Introduction
Human gliomas, which originate from neural
mesenchymal stem cells, currently account for 40–50% of nervous
system tumors (1). In 2007, the
World Health Organization classified astrocytomas into the
following grades: Grade I-II, well-differentiated low-grade diffuse
astrocytoma; grade III, anaplastic astrocytoma; and grade IV,
glioblastoma multiforme (2).
Despite the use of aggressive surgery in combination with
radiotherapy, chemotherapy and biological therapy, gliomas remain
difficult to treat, and the median survival of patients with glioma
is 12–15 months (3). Therefore, it
is necessary to elucidate mechanisms underlying the development of
glioma and to discover novel targets for the treatment of
glioma.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that function as negative regulators of gene expression.
miRNAs regulate expression at the post-transcriptional level by
binding to complimentary sequences in the 3′-untranslated regions
(3′-UTRs) of mRNAs (4,5). Aberrant miRNA expression, and loss of
the dynamic balance between oncogene and tumor suppressor gene
expression, may lead to the formation and development of tumors
(6).
Hsa-miR-495 is located at chromosome 14q32.31 in the
human genome, the expression of which varies in different tumor
types. Downregulation of hsa-miR-495 has been reported in numerous
human malignancies, including non-small cell lung cancer, gastric
cancer and acute myeloid leukemia (7–9).
However, the function and molecular mechanisms of hsa-miR-495 in
glioma remain unclear.
The present study aimed to explore the biological
function and molecular mechanism of hsa-miR-495 in glioma. As
hypothesized, hsa-miR-495 was downregulated in glioma tissues and
cell lines. Notably, hsa-miR-495 was revealed to have vital roles
in glioma cell proliferation and invasion by directly targeting
v-myb avian myeloblastosis viral oncogene homolog (MYB), resulting
in the inhibition of glioma cell invasion. These results indicated
that upregulation of hsa-miR-495 may be considered a potential
future therapeutic strategy for the treatment of glioma.
Materials and methods
Patient samples and cell lines
A total of 32 samples of paired human glioma and
matched normal brain tissues were collected from adult patients
that were admitted and diagnosed at The Second Affiliated Hospital,
Harbin Medical University (Harbin, China). The 32 patients with
glioma were comprised of 18 male patients and 14 female patients,
including 12 low-grade glioma (5 grade I and 7 grade II) and 20
high-grade glioma (8 grade III and 12 grade IV). The age range of
the patients was 45–78 years old, with a median age of 62. Tissue
samples were immediately snap-frozen in liquid nitrogen and were
stored at −80°C. Four human glioma cell lines (A172, U87, U251 and
U373) and a normal neuronal primary human fetal glial (PHFG) cell
line were purchased from the American Type Culture Collection
(Manassas, VA, USA). The cells were maintained in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere containing 5% CO2, and
1% antibiotics (100 µ/ml penicillin and 100 mg/ml
streptomycin sulfates) were added. The present study was approved
by the Institutional Review Board of The Second Affiliated
Hospital, and written informed consent was obtained from all
patients.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the patient tissues and
cell lines, subsequent to mincing and homogenization of the
tissues, using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Total RNA (1 µg) was reverse transcribed using oligo (dT) 18
as a stem-loop RT primer (Qiagen GmbH, Hilden, Germany) for the RT
of miRNA, and using random primers (Invitrogen; Thermo Fisher
Scientific, Inc.) for the RT of mRNA. The RT protocol was as
follows: 60°C for 5 min, 4°C for 2 min, 50°C for 1 h, 85°C for 5
min, and finally maintained at 4°C. qPCR was performed using a
Roche Lightcycler 480 Real-Time PCR system (Roche Diagnostics,
Indianapolis, IN, USA). The total volume of the qPCR reaction was
20 µl, which contained cDNA template 1 µl, 2X
QuantiTect SYBR Green PCR Master Mix (10 µl), 10X reverse
miScript SYBR Green PCR kit Universal Primer (2 µl), forward
primer (0.5 µl) and RNase-free water (6.5 µl). The
qPCR cycling conditions were as follows: 95°C for 10 min, followed
by 45 cycles of 94°C for 20 sec, 60°C for 30 sec and 72°C for 30
sec. The specific primers for hsa-miR-495 and U6 were as follows:
Hsa-miR-495, forward 5′-AGAAGTTGCCCATGTTAT-3′, reverse miScript
SYBR Green PCR kit Universal Primer (Qiagen GmbH); and U6, forward
5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-ACGCTTCACGAATTTGCGT-3′. The
specific primers (Invitrogen; Thermo Fisher Scientific, Inc.) for
MYB and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were as
follows: MYB, forward 5′-TGACGAGGATGATGAGGACTTT-3′, reverse
5′-ATCTGTTCGATTCGGGAGATAA-3′; and GAPDH, forward
5′-ACATCAAGAAGGTGGTGAAGCAGG-3′, reverse
5′-CGTCAAAGGTGGAGGAGTGGGT-3′. All experiments were performed in
triplicate. Relative expression levels were calculated according to
the comparative Cq method (10).
