Introduction
Fatty acid-binding protein 4 (FABP4), also referred
to as aP2 and adipocyte FABP, is expressed at high levels in
adipocytes and macrophages, and is a cytosolic lipid chaperone,
which regulates cellular lipid metabolism and the reception of
lipid signals (1), which are
actively involved in atherogenic processes. FABP4-deficiency has
been shown to protect against atherosclerosis (AS) in
apolipoprotein E-deficient mice, and FABP4 knockout animal models
show reduced lipolysis, which offers protection from the
development of AS (2).
Investigations in bone marrow transplantation have also
demonstrated that this atheroprotective effect of FABP4 is
predominantly associated with its actions in macrophages (3). Thus, it is clear that the involvement
of FABP4 is significant in local macrophage responses in
atherogenesis, however, the precise cellular and molecular
mechanisms underlying these phenomena remain to be fully
elucidated. Previous studies have reported the presence of FABP4 in
macrophages, and have shown that the expression of FABP4 can be
induced by oxidized low density lipoprotein (oxLDL), peroxisome
proliferator-activated receptor γ (PPARγ) agonists and the
differentiation of monocytes to macrophages (4–6).
Homocysteine (Hcy) is a homologue of the amino acid,
cysteine, differing by an additional methylene bridge (–CH2–).
Accumulating evidence has suggested that hyperhomocysteinemia
(HHcy) is an independent risk factor of AS (7). However, the mechanisms responsible,
which may be multifactorial, remain to be fully elucidated
(8). Increasing evidence has
indicated that Hcy may be involved in the disturbance of the
expression of AS-associated genes through the interference of DNA
methylation (9). The metabolism of
Hcy is specifically involved in the methionine cycle for
transmethylation reactions, therefore, its elevation has been
implicated in the interference of DNA methylation modification and
the epigenetic regulation of genes (10).
DNA methylation, the addition of a methyl group to
form 5-methylcytosine in the context of a CpG dinucleotide via
methyltransferase, refers to a form of epigenetic modification. It
has been demonstrated that aberrant DNA methylation is involved in
Hcy-associated pathology, which exhibits global hypomethylation,
however, hypermethylation occurs in certain genes. DNA
hypermethylation leads to gene silencing, whereas hypomethylation
results in gene activation in the regulatory region, together with
altered binding profiles of transcription factors (11). Chronic human diseases, including
AS, are either caused or affected by abnormal DNA methylation
(12). Previous compelling
evidence has indicated that Hcy epigenetically regulates certain
specific targets, including 15-lipoxygenase, extracellular
superoxide dismutase, estrogen receptor α, cyclin A and PPARs by
inhibiting DNA meth-ylation, which in turn favors early AS
(13,14). Therefore, the interference of DNA
methylation may be an important mechanism of Hcy-induced AS.
However, whether Hcy can affect the DNA methylation and expression
of FABP4 in macrophages, and further regulate cellular lipid
metabolism remains to be elucidated.
Therefore, the present study hypothesized that Hcy,
at a relevant concentration, affects the level of FABP4 DNA
methylation and accelerates the development of AS through the
accumulation of cholesterol in macrophages. Investigation to
confirm this hypothesis may elucidate a potential mechanism against
AS, in which epigenetic gene silencing is a feature.
Materials and methods
THP-1 cell culture and cell
treatment
Human monocytic leukemia THP-1 cells (West China
School of Preclinical and Forensic Medicine, Sichuan University,
Sichuan, China) were cultured in RPMI 1640 medium supplemented with
15% (v/v) heat-inactivated fetal bovine serum (GE Healthcare Life
Sciences, Logan, UT, USA) in a humidified atmosphere of 5%
CO2 at 37°C. The monocytes were initially differentiated
into macrophages by the addition of 500 nM phorbol 12-myristate
13-acetate (PMA; Sigma-Aldrich, St. Louis, MO, USA) for 48 h,
following which the macrophages were incubated (37°C, 5%
CO2) in medium with different concentrations of Hcy and
its antagonist for 24 h. In the present study, the cells were
divided into five groups: Cells (2×106) were exposed to
different concentrations of Hcy (50, 100, 200 and 500 µM),
as the Hcy group, others were incubated with Hcy (100 µM)
and folic acid (FA; 30 µM) and vitamin B12
(VB12; 30 µM), as the treatment group, and the
remainder were treated without Hcy, as a control group. The cells
in all groups were cultured for another 24 h, and all cells were
used for subsequent analysis.
Construction of the pcDNA3.1-EGFP/FABP4
recombinant plasmid and its transfection into THP-1
monocyte-derived macrophages
The human FABP4 gene was amplified from the
cDNA of THP-1 macrophages. The human FABP4 gene and the
pcDNA3.1-EGFP vector were double-digested using EcoRI and
HandIII restriction endonucleases (Takara Bio Inc., Otsu,
Japan), and the FABP4 gene fragment was then inserted into
the pcDNA3.1-EGFP cloning vector to construct the recombinant
plasmid, pcDNA3.1-EGFP/FABP4. The recombinant pcDNA3.1-EGFP/FABP4
plasmid was identified using restriction endonuclease digestion
analysis and DNA sequencing. According to the manufacturer's
protocol, the THP-1 macrophages grown in 6-well plates were
transfected with the recombinant plasmid (pcDNA3.1-EGFP/FABP4) and
empty plasmid (pcDNA3.1-EGFP), mediated by liposome reagent
(Lipofectamine 2000; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) as the recombinant plasmid group and the empty
plasmid control group, respectively. The untransfeced group was set
as the normal control group. The levels of green fluorescence in
the cells were observed using fluorescence microscopy, and the
expression levels of FABP4 were detected using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses.
RT-qPCR for the mRNA expression of
FABP4
Total RNA was extracted from the cultured
macrophages using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), following which the RNA was reverse transcribed
into cDNA using a RevertAid first strand cDNA synthesis kit (MBI
Fermentas, Inc., Vilnius, Lithuania), according to the
manufacturer's protocol. The cDNAs (2 µl) were amplified
using a SYBR Green PCR kit (MBI Fermentas, Inc.). Primer Premier 5
software (Premier Biosoft, Palo Alto, CA, USA) was used to design
the primers. The primer nucleotide sequences of FABP4 (NM_001442.2;
https://www.ncbi.nlm.nih.gov/nuccore/NM_001442.2)
were as follows: Forward 5′-TACTGAGATTTCCTTCATACTGGGC-3′ and
reverse 5′-GCTCTCTCATAAACTCTCGTGGAAG-3′, with a length of 372 bp.
