RFP, red fluorescent protein; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; RNCMs, rat neonatal cardiomyocytes. |
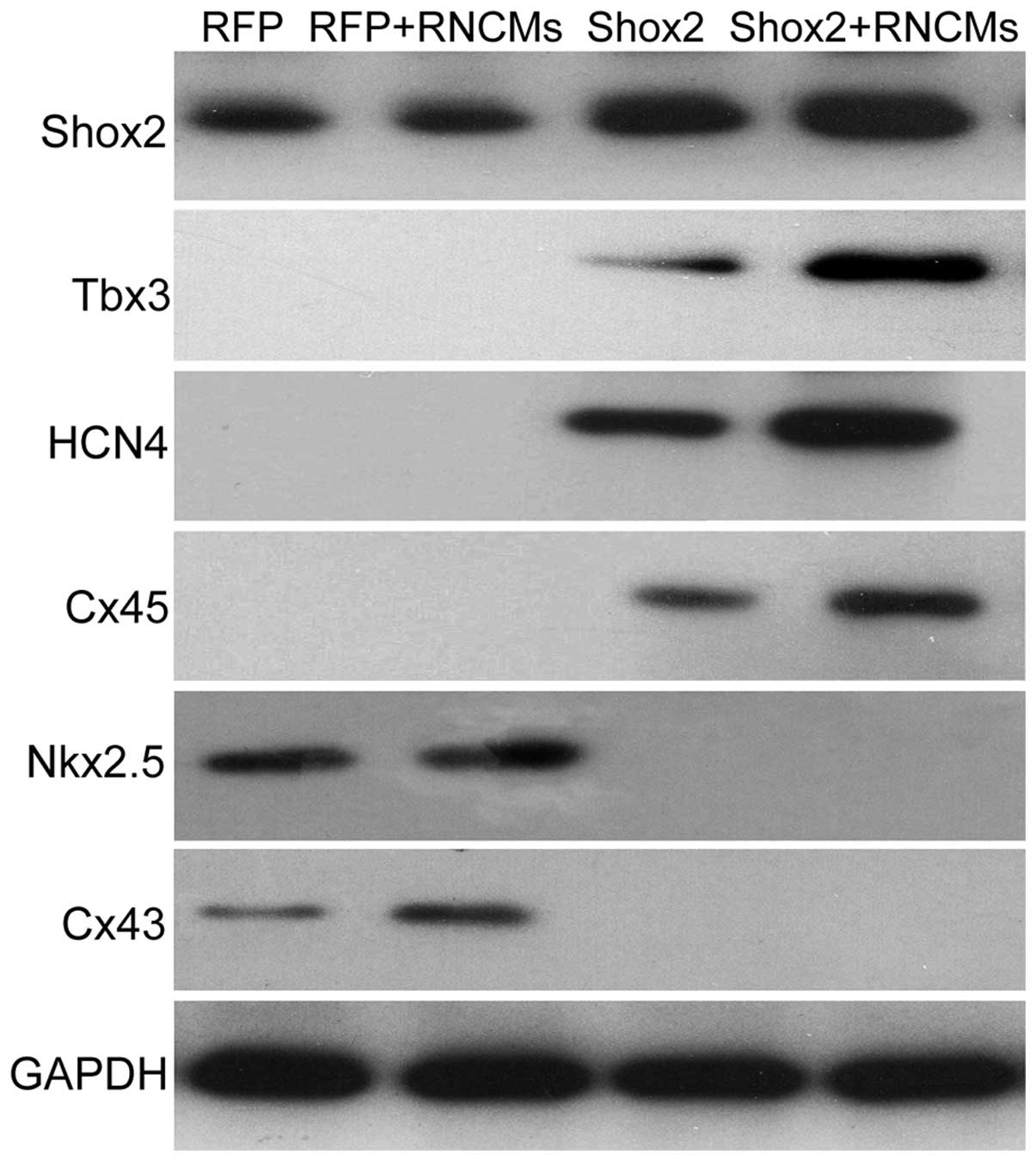 | Figure 3Shox2, Tbx3, HCN4, Cx45, Nkx2.5 and
Cx43 protein expression were examined using western blotting.