Cell transfection
The hsa-miR-495 oligonucleotide and MYB-specific
small interfering (si)RNA molecules were chemically synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were
as follows: Hsa-miR-495 mimics, 5′-UGUGACGAAACAAACAUGGUGCACU-3′;
and hsa-miR-495 mimics negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′. Four MYB-specific siRNA sequences were
synthesized, as follows: 5′-CGUUGGUCUGUUAUUGCCAAGCACU-3′;
5′-CAGAUGACUGGAAAGUUAUUGCCAA-3′; 5′-GGAAGGUUAUCUGC-AGGAGUCUUCA-3′
and 5′-GCUUCCAGAAGAACAGUCAUUUGAU-3′. The following siRNA sequence
was used as a siRNA negative control: 5′-UUCUCCGAACGUGUCACGUU-3′.
Transfection was performed using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells were counted and seeded onto plates
1 day prior to transfection, in order to ensure suitable cell
confluence. Hsa-miR-495 oligonucleotide and MYB-specific siRNA were
used at final concentrations of 10 nmol/l, and were transfected in
antibiotic-free Opti-MEM medium (Invitrogen; Thermo Fisher
Scientific, Inc.).
Cell proliferation assay
A172 and U87 cells were incubated in 10% Cell
Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) diluted in normal culture medium at 37°C, until
visible color conversion was detected. Proliferation levels were
determined 0, 24, 48 and 72 h post-transfection. CCK-8 solution (10
µl) was added to each well, and absorbance was measured at
450 nm using a micro-plate reader (Model 550; Bio-Rad Laboratories,
Inc., Hercules, CA, USA) after incubation for 2 h at 37°C. All
experiments were performed in triplicate.
Cell invasion assay
Cell invasion was determined using a Transwell
assay. For the Transwell assay, the appropriate hsa-miR-495
oligonucleotide and MYB-specific siRNA molecules were transfected
into A172 and U87 cells, according to the manufacturer's protocol.
Subsequent to incubation for 48 h, 5×104 cells were
seeded into the upper Matrigel-coated invasion chambers (BD
Biosciences, San Jose, CA, USA) in serum-free DMEM. DMEM
supplemented with 10% FBS was added to the lower chambers. After 24
h, the invading cells were fixed using 95% ethanol and were stained
with 0.1% crystal violet. A DM4000B microscope was used (Leica,
Microsystems, Inc., Buffalo Grove, IL, USA). The assay was repeated
in three independent experiments.
Bioinformatics
MiRanda (http://www.microrna.org) and miRcode (http://www.mircode.org) were used to predict the
binding sites between hsa-miR-495 and MYB.