The gene expression of FABP4 was normalized to that of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse 5′-AGGGGCCATCCACAGTCTTC-3′.
The RT-qPCR reaction was performed using an FTC-3000 real-time PCR
detection system (Funglyn, Toronto, Canada), according to the
manufacturer's protocol, as follows: 94°C for 10 min, 50 cycles at
94°C for 15 sec, at the annealing temperature of 53°C for 30 sec
and 72°C for 30 sec. The RNA level of the gene was acquired from
the value of the quantification cycle (Cq) of the RT-qPCR
associated with that of GAPDH using the following formula: ΔCq, ΔCq
= CqGAPDH − Cqgene (15). The final results were expressed as
N-fold differences in the target gene expression, relative to the
calibrator, termed 'Ntarget', which was determined as
follows: Ntarget = 2ΔCqsample −
ΔCqcalibrator, where the ΔCq values of the calibrator and
sample were determined by subtracting the Cq value of the target
gene from the Cq value of GAPDH, as previously described (11).
Western blot analysis for FABP4
The cultured macrophages were harvested by scraping
with a plastic scraper and washing with phosphate-buffered saline
(PBS). The protein was extracted using a protein extraction kit
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), and protein
concentration was determined using a Bicinchoninic Acid Protein
Assay kit (Nanjing KeyGen Biotech Co., Ltd.). Equal quantities of
protein (80 µg) and a known molecular weight marker were
loaded onto 12% sodium dodecyl sulfate-polyacrylamide gels (Nanjing
KeyGen Biotech Co., Ltd.) and were transferred onto a
polyvinylidene fluoride membrane (EMD Millipore, Boston, MA, USA)
by electrophoresis at 300 mA for 50 min at 4°C. The membrane was
then blocked in 10 ml 5% skimmed milk for 2 h at room temperature
with gentle agitation on a platform shaker. The blocked membranes
was respectively incubated with a rabbit monoclonal anti-FABP4
antibody (cat. no. MABS172; 1:400 dilution; EMD Millipore) and
β-actin antibody (cat. no. TA-09; 1:2,000 dilution; ZSGB-BIQ,
Beijing, China) in 10 ml primary antibody dilution buffer overnight
at 4°C. The membrane was then washed three times for 10 min/wash
with PBS-0.1% Tween-20 (PBST) and incubated with secondary antibody
(cat. no. ab6721; goat anti-rabbit horseradish peroxidase-IgG;
Abcam, Cambridge, MA, USA) in PBS at 1:2,000 dilution for 2 h at
room temperature. Following being washed three times with PBST, the
FABP4 on the membrane was incubated with enhanced chemiluminescence
(ECL; Elabscience Biotechnology Co., Ltd., Wuhan, China) for 1 min
at room temperature, following which the excess ECL solution was
drained, and the membrane was wrapped in a plastic wrap and exposed
to X-ray film (Kodak, Shanghai, China). The densities of the bands
were quantified and the relative values were normalized to that of
β-actin using the following formula: Associated value =
experimental densitometry value / β-actin value).
Nested touchdown methylation specific-PCR
(ntMSP) analysis of FABP4 DNA methylation
Genomic DNA from the macrophages was isolated using
a Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA)
and modified using an EZ DNA Methylation-Gold™ kit (Zymo Research
Corp., Orange, CA, USA), following which it was used as a template
for ntPCR. Following bisulfite modification of the genomic DNA,
unmethylated cytosine residues were converted into uracil. The
FABP4 DNA methylation primers for MSP amplification were
designed using the MethPrimer bioinformatics program (http://www.urogene.org/methprimer/index.html). The
inner and outer primers were designed and synthesized based on the
DNA sequences of FABP4 (Table
I).
 | Table IInner and outer primers used for
FABP4 DNA methylation analysis. |
Table I
Inner and outer primers used for
FABP4 DNA methylation analysis.
| FABP4 | Primer sequence
(5′→3′) | Temperature
(°C) | Product size
(bp) |
|---|
| Outer | Forward:
ATTTTATTAGGGAGAGAAGGAAAAA | 71.1 | 414 |
| Reverse:
TCACATCTCAAAATCTCAAAACTAAC | | |
| Methylated | Forward:
AAGTTGGAAGTTTTTTTTGTTAACG | 69.3 | 137 |
| Reverse:
CCTTTACCTATATTTACTTCTTTCGAA | | |
| Unmethylated | Forward:
AGTTGGAAGTTTTTTTTGTTAATGG | 69.2 | 134 |
| Reverse:
TTTACCTATATTTACTTCTTTCAAA | | |
To reduce mispriming and to increase efficiency,
ntMSP was used for amplification, which comprised a two-step PCR
amplification procedure following the standard sodium bisulfite DNA
modification. In accordance with the specification requirements of
DNA remodified sodium, 4 µl of modified DNA was obtained, 1
µl upstream and 1 µl downstream of each primer, using
Go Taq Colorless Master mix (×2; 12.5 µl; Promega), with
nuclease-free water added to 25 µl. The samples were
subjected to the following steps in a thermal cycler: 94°C for 5
min, 94°C for 30 sec, 74°C for 30 sec and 72°C for 1 min, for 30
cycles, decreasing 0.5°C every cycle, 94°C for 30 sec, for 59°C 30
sec and 72°C for 1 min, 20 cycles, and 72°C for 7 min. The PCR
products were separated by electrophoresis on 2% agarose gel. The
bands were visualized under ultraviolet illumination and calculated
using the following formula: Methylation (%) = methylation /
(methylation + unmethylation) × 100%. Each sample was examined
using ntMSP analysis three times.