Similar results were obtained in three independent experiments.
Shox2, Short stature homeobox 2; Tbx3, T box 3; HCN4,
hyperpolarization-activated cyclic nucleotide-gated cation channel;
Cx45, connexin 45; Cx43, Connexin 43; RFP, red fluorescent protein;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; RNCMs, rat
neonatal cardiomyocytes. |
The effects of Shox2 on the expression changes of
two working myocardium markers Nkx2.5 and Cx43 in cMSCs co-cultured
with RNCMs were also examined. As shown in Fig. 2, Nkx2.5 and Cx43 mRNA were
downregulated in the Shox2 overexpression cMSCs compared with the
negative control group, and the difference was statistically
significant in the co-culture group (P<0.05). By contrast, the
expression of Nkx2.5 and Cx43 were significantly increased in
RFP-cMSCs co-cultured with RNCMs at the mRNA and protein levels
(Figs. 2 and 3, P<0.05), in concordance with results
obtained by Li et al (9).
Shox2-transfected cMSCs drive the rate of
co-cultured RNCMs
It was then evaluated whether overexpression of
Shox2 in cMSCs can result in a pacemaker phenotype. After 5 days in
co-culture, when a syncytium was established, the beat rates were
counted. The mean rate was (167±38 bpm, n=6) in RNCMs co-cultured
with Shox2-transfected cMSCs, which was markedly higher than the
rate obtained from RNCMs co-cultured with control cells (79±15 bpm,
n=5) (Fig. 1B).
Discussion
In the present study, cMSCs overexpressing Shox2
were co-cultured with RNCMs. The results demonstrated the
functional role of Shox2 in pacemaker cell differentiation, which
indicated that overexpression of Shox2 in cMSCs can greatly enhance
the pacemaker phenotype in a co-culture model in vitro.
In our previous study, it was demonstrated that HCN4
transfected cMSCs can induce spontaneous activity in vivo;
however, the spontaneous rates were lower than those in the normal
SAN cells(3). It was hypothesized
that this may be due to engrafted HCN4 gene loss in the host heart
microenvironment (3–5). In our previous study, it was
confirmed that overexpression of Shox2 in cMSCs could upregulate
HCN4 expression, and its level was significantly increased by
co-culture induction. This was accompanied by altered expression of
several other genes essential for SAN formation. Previous studies
have demonstrated high expression levels of early transcription
factor Tbx3 and low levels of conductance gap junction protein Cx45
in the SAN (9,16,17).
By contrast, the expression of transcription factor Nkx2.5 and high
conductance gap junction protein Cx43 is widely observed in working
myocardium but not in SAN (7,9).
Increased Tbx3, HCN4 and Cx45 expression, and loss of Nkx2.5 and
Cx43 expression indicated the formation of SAN-like cells (6,7,16,17).
In this study, it was indicated that Tbx3 and Cx45 were
significantly upregulated in mShox2-RFP transfected cMSCs.
Additionally, co-culturing with RNCMs enhanced this effect and it
was accompanied by the downregulated expression of Nkx2.5 and Cx43.
Co-culturing with RNCMs was shown to provide a model mimicking the
physiological microenvironment of the heart and constructed
Shox2-cMSCs were able to differentiate into SAN like cells when
co-cultured with RNCMs.
Genetically, SAN is a complicated and tightly
regulated process including a variety of signaling molecules. Tbx3
is a member of the T-box family, particularly expressed in the
cardiac conduction system, including in the SAN, and is crucial
during heart embryogenesis. Previous studies have suggested that
Tbx3 can repress the expression of chamber-specific genes, such as
Cx40, Cx43 and Nppa, and promote the pacemaker phenotype (16). By contrast, in Tbx3 mutants these
markers may span the entire SAN and cause lethal arrhythmias: sinus
pauses, bradycardia, atrioventricular block and sudden death
(18). In this study, it was
demonstrated that Tbx3 was significantly upregulated in mShox2
transfected cMSCs and co-culturing with RNCMs enhanced this effect.