Dual luciferase assays
MYB mRNA 3′-UTRs containing the predicted
hsa-miR-495 binding sites were amplified from human cDNA using PCR
and were inserted into psiCHECK-2 luciferase reporter vectors
(Promega Corporation, Madison WI, USA), in order to obtain
constructed plasmids containing wild-type MYB mRNA 3′-UTR
(psiCHECK-2-HMGB1-WT-3′-UTR). MYB mRNA 3′-UTRs containing mutation
sequences in the putative binding site
(psiCHECK-2-HMGB1-MUT-3′-UTR) were chemically synthesized by
Shanghai GenePharma Co., Ltd. The MYB-WT or MYB-MUT recombinant
construct plasmids and hsa-miR-495 mimics or negative control
mimics were co-transfected into A172 and U87 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Firefly and Renilla luciferase activities were measured 24 h
post-transfection using the Dual Luciferase Assay (Promega
Corporation) according to the manufacturer's protocol. All
luciferase assays were carried out in triplicate. Luciferase
activity was detected using the Modulus™ Single Tube Multimode
Reader (Bio-Systems International, Beloit, WI, USA).
Western blot analysis
Cells were lysed in ice-cold cell lysis buffer [50
mM Tris, pH 8.0; 120 mM NaCl; 0.5% NP-40; 50 mM NaF; 1 mM
phenylmethylsulphonyl fluoride; 20 µM sodium orthovanadate;
1X protease inhibitors (Invitrogen; Thermo Fisher Scientific,
Inc.); 1X phosphatase inhibitors (Invitrogen; Thermo Fisher
Scientific, Inc.)], and protein concentration was measured using a
bicinchoninic acid protein assay kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA). Proteins were separated by 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and were
electrotransferred onto polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 5%
non-fat dried milk for 2 h at 37°C, and were then incubated with
mouse anti-MYB (1:500; ab17851; Abcam, Cambridge, UK) and mouse
anti-GAPDH (1:5,000; ab8245; Abcam) antibodies overnight at 4°C.
Subsequently, the membranes were incubated with a horseradish
peroxidase-labeled rabbit anti-mouse immunoglobulin G secondary
antibody (1:10,000; ab6728; Abcam) for 1 h at room temperature.
Positive bands were detected using an enhanced chemiluminescence
kit (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were repeated at least three times.
Statistical analysis was performed using SPSS 17.0 (SPSS Inc.,
Chicago, IL, USA). Differences were assessed using Student's
two-tailed t-test, while the CCK-8 data were examined by analysis
of variance. Data are presented as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Hsa-miR-495 is downregulated in glioma
tissues and cell lines
In order to assess the expression levels of
hsa-miR-495 in glioma tissues, qPCR was performed on 32 pairs of
glioma tissues and matched normal brain tissues. As presented in
Fig. 1A, the expression levels of
hsa-miR-495 were downregulated in glioma tissues compared with in
the matched normal brain tissues. Furthermore, hsa-miR-495
expression levels were downregulated in the glioma cell lines
(A172, U251, U373, and U87) compared with in the normal neuronal
cell (PHFG) (Fig. 1B). These
results indicate that hsa-miR-495 may have a role as a tumor
suppressor gene in glioma. Each experiment was performed in
triplicate.