Assessment of intracellular total
cholesterol (TC)
The macrophages were scraped off the culture plate
using a rubber scraper and washed in PBS three times, following
which they were re-suspended in 0.5 ml PBS (pH 7.4) and lysed by
ultrasound for 1 min. The TC was determined, according to the
manufacturer's protocol of the Total cholesterol kit-CHO (Beijing
BHKT Clinical Reagent Co., Ltd, Beijing, China). TC concentrations
were expressed as mM/l.
Statistical analysis
Experiments were performed at least three times and
the results are expressed as the mean ± standard error of the mean.
The data were analyzed using one-way analysis of variance, and
additional analysis was performed using the Student-Newman-Keuls
test for multiple comparisons within different groups. GraphPad
Prism 5.01 software was used for analysis (version 5.01, GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
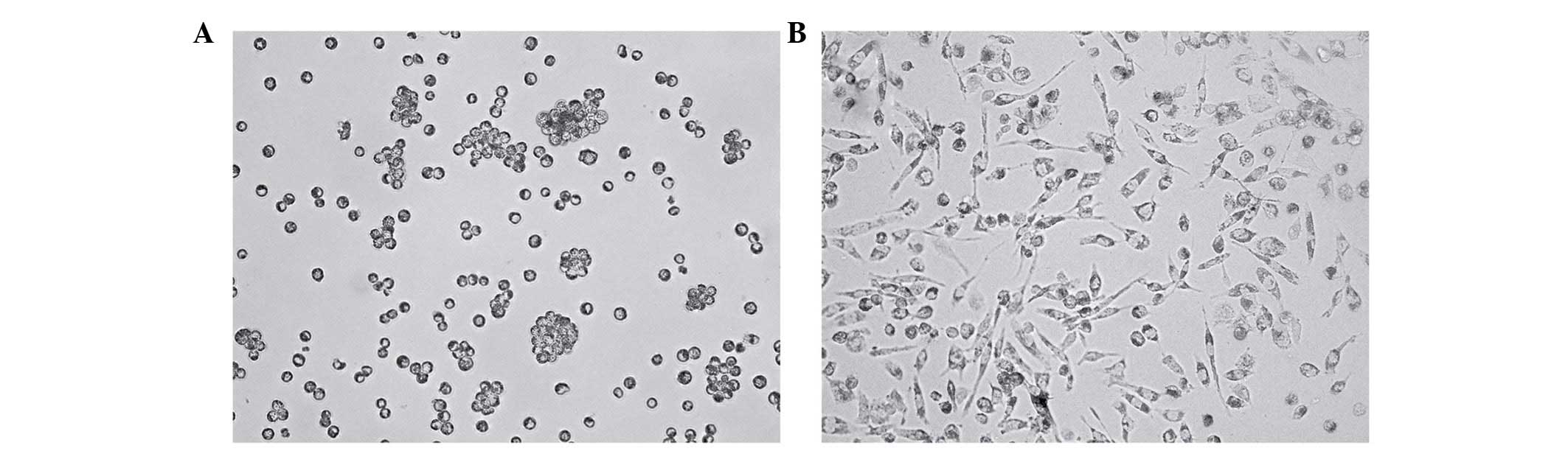
Formation of macrophages derived from
activated THP-1 monocytes
The THP-1 monocytes were observed to grow in
suspension and not adhere to the plastic surfaces of the culture
flasks (Fig. 1A). For the
induction of terminal differentiation into macrophages, the THP-1
cells were cultured in the presence of 500 nM PMA for 2 days, and
then co-cultured with 50, 100, 200 and 500 mM Hcy or 100 mM
Hcy+VB12+FA for 24 h. Following 72 h of culture, the
cells had adhered and spread across the bottom of the dish, and had
the morphological characteristics of macrophages (Fig. 1B).
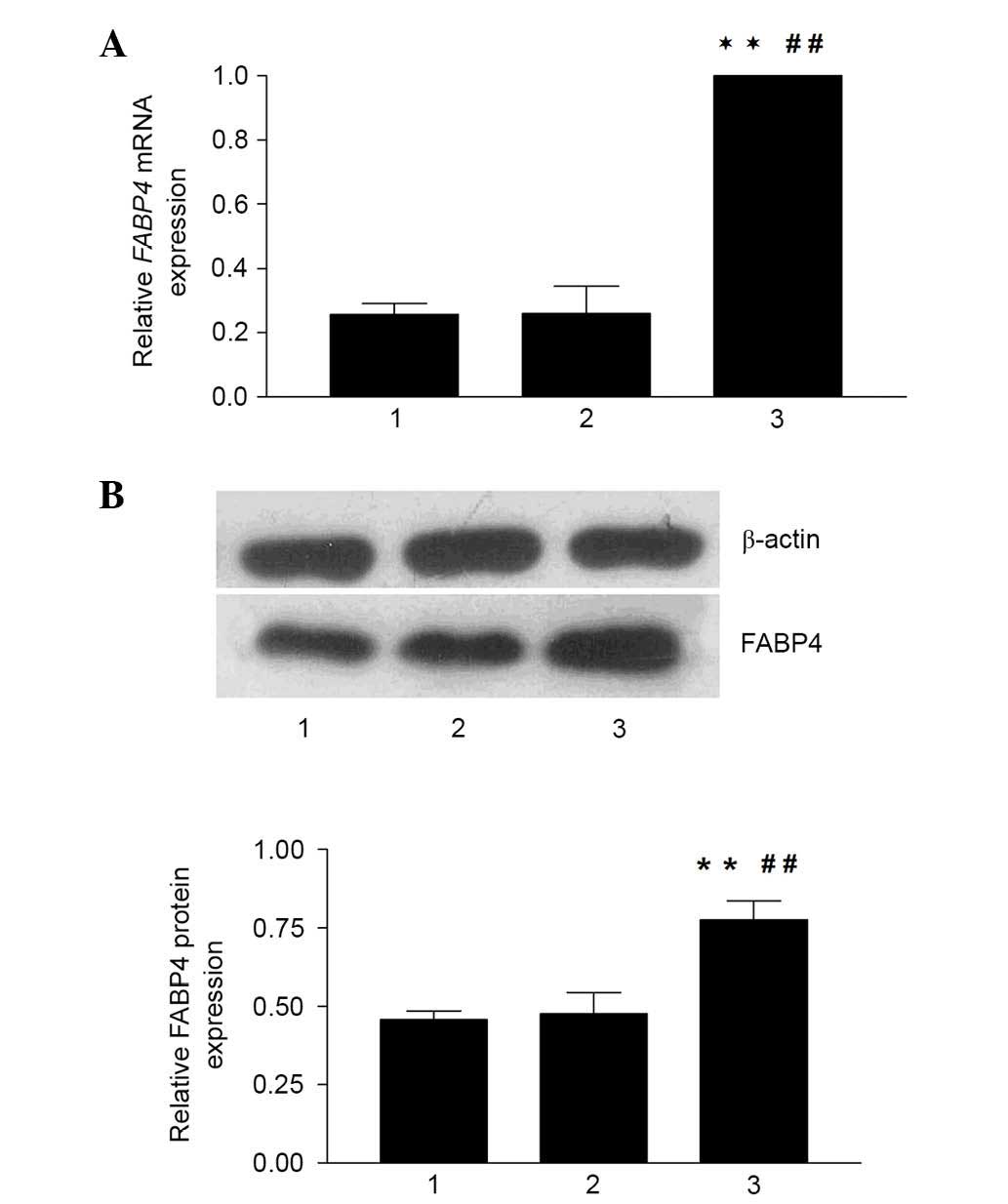
Hcy-induced upregulation of the mRNA and
protein expression levels of FABP4
To investigate whether the regulation of FABP4
occurred in the context of Hcy, the present study examined the mRNA
and protein expression levels of FABP4 using RT-qPCR and western
blot analyses in macrophages exposed to Hcy for 24 h. The data
demonstrated that the expression of FABP4 was substantially
upregulated following incubation with Hcy for 24 h at various
concentrations (between 50 and 500 µM), and the mean mRNA
expression levels of FABP4 in the experimental groups (50,
100, 200 and 500 mM Hcy, and 100+VB12+FA) were 10.84,