In addition to the downregulation of Tbx3 in heart tissue with
Shox2 null mutation as reported by Espinoza-Lewis et al
(6), it was hypothesized in the
present study that Shox2 may act earlier than Tbx3 in pacemaker
differentiation. In addition, Cx45 was also upregulated in
Shox2-transfected cMSCs in this study, and the difference became
greater when co-cultured with RNCMs. Cx45 is a member of the
connexin gene family that can form gap junctions with low
conductance preventing the suppressing hyperpolarizing influence of
the atrium, and is seen as another marker of SAN (19,20).
Therefore, it was suggested that Shox2 can promote pacemaker
differentiation by enhancing the expression of Tbx3 and Cx45 in
vitro.
In this study, another cardiac transcription factor
Nkx2.5 was also examined, which is critical for working myocardium
differentiation but is not expressed in SAN (7,9).
Previous studies have shown that ectopic expression of Nkx2.5 can
suppress the formation of SAN in the following ways: i)
Histologically, hypoplastic SAN is observed to be attributed to
reduced cell proliferation, thinned atrial wall and thickened
ventricular wall; ii) functionally, overexpression of Nkx2.5 in the
heart results in a reduced heartbeat rate; and iii) genetically,
Nkx2.5 can induce differentiation of working myocardium, resulting
in downregulation of HCN4 and Tbx3, and ectopic expression of Cx40
and Nppa in the SAN region (7).
These changes in histology, function and genetics are consistent
with the findings observed in Shox2-deficient embryonic hearts
(6). Recently, it has been
demonstrated that the expression patterns of Nkx2.5 and Shox2 are
mutually exclusive during SAN formation (14). Indeed, Shox2 injected in mice and
Xenopus embryos may lead to a downregulation of Nkx2.5 (6), and it was also indicated that
overexpression of Shox2 in cMSCs in vitro inhibited the
expression of Nkx2.5 when co-culturing with RNCMs. Thus, Shox2 may
act as an Nkx2.5 repressor to regulate SAN differentiation. The
effect of Shox2-overexpression on Nkx2.5 causes cells to
differentiate into pacemaker-like cells rather than working
myocardium. Taking Tbx3 and HCN4 as downstream of Nkx2.5, it may be
possible to improve pacemaker function by affecting the levels of
Shox2, Nkx2.5, Tbx3, HCN4, Cx43 and Cx45. In this network, Shox2
firstly inhibits Nkx2.5 expression and activates the pacemaker
differentiation program. Then, Tbx3, HCN4 and Cx45 are in turn
expressed. In addition, the activation of Tbx3 further inhibits the
expression of Cx43.
In conclusion, the results indicated that
overexpression of Shox2 can regulate the differentiation of cMSCs
into pacemaker-like cells and promote pacemaker function in
vitro. This offers a good model for the development of
biological pacemakers. Additionally, this study provides a basis
for future in vivo experiments in dogs using
Shox2-transfected cMSCs, and insight into future gene-targeted and
regenerative therapeutic strategies for SAN dysfunction in
humans.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 81270246).
References
|
1
|
Miake J, Marbán E and Nuss HB: Biological
pacemaker created by gene transfer. Nature. 19:132–133. 2002.
View Article : Google Scholar
|
|
2
|
Tong S, Yao Q, Wan Y, Zhou J, Shu M, Zhong
L, Li Y, Zhang Q, Yindai J and Song Z: Development of functional I
f channels in mMSCs after transfection with mHCN4: Effects on cell
morphology and mechanical activity in vitro. Cardiology.
112:114–121. 2009. View Article : Google Scholar
|
|
3
|
Jun C, Zhihui Z, Lu W, Yaoming N, Lei W,
Yao Q and Zhiyuan S: Canine bone marrow mesenchymal stromal cells
with lentiviral mHCN4 gene transfer create cardiac pacemakers.