Hsa-miR-495 inhibits glioma cell
proliferation and invasion in vitro
Subsequently, the functional roles of hsa-miR-495 in
A172 and U87 cells were explored. qPCR was performed to confirm
that hsa-miR-495 expression was restored in A172 and U87 cells
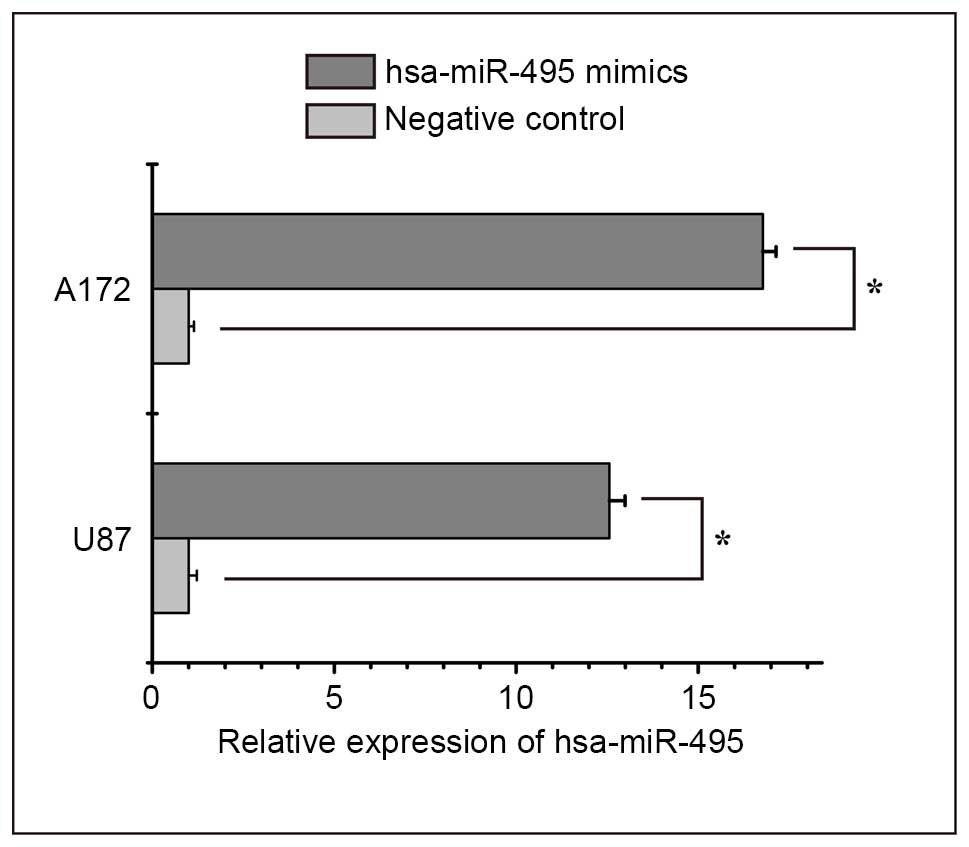
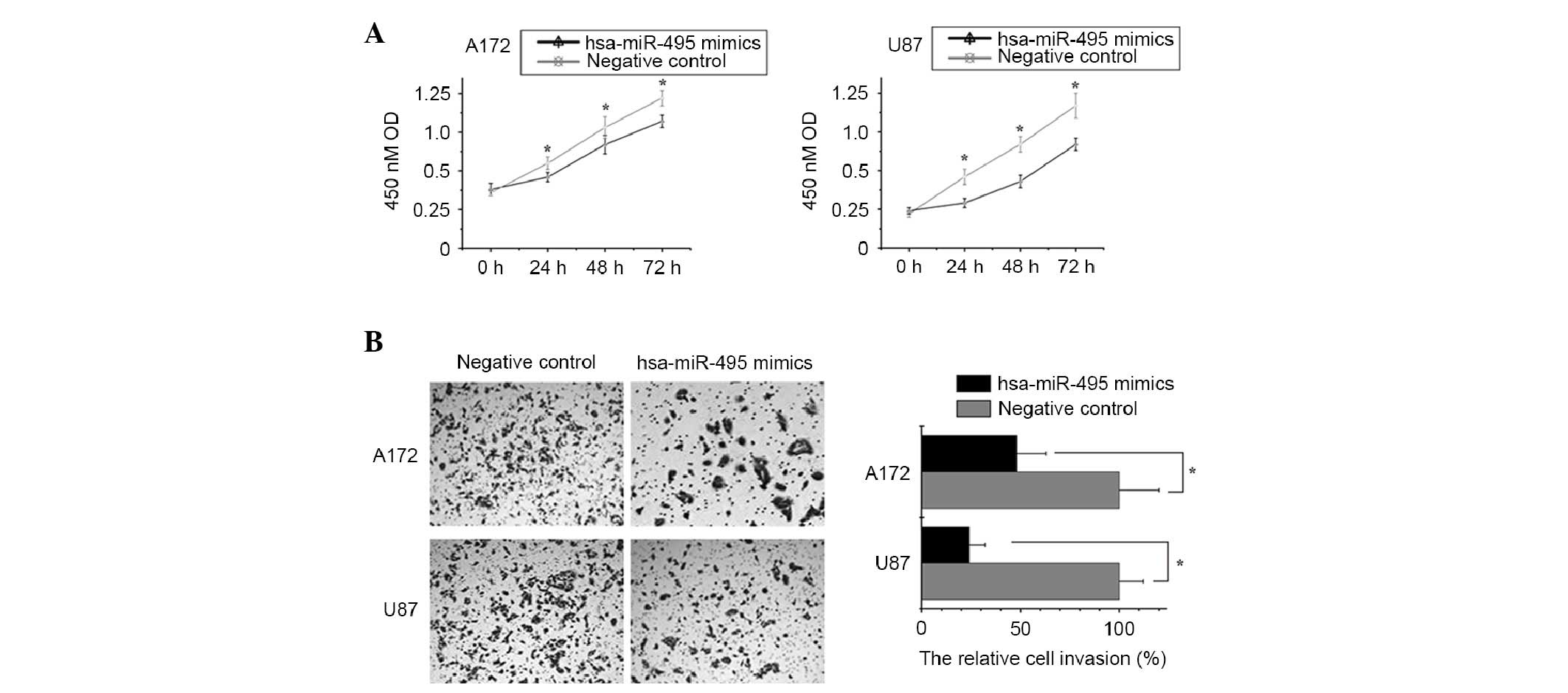
post-transfection with hsa-miR-495 mimics (Fig. 2). CCK-8 and Transwell assays
indicated that increased expression levels of hsa-miR-495 markedly
inhibited cell proliferation and invasion in A172 and U87 cells
compared with in the control group (Fig. 3). These results suggest that
hsa-miR-495 may function as a tumor suppressor by inhibiting cell
proliferation and invasion in glioma.
Hsa-miR-495 exerts its effects via
negative regulation of MYB in glioma
To explore the downstream targets of hsa-miR-495,
bioinformatic analysis was performed to identify potential targets.
MYB was one of the putative genes identified by bioinformatics
prediction, its mRNA 3′-UTR contains a complementary site for the
seed region of hsa-miR-495 (Fig.
4A). Furthermore, MYB has been identified as an oncogene, which
is associated with progression and development of various types of
cancer (11–14).
In order to determine whether MYB was directly
regulated by hsa-miR-495, the 3′-UTR fragment of MYB, containing a
putative or mutant binding site, was cloned into psiCHECK-2 to
construct recombinant plasmids (psiCHECK-2-HMGB1-WT-3′-UTR and
psiCHECK-2-HMGB1-MUT-3′-UTR). Subsequently, a luciferase reporter
assay was performed on A172 and U87 cells. As presented in Fig. 4B, relative luciferase activity in
the psiCHECK-2-HMGB1-WT-3′-UTR group was significantly decreased
post-transfection of A172 and U87 cells with hsa-miR-495 mimics
(P<0.05), whereas no notable reduction was detected in the
psiCHECK-2-HMGB1-MUT-3′-UTR group. qPCR and western blot analysis
were performed to examine the inhibitory effects of hsa-miR-495 on
endogenous MYB expression in glioma cells. As shown in Fig. 4C, increased hsa-miR-495 expression
markedly inhibited the expression of MYB in A172 and U87 cells.
These data suggest that hsa-miR-495 can inhibit MYB expression by
directly targeting its mRNA 3′-UTR in glioma.