11.46, 5.89, 3.45 and 1.41 times higher, compared with that in the
control group, respectively (Fig.
2A). The highest level of expression was observed at the
concentration of 100 mM Hcy (P<0.01). In addition, the relative
mRNA expression level of FABP4 in the 100+VB12+FA
group decreased significantly (87.69%) compared with the 100
µM Hcy group (P<0.01). The effects of Hcy on the protein
levels of FABP4 (Fig. 2B) were
measured using western blot analysis, which showed similar results
to the mRNA levels. The protein expression levels of FABP4 in the
100 and 200 µM Hcy groups were 3.27 and 2.64 times higher,
compared with that in the control group (P<0.05), respectively.
FA and VB12 also affected the protein expression of
FABP4 significantly, and the relative protein expression of FABP4
in the 100+VB12+FA group was significantly decreased by
52.52%, compared with that in the 100 µM Hcy group
(P<0.05). However, as the concentrations of Hcy increased, no
dose-dependent increase in the mRNA or protein levels of FABP4 were
observed. These results indicated that Hcy affected the expression
of FABP4 at the transcriptional and translational levels.
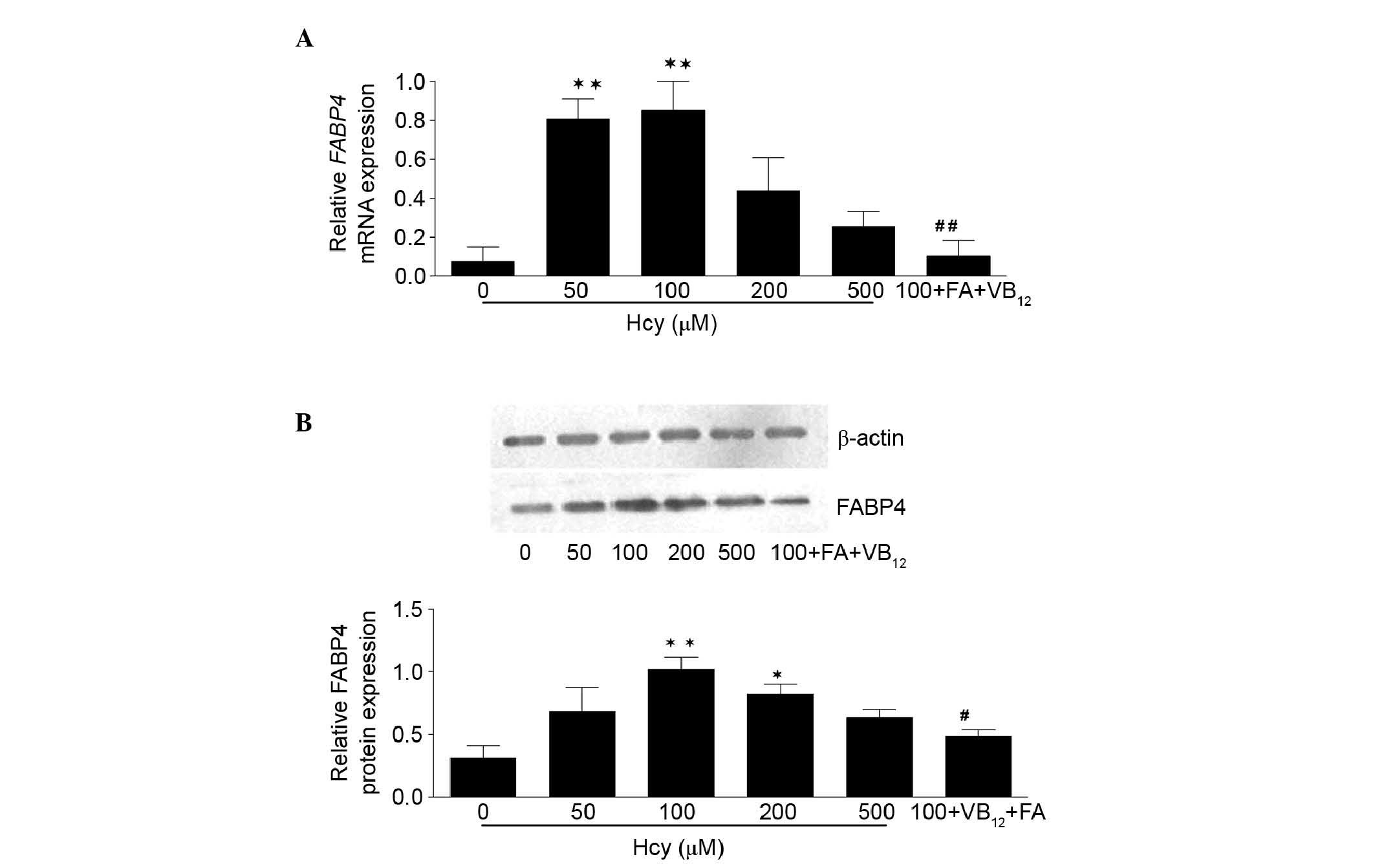 | Figure 2mRNA and protein expression levels of
FABP4 in THP-1 monocyte-derived macrophages. (A) mRNA expression
levels of FABP4, acquired from the Cq values of reverse
transcription-quantitative polymerase chain reaction analysis,
relative to GAPDH and expressed as N-fold differences in the
expression of FABP4 relative to the calibrator, termed
'Ngene'. Data are expressed as the mean ± standard error of the
mean. (B) Effect of Hcy on the protein levels of FABP4 were
measured by western blotting. The immunoblots were analyzed by
densitometry. Protein samples were prepared from THP-1
monocyte-derived macrophages. Protein signals were visualized by
electrochemiluminescence using Quantity One software. Relative
values were normalized to the corresponding β-actin band
intensities, The protein levels of FABP4 in the macrophages
incubated with Hcy were significantly increased, similar to the
mRNA levels of FABP4. These results suggested that the
macrophages cultured with Hcy for 24 h produced increased FABP4.
