Introduction
Breast cancer is one of the types of cancer, which
readily metastasizes to bone. Coleman (1) reported that between 65 and 75% of
patients with advanced breast cancer develop bone metastases. Bone
metastases are usually accompanied by pain, pathological fractures,
nerve compression syndromes and hypercalcemia (2). Histomorphological analyses of bone
metastases have revealed two types of lesions, osteolytic and
osteogenetic. In bone metastasis of breast cancer, 80% of stage IV
cases are found to be osteolytic and are accompanied by increased
osteoclast activity (3). The
process of osteoclastic bone resorption leads to the release of
several cytokines, including transforming growth factor-β (TGF-β)
and insulin-like growth factors, which stimulate the proliferation
and invasion of tumor cells, thus promoting a 'vicious cycle' of
tumor metastasis and bone destruction (4).
In our previous studies, it was found that matrix
metalloproteinases (MMPs) are important in the development and
expansion of tumor cells in bone metastasis and skeletal osteolysis
(5,6). The degradation of extracellular
matrix (ECM) by MMPs facilitates tumor cell invasion and
proliferation in the metastatic environment (7–9).
Among all MMP members, MMP1, 2, 3, 9 and 13 have been reported to
correlate with tumor metastasis (10,11).
Lee et al demonstrated that the inhibition of MMP2 and MMP9
undermines the capability of bone degradation by tumor metastasis
(12,13). MMP2 is secreted predominantly by
fibroblasts and osteoblasts (14,15),
and is involved in the activation of MMP13 (16) and degradation of the basement
membrane (17). MMP9 is produced
principally by osteoclasts (15)
and cells of the immune system, including macrophages and
neutrophils, which have been reported to be important for tumor
growth (10,18). MMP2 and MMP9 are able to cleave
collagen type I, IV and V, and are important in the degradation of
bone matrix (19).
Although the majority of the studies have focussed
on host-derived MMPs, there have been few reports on the
interrelation between MMPs and metastatic tumor cells. Therefore,
the present study investigated the expression of MMPs in osteolytic
bone metastasis nests originating from human breast cancer
cells.
Materials and methods
Cell culture
Human MDA-MB-231 breast cancer cells were supplied
by Professor Xiangzhi Li (Shandong University, Jinan, China). These
cells were grown in RPMI 1640 media supplemented with 10% fetal
bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, 0.02 mM
non-essential amino acids and 1% streptomycin/penicillin at 37°C in
a 5% CO2 environment. All cells were cultured in 25
cm2 cell-culture flasks to 70–80% confluence.
Animal model of breast cancer bone
metastasis and tissue preparation
All animal experiments were performed under the
Guidelines for Animal Experimentation of Shandong University. The
animal model of human breast cancer bone metastasis was established
through intracardiac injection of the MDA-MB-231 human breast
cancer cells into 5-week-old BALB/c nu/nu female mice (Vital River
Laboratory Animal Technology Co. Ltd., Beijing, China) under
anesthesia. On the day of injection, the flask-cultured MDA-MB-231
cells were trypsinized, counted with a hemocytometer, and diluted
to a concentration of 2×106 cells/ml in ice-cold Hank's
balanced salt solution. Following anesthetization of the mice with
8% chloral hydrate (400 mg/100 g body weight), a 0.1 ml dilution
(2×105 cells) was injected intracardially into the left
ventricle of each mouse (n=10), using a 1 ml syringe, similar to a
previously published method (5,6). The
mice were housed in micro-isolator solid-bottomed, polycarbonate
cages under standard laboratory conditions with a 12-h light/dark
cycle and a constant temperature of 20°C and humidity of 48%. All
mice were maintained on a standard commercial diet with autoclaved
water available ad libitum. At 4 weeks post-injection, upon
confirmation of an visible bone metastasis in the tibiae through
soft X-ray analysis, the mice were anesthetized with an
intraperitoneal injection of 10% chloral hydrate (400 mg/100 g body
weight) and fixed with 4% formaldehyde in 0.1 M phosphate buffer
(pH 7.4) by transcardial perfusion, and then the tibiae were
removed for histological processing. Briefly, the samples were
decalcified with 10% EDTA-2Na solution for 3 weeks at 4°C. The
specimens were dehydrated using an ascending ethanol series and
then embedded in paraffin using standard procedures. Serial
sections of 5 µm in thickness were prepared for
histochemical analysis.
Histological examination
Hematoxylin and eosin (H&E) staining was
performed to investigate the morphology of tibia in both groups.
Following dewaxing and hydration, the prepared sections were
immersed in Erthlich's haematoxylin for 15 min. The sections were
then washed with distilled water and differentiated in 1% HCl in
70% alcohol for 1 min and washed again for 2 mins. Following this,
the sections were stained with 1% eosin for 10 min and washed with
distilled water. Subsequently, all sections were dehydrated and
mounted. The stained sections were observed and then digital images
were obstained using a light microscope (Olympus BX-53; Olympus
Corportation, Tokyo, Japan).