Knockdown of MYB inhibits cell
proliferation and invasion in glioma
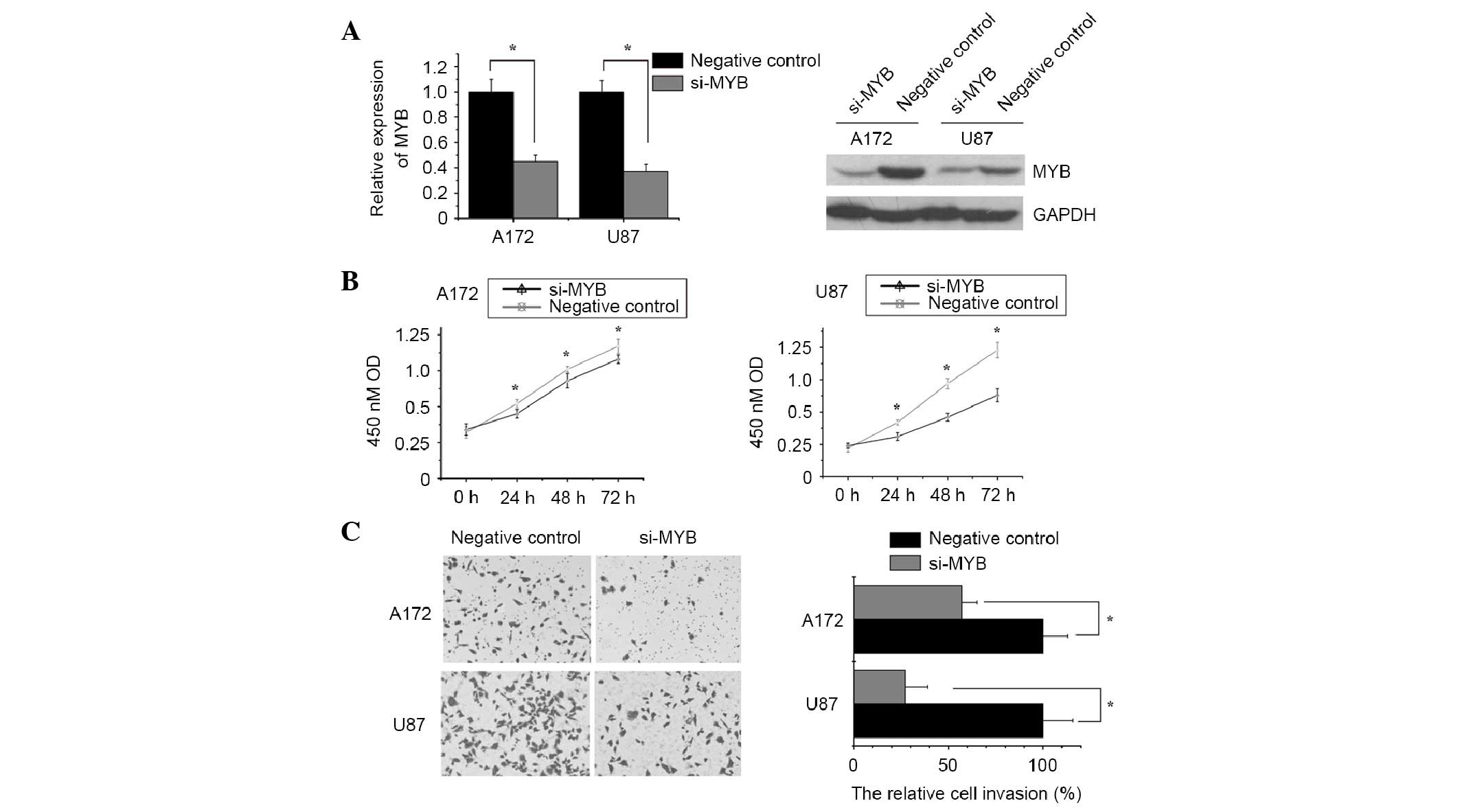
To confirm the functional roles of MYB in glioma
cells, MYB expression was knocked down in A172 and U87 cells by
MYB-specific siRNA (5′-CAGAUGACUGGAAAGUUAUUGCCAA-3′ was selected as
the optimum MYB-specific siRNA, thus was used for further
experiments). The knockdown was confirmed by qPCR and western
blotting (Fig. 5A). CCK-8 and
Transwell assays indicated that knockdown of MYB expression in A172
and U87 cells was able to significantly inhibit cell proliferation
and invasion compared with the control group (Fig. 5B and C), which was similar to the
effects of hsa-miR-495 overexpression. These results strongly
indicate that hsa-miR-495 acts as a tumor suppressor gene in glioma
via the negative regulation of MYB.