*P<0.05 and **P<0.01, vs. control;
#P<0.05 and ##P<0.01, vs. 100 µM
Hcy. FABP4, fatty acid binding protein 4; Hcy, homocysteine;
100+FA+VB12, 30 µM folate+30 µM vitamin
B12, dissolved in 100 µM Hcy. |
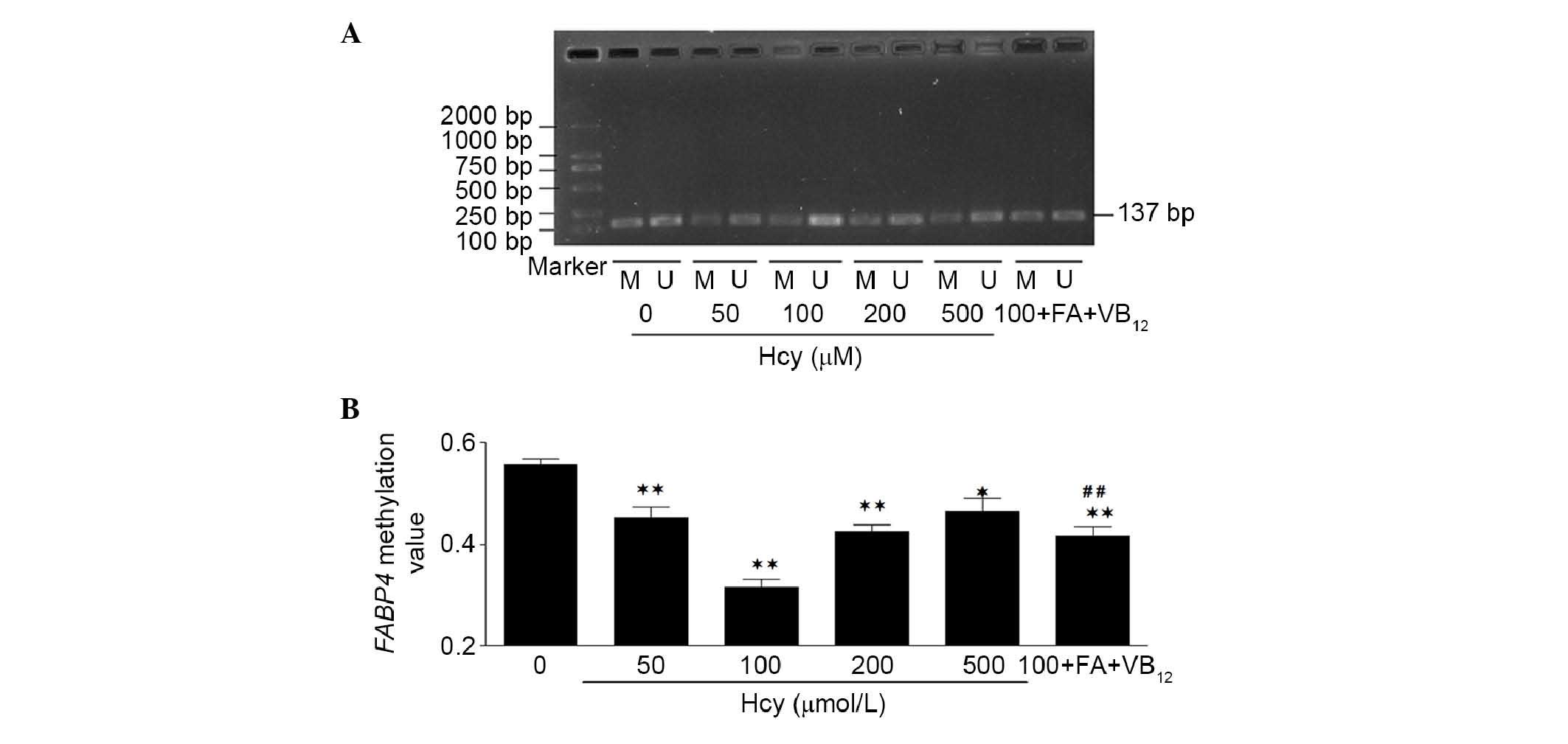
Hcy-induced FABP4 DNA hypomethylation in
THP-1 macrophages
DNA methylation patterns have been considered a
useful molecular marker for AS, and Hcy can have a global effect on
DNA methylation and activate various target genes (16). To determine the effects of Hcy on
FABP4 DNA methylation and investigate DNA methylation in the
promoter region of FABP4, the THP-1 monocyte-derived
macrophages in the present study were treated with Hcy at various
concentrations for 24 h, and the DNA methylation status of the
FABP4 promoter region of the THP-1 macrophages was
determined via ntMSP. As shown in Fig.