Cytotherapy. 14:529–539. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu W, Yaoming N, Boli R, Jun C, Changhai
Z, Yang Z and Zhiyuan S: mHCN4 genetically modified canine
mesenchymal stem cells provide biological pacemaking function in
complete dogs with atrioventricular block. Pacing Clin
Electrophysiol. 36:1138–1149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai J, Yi FF, Li YH, Yang XC, Song J,
Jiang XJ, Jiang H, Lin GS and Wang W: Adenoviral gene transfer of
HCN4 creates a genetic pacemaker in pigs with complete
atrioventricular block. Life Sci. 80:1476–1753. 2007. View Article : Google Scholar
|
|
6
|
Espinoza-Lewis RA, Yu L, He F, Liu H, Tang
R, Shi J, Sun X, Martin JF, Wang D, Yang J and Chen Y: Shox2 is
essential for the differentiation of cardiac pacemaker cells by
repressing Nkx2-5. Dev Biol. 327:376–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Espinoza-Lewis RA, Liu H, Sun C, Chen C,
Jiao K and Chen Y: Ectopic expression of Nkx2.5 suppresses the
formation of the sinoatrial node in mice. Dev Biol. 356:359–369.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang MG, Tung L, Sekar RB, Chang CY,
Cysyk J, Dong P, Marbán E and Abraham MR: Proarrhythmic potential
of mesenchymal stem cell transplantation revealed in an in vitro
coculture model. Circulation. 113:1832–1841. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Li B, Zhang C, Zhang J, Zeng M and
Zheng Z: Effect of NRG-1/ErbB signaling intervention on the
differentiation of bone marrow stromal cells into sinus node-like
cells. J Cardiovasc Pharmacol. 63:434–440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang T, Xu Z, Jiang W and Ma A:
Cell-to-cell contact induces mesenchymal stem cell to differentiate
into cardiomyocyte and smooth muscle cell. Int J Cardiol.
109:74–81. 2006. View Article : Google Scholar
|
|
11
|
Wen L, Zhang C, Nong Y, Yao Q and Song Z:
Mild electrical pulse current stimulation upregulates S100A4 and
promotes cardiogenesis in MSC and cardiac myocytes coculture
monolayer. Cell Biochem Biophys. 65:43–55. 2013. View Article : Google Scholar
|
|
12
|
Kim MO, Jung H, Kim SC, Park JK and Seo
YK: Electromagnetic fields and nanomagnetic particles increase the
osteogenic differentiation of human bone marrow-derived mesenchymal
stem cells. Int J Mol Med. 35:153–160. 2015.
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-timequantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Liu H, Chen CH, Ye W, Espinoza-Lewis RA,
Hu X, Zhang Y and Chen Y: Phosphorylation of Shox2 is required for
its function to control sinoatrial node formation. J Am Heart
Assoc. 3. pp. e0007962014, View Article : Google Scholar
|
|
15
|
Valiunas V, Doronin S, Valiuniene L,
Potapova I, Zuckerman J, Walcott B, Robinson RB, Rosen MR, Brink PR
and Cohen IS: Human mesenchymal stem cells make cardiac connexins
and form functional gap junctions. J Physiol. 555:617–623. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hoogaars WM, Engel A, Brons JF, Verkerk
AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P,
et al: Tbx3 controls the sinoatrial node gene program and imposes
pacemaker function on the atria. Genes Dev. 21:1098–1112. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wiese C, Grieskamp T, Airik R, Mommersteeg
MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF,
Kispert A and Christoffels VM: Formation of the sinus node head and
differentiation of sinus node myocardium are independently
regulated by Tbx18 and Tbx3. Circ Res. 104:388–397. 2009.
View Article : Google Scholar
|
|
18
|
Frank DU, Carter KL, Thomas KR, Burr RM,
Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels
VM and Moon AM: Lethal arrhythmias in Tbx3-deficient mice reveal
extreme dosage sensitivity of cardiac conduction system function
and homeostasis. Proc Natl Acad Sci USA. 109:E154–E163. 2012.
View Article : Google Scholar :
|
|
19
|
Söhl G and Willecke K: Gap junctions and
the connexin protein family. Cardiovasc Res. 62:228–232. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Desplantez T, Dupont E, Severs NJ and
Weingart R: Gap junction channels and cardiac impulse propagation.
J Membr Biol. 218:13–28. 2007. View Article : Google Scholar : PubMed/NCBI
|