Discussion
Glioma is one of the most devastating types of
cancer, since it often exhibits aggressive behavior and cannot be
cured by current therapeutic strategies (15). Glioma often develops as a result of
genetic alterations that accumulate throughout tumor progression
(16). Therefore, elucidation of
the molecular mechanisms underlying glioma progression, in
particular those associated with cellular proliferation and
invasion, is essential to improve understanding regarding the
prevention and treatment of glioma (17).
It has previously been reported that miRNAs have an
important role in the proliferation and invasion of glioma. For
example, hsa-miR-25 inhibits glioma cell proliferation by targeting
cyclin-dependent kinase inhibitor 1C (18); hsa-miR-383 regulates proliferation,
migration, invasion and apoptosis in human glioma (19); and hsa-miR-320 inhibits cell
proliferation by targeting E2F transcription factor 1 in glioma
(20). Previous studies have
demonstrated that hsa-miR-495 expression is often downregulated in
several types of human cancer, including non-small cell lung
cancer, gastric cancer, and acute myeloid leukemia, thus indicating
that hsa-miR-495 may function as a tumor suppressor gene (7–9).
The present study demonstrated that hsa-miR-495
expression was downregulated in glioma tissues and cell lines.
Consistent with previous reports, these data suggested that
hsa-miR-495 may function as a tumor suppressor gene in glioma by
inhibiting cell growth and invasion in A172 and U87 cells.
Furthermore, bioinformatics and a luciferase reporter assay
revealed that hsa-miR-495 could directly target MYB and negatively
regulate its expression.
MYB is a well-acknowledged oncogene in several types
of human cancer. Knockdown of MYB contributes to inhibit malignant
transformation by regulating genes that participate in numerous
aspects of tumorigenesis, including cell growth arrest, invasion
inhibition and apoptosis induction (21–24).
In the present study, knockdown of MYB expression significantly
decreased the proliferation and invasion of A172 and U87 cells,
which was similar to the effects of hsa-miR-495 overexpression.
These data revealed that hsa-miR-495 suppressed glioma development
by targeting MYB, thus providing a potential mechanism underlying
post-transcriptional control of glioma.
In conclusion, hsa-miR-495 was downregulated in
glioma tissues and cell lines, and acts as a tumor suppressor gene
in glioma via the negative regulation of MYB. Therefore,
hsa-miR-495 may be considered a potential therapeutic biomarker for
the future treatment of glioma.
Acknowledgments
The present study was supported by the Heilongjiang
Province Scientific Research Foundation for Returness (grant no.