3, an increase in the concentration of Hcy between 50 and 100
mM, and a further increase to 500 mM, led to statistically
significant differences in the levels of FABP4 promoter
region DNA methylation, compared with the control group. The
methylation changes in the 100 mM group were the most marked. It
was also found that, in the experimental groups (50, 100, 200 and
500 mM Hcy, and 100+VB12+FA), the levels of FABP4
DNA methylation decreased significantly, by 18.84, 43.38, 23.65,
16.50 and 25.28%, respectively. The level of FABP4 DNA
methylation in the 100+VB12+FA group increased
significantly (31.97%), compared with the 100 mM Hcy group
(P<0.01; Fig. 3). These results
indicated that Hcy decreased the level of FABP4 promoter
region DNA methylation.
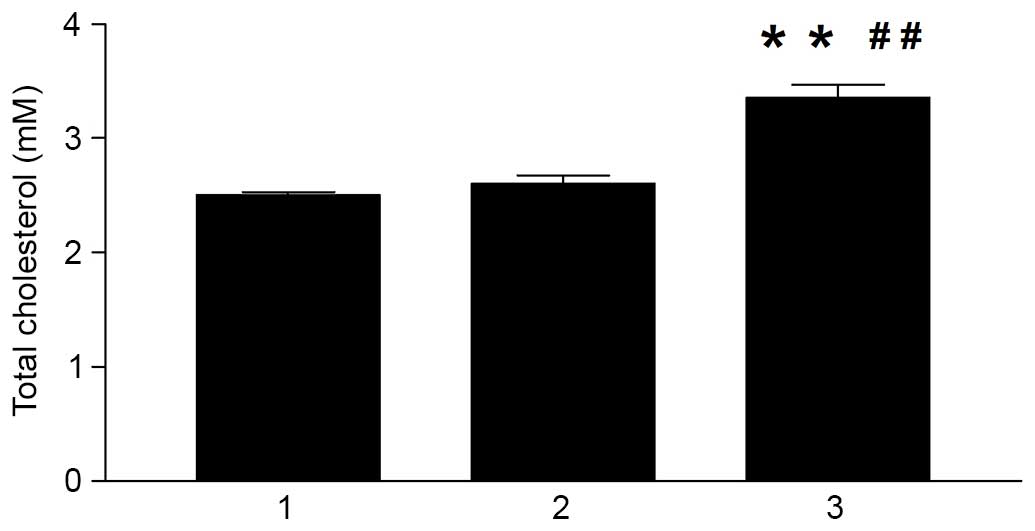
Hcy-induced cellular TC accumulation in
THP-1 macrophages
The present study also assessed whether Hcy
accelerated cholesterol accumulation in the macrophages. The human
THP-1 macrophages were treated with increasing concentrations of
Hcy, and intracellular TC accumulation was assessed using a Total
Cholesterol kit. Hcy significantly increased intracellular TC
accumulation, compared with the control group, and the highest
level was observed at a concentration of 100 µM Hcy
(P<0.01). However, the Hcy-induced increase in intracellular TC
accumulation did not occur in a dose-dependent manner (Fig. 4).
Identification of the inserted
constructed recombinant pcDNA3.1-EGFP/FABP4 plasmid
The recombinant vector encoding FABP4 was
constructed, and it was confirmed that the human FABP4
fragment had been successfully inserted into the pcDNA3.1-EGFP
fluorescent eukaryotic expression vector. The FABP4 PCR products
were 412 bp on the 1.5% agarose gel (Fig. 5A). The 412 bp fragment encoding
human FABP4 was inserted into the pcDNA3.1-EGFP plasmid
between the EcoRI and HindIII sites. The successfully
constructed recombinant plasmid, pcDNA3.1-EGFP/FABP4, was cut into
two fragments using EcoRI and HindIII (Fig. 5B). In addition, the results of the
DNA sequencing were identical to the FABP4 sequence reported
on GenBank confirming the results.
Transient FABP4 recombinant transfection
induces overexpression of the mRNA and protein levels of FABP4 in
THP-1 macrophages
The recombinant expression plasmid containing the
FABP4 gene was successfully transfected into the THP-1
macrophages, and the effective mRNA and protein expression of FABP4
were also confirmed using RT-qPCR and western blot analyses,
respectively. The data demonstrated that the expression of FABP4
was significantly upregulated in the recombinant plasmid group,
with the mean mRNA expression of FABP4 in the recombinant
plasmid group being 3.90 and 3.85 times higher than those in the
normal control group and empty plasmid control group, respectively
(P<0.01; Fig. 6A). The protein
levels of FABP4 were measured using western blot analysis, which
showed similar results to those observed for mRNA expression
(Fig. 6B). The protein expression
of FABP4 in the recombinant plasmid group was higher, compared with
the normal control group and empty plasmid control group, (69.79
and 63.36%, respectively; P<0.01). Therefore, the
pcDNA3.1-EGFP/FABP4 recombinant fluorescent eukaryotic expression
vector was successfully constructed and effectively expressed in
the THP-1 macrophages, which provided an experimental basis for
further experiments.
Effect of the FABP4-specific recombinant
on intracellular TC contents
The present study subsequently investigated whether
FABP4 accelerated cholesterol accumulation in the macrophages. The
human THP-1 macrophages were transfected with the pcDNA3.1-EGFP
plasmid and pcDNA3.1-EGFP/FABP4 recombinant plasmid, respectively,
and TC levels were examined. As shown in Fig. 7, intracellular TC accumulation in
the recombinant plasmid group was significantly higher, compared
with that in the normal control group and empty plasmid control
group (P<0.01). These results indicated that the overexpression
of FABP4 significantly increased intracellular TC accumulation.
These observations were consistent with a previous report which
suggested a critical role for FABP4 in macrophage lipid
accumulation (17).
Discussion
Previous reports have demonstrated that elevated
levels of Hcy have been associated with a number of disease states,
including AS (18,19). Despite the important contribution
of Hcy to accelerated AS, the specific molecular mechanisms in
response to Hcy within macrophages, which is important in
atherogenesis, remain to be fully elucidated.
FABP4 is a 15 kD cytoplasmic protein binding
hydrophobic ligands. Deficiency of FABP4 enhances CD36-mediated
lipoprotein entry and, at the same time, activates ABCA1-dependent
lipid efflux to a greater extent, thereby lowering intracellular
lipid contents (20). In the
present study, it was found that the exposure of THP-1 macrophages
to Hcy was associated with a significant elevation in the
expression of FABP4, reaching its maximum level when 100 µM
Hcy was used. It was also found that the enhanced expression of
FABP4 was paralleled with increases in intracellular TC levels. The
present study demonstrated that the exposure of macrophages to Hcy
appeared to increase intracellular FAPB4 levels and lipid contents,
similar to observations in the transformation of macrophages into
foam cells in the presence of ox-LDL stimulation (21). In addition, Hcy can be recycled
into methionine or converted into cysteine with the assistance of
FA and VB12 (22), and
mild doses of Hcy inhibitors, including VB12 and FA,
markedly attenuated FABP4-induced macrophage lipid accumulation.
These findings indicated that the Hcy inhibitors suppressed
exclusively pathological excessive lipid accumulation in the
macrophages.
To further understand the role of FABP4, the present
study constructed the pcDNA3.1-EGFP/FABP4 recombinant vector and
successfully transduced the FABP4 gene into macrophages
derived from THP-1 monocytes. The increased expression of FABP4
from the transfected macrophages was confirmed using RT-qPCR and
western blot analyses. In addition, cellular TC accumulation was
examined, and it was found that the intracellular TC content in the
FABP4 recombinant plasmid group was also significantly increased.
Thus, the increase in cellular TC accumulation was accelerated by
the enhanced expression of FABP4, either directly or indirectly.
These results suggested that FABP4 may be a potential therapeutic
target for the treatment of AS.
Previous population studies have revealed that
genetic variations at the FABP4 locus in humans lead to
lowered serum triglyceride levels, and a markedly reduced risk of
coronary heart disease (23,24).
However, the exact molecular mechanisms remain to be elucidated. In
our previous study, Hcy was found to be involved in the disturbance
of the expression of the LDL-receptor promoter region and global
genome DNA hypomethylation through the interference of DNA
methylation (25–27). Increasing evidence also suggests
that aberrant DNA methylation around promoter regions can lead to
the development of AS (28).
Therefore, the present study investigated the epigenetic mechanisms
by which the elevated levels of FAPB4 and lipids were affected by
Hcy. The resulting data showed tha thet FABP4 promoter was
hypomethylated following exposure to Hcy. These results indicated a
causal association between the expression of FABP4 and the changes
in DNA methylation, and the epigenetic mechanism of Hcy-induced AS
was ascertained.
In conclusion, the results of the present study
demonstrated that Hcy not only promoted the secretion of FABP4, but
also induced FABP4 gene DNA hypomethylation in cultured
human THP-1 monocyte-derived macrophages. The invasion of monocytes
into the arterial wall and their subsequent differentiation into
cholesterol-laden macrophages is a central feature of AS (29). Therefore, the present study
examined the effects of Hcy on FABP4, and demonstrated a causal
molecular link between Hcy and FABP4 in macrophage lipid
accumulation. The most notable finding of the present study was
that, Hcy was a stimulator of FABP4 DNA methylation in
cultured macrophages, a critical cell in atherogenesis, which
induced FABP4 DNA hypomethylation. The results of the
present study also confirmed the mechanism that the overexpression
of FABP4 accelerates TC accumulation, and thereby promotes the
atherosclerotic process, Further investigations, including the
analysis of lipid trafficking in macrophages from FABP4-deficient
mice and inhibition of lipoprotein transporters by specific small
interfering RNA or antibodies, are required to further elucidate
the mechanisms.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 80160105, 81360052,
81460121, 81360027, 81360073 and 81260063) and the Advantages
Besides Construction Projects of Ningxia Medical University 2014
(grant nos. XY201415 and Ningxia Medical University [2014]
062).
References
|
1
|
Furuhashi M and Hotamisligil GS: Fatty
acid-binding proteins: Role in metabolic diseases and potential as
drug targets. Nat Rev Drug Disc. 7:489–503. 2008. View Article : Google Scholar
|
|
2
|
Cabré A, Babio N, Lázaro I, Bulló M,
Garcia-Arellano A, Masana L and Salas-Salvadó J: FABP4 predicts
atherogenic dyslipidemia development. The PREDIMED study.
Atherosclerosis. 222:229–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furuhashi M, Ishimura S, Ota H and Miura
T: Lipid chaperones and metabolic inflammation. Int J Inflam.
2011:6426122011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fu Y, Luo N and Lopes-Virella MF: Oxidized
LDL induces the expression of ALBP/aP2 mRNA and protein in human
THP-1 macrophages. J Lipid Res. 41:2017–2023. 2000.PubMed/NCBI
|
|
5
|
Pelton PD, Zhou L, Demarest KT and Burris
TP: PPARgamma activation induces the expression of the adipocyte
fatty acid binding protein gene in human monocytes. Biochem Biophys
Res Commun. 261:456–458. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu Y, Luo N, Lopes-Virella MF and Garvey
WT: The adipocyte lipid binding protein (ALBP/aP2) gene facilitates
foam cell formation in human THP-1 macrophages. Atherosclerosis.
165:259–269. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tehlivets O: Homocysteine as a risk factor
for atherosclerosis: Is its conversion to s-adenosyl-L-homocysteine
the key to deregulated lipid metabolism? J Lipids. 2011:7028532011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li L, Xie J, Zhang M and Wang S:
Homocysteine harasses the imprinting expression of IGF2 and H19 by
demethylation of differentially methylated region between IGF2/H19
genes. Acta Biochim Biophys Sin (Shanghai). 41:464–471. 2009.
View Article : Google Scholar
|
|
9
|
Yi-Deng J, Tao S, Hui-Ping Z, Jian-Tuan X,
Jun C, Gui-Zhong L and Shu-Ren W: Folate and ApoE DNA methylation
induced by homo-cysteine in human monocytes. DNA Cell Biol.