LC2013C18).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Osborn AG, Salzman KL, Thurnher MM, Rees
JH and Castillo M: The new World Health Organization Classification
of Central Nervous System Tumors: What can the neuroradiologist
really say? AJNR Am J Neuroradiol. 33:795–802. 2012. View Article : Google Scholar
|
|
3
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim VN: Small RNAs: Classification,
biogenesis, and function. Mol Cells. 19:1–15. 2005.PubMed/NCBI
|
|
6
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chu H, Chen X, Wang H, Du Y, Wang Y, Zang
W, Li P, Li J, Chang J, Zhao G and Zhang G: MiR-495 regulates
proliferation and migration in NSCLC by targeting MTA3. Tumour
Biol. 35:3487–3494. 2014. View Article : Google Scholar
|
|
8
|
Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu
G, Liu Z, Tu Y, Peng G, et al: MiR-495 and miR-551a inhibit the
migration and invasion of human gastric cancer cells by directly
interacting with PRL-3. Cancer Lett. 323:41–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang X, Huang H, Li Z, He C, Li Y, Chen
P, Gurbuxani S, Arnovitz S, Hong GM, Price C, et al: MiR-495 is a
tumor-suppressor microRNA down-regulated in MLL-rearranged
leukemia. Proc Natl Acad Sci USA. 109:19397–19402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang W, Wu S, Shi Y, Miao Y, Luo X, Ji M,
Yao K and He J: c-MYB regulates cell growth and DNA damage repair
through modulating MiR-143. FEBS Lett. 589:555–564. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang J, Liu X, Xue H, Qiu B, Wei B and
Sun K: MicroRNA-103a inhibits gastric cancer cell proliferation,
migration and invasion by targeting c-Myb. Cell Prolif. 48:78–85.
2015. View Article : Google Scholar
|
|
13
|
Yang K, He M, Cai Z, Ni C, Deng J, Ta N,
Xu J and Zheng J: A decrease in miR-150 regulates the malignancy of
pancreatic cancer by targeting c-Myb and MUC4. Pancreas.
44:370–379. 2015.
|
|
14
|
Mets E, Van der Meulen J, Van Peer G,
Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De
Moerloose B, Benoit Y, et al: MicroRNA-193b-3p acts as a tumor
suppressor by targeting the MYB oncogene in T-cell acute
lymphoblastic leukemia. Leukemia. 29:798–806. 2015. View Article : Google Scholar
|
|
15
|
Claes A, Idema AJ and Wesseling P: Diffuse
glioma growth: A guerilla war. Acta Neuropathol. 114:443–458. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huse JT and Holland EC: Targeting brain
cancer: Advances in the molecular pathology of malignant glioma and
medulloblastoma. Nat Rev Cancer. 10:319–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoelzinger DB, Demuth T and Berens ME:
Autocrine factors that sustain glioma invasion and paracrine
biology in the brain micro-environment. J Natl Cancer Inst.
99:1583–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Gong X, Tian K, Chen D, Sun J,
Wang G and Guo M: MiR-25 promotes glioma cell proliferation by
targeting CDKN1C. Biomed Pharmacother. 71:7–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu D, Ma P, Gao G, Gui Y, Niu X and Jin B:
MicroRNA-383 expression regulates proliferation, migration,
invasion, and apoptosis in human glioma cells. Tumour Biol.
36:7743–7753. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun JY, Xiao WZ, Wang F, Wang YQ, Zhu YH,
Wu YF, Miao ZL and Lin YC: MicroRNA-320 inhibits cell proliferation
in glioma by targeting E2F1. Mol Med Rep. 12:2355–2359.
2015.PubMed/NCBI
|
|
21
|
Cai W, Li Q, Yang Z, Miao X, Wen Y, Huang
S and Ouyang J: Expression of p53 upregulated modulator of
apoptosis (PUMA) and C-myb in gallbladder adenocarcinoma and their
pathological significance. Clin Transl Oncol. 15:818–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen RX, Xia YH, Xue TC and Ye SL:
Transcription factor c-Myb promotes the invasion of hepatocellular
carcinoma cells via increasing osteopontin expression. J Exp Clin
Cancer Res. 29:1722010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu H, Wang Y, Huang Y, Shi H, Xue Q, Yang
S, He S and Wang H: Expression and prognostic role of c-Myb as a
novel cell cycle protein in esophageal squamous cell carcinoma.
Clin Transl Oncol. 15:796–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang R, Li J, Yue M, Liu Z, Feng S, Tang S
and Wang T: A correlation analysis of miRNA-34a and its predicted
target genes in leukemia. Mol Med Rep. 9:1283–1288. 2014.PubMed/NCBI
|