26:737–744. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma S, Zhang H, Sun W, Gong H, Wang Y, Ma
C, Wang J, Cao C, Yang X, Tian J and Jiang Y: Hyperhomocysteinemia
induces cardiac injury by up-regulation of p53-dependent Noxa and
Bax expression through the p53 DNA methylation in ApoE(−/−) mice.
Acta Biochim Biophys Sin (Shanghai). 45:391–400. 2013. View Article : Google Scholar
|
|
11
|
Deng G, Nguyen A, Tanaka H, Matsuzaki K,
Bell I, Mehta KR, Terdiman JP, Waldman FM, Kakar S, Gum J, et al:
Regional hypermethylation and global hypomethylation are associated
with altered chromatin conformation and histone acetylation in
colorectal cancer. Int J Cancer. 118:2999–3005. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Subramanyam MA, Diez-Roux AV, Pilsner JR,
Villamor E, Donohue KM, Liu Y and Jenny NS: Social factors and
leukocyte DNA methylation of repetitive sequences: The multi-ethnic
study of atherosclerosis. PLoS One. 8:e540182013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma P, Senthil kumar RD, Brahmachari V,
Sundaramoorthy E, Mahajan A, Sharma A and Sengupta S: Mining
literature for a comprehensive pathway analysis: A case study for
retrieval of homocysteine related genes for genetic and epigenetic
studies. Lipids Health Dis. 5:12006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yideng J, Zhihong L, Jiantuan X, Jun C,
Guizhong L and Shuren W: Homocysteine-mediated PPARalpha, gamma DNA
methylation and its potential pathogenic mechanism in monocytes.
DNA Cell Biol. 27:143–150. 2008. View Article : Google Scholar
|
|
15
|
Jiang Y, Zhang H, Sun T, Wang J, Sun W,
Gong H, Yang B, Shi Y and Wei J: The comprehensive effects of
hyperlipidemia and hyperhomocysteinemia on pathogenesis of
atherosclerosis and DNA hypomethylation in ApoE−/− mice. Acta
Biochim Biophys Sin. 44:866–875. 2012. View Article : Google Scholar
|
|
16
|
Liang Y, Yang X, Ma L, Cai X, Wang L, Yang
C, Li G, Zhang M, Sun W and Jiang Y: Homocysteine-mediated
cholesterol efflux via ABCA1 and ACAT1 DNA methylation in THP-1
monocyte-derived foam cells. Acta Biochim Biophys Sin (Shanghai).
45:220–228. 2013. View Article : Google Scholar
|
|
17
|
Wang XQ, Yang K, He YS, Lu L and Shen WF:
Receptor mediated elevation in FABP4 levels by advanced glycation
end products induces cholesterol and triacylglycerol accumulation
in THP-1 macrophages. Lipids. 46:479–486. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Veeranna V, Zalawadiya SK, Niraj A,
Pradhan J, Ference B, Burack RC, Jacob S and Afonso L: Homocysteine
and reclassification of cardiovascular disease risk. J Am Coll
Cardiol. 58:1025–1033. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang AN, Zhang HP, Sun Y, Yang XL, Wang N,
Zhu G, Zhang H, Xu H, Ma SC, Zhang Y, Li GZ, Jia YX, Cao J and
Jiang YD: High-methionine diets accelerate atherosclerosis by
HHcy-mediated FABP4 gene demethylation pathway via DNMT1 in
ApoE(−/−) mice. FEBS Lett. 589:3998–4009. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Makowski L, Brittingham KC, Reynolds JM,
Suttles J and Hotamisligil GS: The fatty acid-binding protein, aP2,
coordinates macrophage cholesterol trafficking and inflammatory
activity. Macrophage expression of aP2 impacts peroxisome
proliferator-activated receptor gamma and IkappaB kinase
activities. J Biol Chem. 280:12888–12895. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu Y, Luo N and Lopes-Virella MF: Oxidized
LDL induces the expression of ALBP/aP2 mRNA and protein in human
THP-1 macrophages. J Lipid Res. 41:2017–2023. 2000.PubMed/NCBI
|
|
22
|
Miller AL: The methionine-homocysteine
cycle and its effects on cognitive diseases. Altern Med Rev.
8:7–19. 2003.PubMed/NCBI
|
|
23
|
Tuncman G, Erbay E, Hom X, De Vivo I,
Campos H, Rimm EB and Hotamisligil GS: A genetic variant at the
fatty acid-binding protein aP2 locus reduces the risk for
hypertriglyceridemia, type 2 diabetes, and cardiovascular disease.
Proc Natl Acad Sci USA. 103:6970–6975. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Furuhashi M, Fuseya T, Murata M, Hoshina
K, Ishimura S, Mita T, Watanabe Y, Omori A, Matsumoto M, Sugaya T,
et al: Local production of fatty acid-binding protein 4 in
epicardial/perivascular fat and macrophages is linked to coronary
atherosclerosis. Arterioscler Thromb Vasc Biol. 36:825–834. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Y, Peng K, Su J, Huang Y, Xu Y and
Wang S: Different effects of homocysteine and oxidized low density
lipoprotein on methylation status in the promoter region of the
estrogen receptor a gene. Acta Biochim Biophys Sin (Shanghai).
39:19–26. 2007. View Article : Google Scholar
|
|
26
|
Yideng J, Jianzhong Z, Ying H, Juan S,
Jinge Z, Shenglan W, Xiaoqun H and Shuren W: Homocysteine-mediated
expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential
pathogenic mechanism in VSMCs. DNA Cell Biol. 26:603–611. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang Y, Sun T, Xiong J, Cao J, Li G and
Wang S: Hyperhomocysteinemia-mediated DNA hypomethylation and its
potential epigenetic role in rats. Acta Biochim Biophys Sin
(Shanghai). 39:657–667. 2007. View Article : Google Scholar
|
|
28
|
Zaina S, Lindholm MW and Lund G: Nutrition
and aberrant DNA methylation patterns in atherosclerosis: More than
just hyperhomocysteinemia? J Nutr. 135:5–8. 2005.
|
|
29
|
Moore KJ and Freeman MW: Targeting innate
immunity for CV benefit. Drug Discov Today Ther Strateg. 5:15–23.
2008. View Article : Google Scholar : PubMed/NCBI
|