Immunohistochemical examinations for
mammaglobin 1 (MGB1), proliferating cell nuclear antigen (PCNA),
MMP2, MMP9 and MMP13
The dewaxed paraffin sections were treated with 0.3%
hydrogen peroxide for 30 min at room temperature, and then
pre-incubated with 1% bovine serum albumin in phosphate-buffered
saline (BSA-PBS) for 20 min at room temperature to reduce
nonspecific binding. Subsequently, the sections were incubated with
the following primary antibodies in BSA-PBS at room temperature for
2 h: SCGB2A2/mammaglobin A polyclonal antibody (MGB1; Proteintech;
Sanying Biotechnology, Wuhan, China; cat. no. 235-1-AP; 1:50),
anti-PCNA (Ab-1) mouse monoclonal antibody (PC10l; Epitomics;
Abcam, Burlingame, CA, USA; cat. no. NA03; 1:50), mouse anti-human
MMP2 monoclonal antibody (EMD Millipore, Billerica, MA, USA; cat.
no. MAB3308; 1:50), mouse MMP9 antibody antigen affinifty-purified
polyclonal goat IgG (EMD Millipore; cat. no. AF909; 1:50) and goat
anti-MMP-13 polyclonal antibody (EMD Millipore; cat. no. AB8120;
1:50). Following rinsing with PBS, the sections were incubated with
the following secondary antibodies for 1 h at room temperature:
Polyclonal swine anti-rabbit immunoglobulin(Ig)/HRP from
DakoCytomation, Denmark (cat. no. Nr.P 0399; 1:100), goat
polyclonal anti-mouse IgG+IgM+IgA-H&L (HRP) from Abcam (cat.
no. ab102448; 1:100), goat polyclonal anti-mouse
IgG+IgM+IgA-H&L (HRP) from Abcam (cat. no. ab102448; 1:100),
peroxidase-conjugated AffiniPure anti-goat++IgG (H+L) from Jackson
Immunoresearch Laboratories, Inc. (West Grove, PA, USA; cat. no.
93894; 1:100) and peroxidase-conjugated AffiniPure anti-goat++IgG
(H+L) from Jackson Immunoresearch Laboratories, Inc. (cat. no.
93894; 1:100). The immune complexes were then visualized using
3,3′-diamino-benzidine tetrahydrochloride (Sigma-Aldrich, St.
Louis, MO, USA) as the substrate. All stained sections were faintly
counterstained with methyl green for assessment using light
microscopy (BX53; Olympus Corporation, Tokyo, Japan). The
immunostaining intensities (optical density; OD) for all sections,
with the exception of PCNA, were analyzed using Image-Pro Plus 6.2
software (Media Cybernetics, Inc., Silver Spring, MD, USA). Areas
exhibiting a positive reaction were manually selected using a
colour cube-based colour separate module in Image-Pro Plus. At
least six sections from each sample were analyzed. All values are
presented as the mean ± standard deviation. The differences in OD
values between the metaphysis and diaphysis for each
immunostaining, and differences in the OD values between MMP2 and
MMP9 in the metaphysis and diaphysis were assessed using Student's
t-test. Differences among the MMP2 immunointensity in
metaphysis group, MMP9 immunointensity in the metaphysis group,
MMP2 immunointensity in the diaphysis group and MMP9
immunointensity in the diaphysis group were analyzed using analysis
of variance (ANOVA). Statistical analysis was performed using
GraphPad Prism, version 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA). P<0.01 was considered to indicate a statistically
significant difference.
Tartrate-resistant acid phosphatase
(TRAP) staining for osteoclast localization
To evaluate the localization of osteoclasts, TRAP
staining was performed, as previously reported (20). In brief, the dewaxed paraffin
sections were submerged in a mixture of 3.0 mg naphthol AS-BI
phosphate, 18 mg red violet LB salt and 100 mM L(+) tartaric acid
(0.36 g) diluted in 30 ml of 0.1 M sodium acetate buffer (pH 5.0)
for 15 min at 37°C. The sections were then faintly counterstained
with methyl green for assessment using light microscopy (BX53;
Olympus Corporation).
In situ detection of apoptosis in breast
cancer bone metastasis
In order to identify the apoptotic status of the
cells in the metastatic tissues, TdT-mediated dUTP nick-end
labeling (TUNEL) analysis was performed using a TACS 2 TdT-blue
label in situ apoptosis detection kit (cat. no. 4811-30-K;
Trevigen, Inc., Gaithersburg, MD, USA). Briefly, the sections were
placed in 1X PBS for 10 min at room temperature following
rehydration in ethanol, and then covered with 50 µl
proteinase K solution and incubated for 15–30 min at 37°C.
Following washing twice in deionized water (2 min per wash), the
sections were immersed in quenching solution for 5 min at room
temperature. Following washing in 1X PBS for 1 min at room
temperature, the sections were immersed in 1X TdT labeling buffer
for 5 min, following which they were covered with 50 µl
labeling reaction mix and incubated at 37°C for 1 h in a humidity
chamber. Subsequently, the sections were immersed in 1X TdT Stop
buffer for 5 min at room temperature to terminate the labeling
reaction. Following washing twice in 1X PBS for 5 min each at room
temperature, the sections were covered in 50 µl
streptavidin-HRP solution and incubated for 10 min at 37°C.
Following washing twice in 1X PBS for 2 min each, the sections were
immersed in TACS-Blue label solution for 4 min, following which the
samples were washed in several changes of deionized water for 2 min
each. Finally, the samples were counterstained using nuclear fast
red. The numbers of PCNA- and TUNEL-positive cells were counted
using Image pro Plus 6.2 software (Media Cybernetics, Inc., Silver
Spring, MD, USA). The positively stained cells were manually
selected. At least six sections from each sample were analyzed. All
values are presented as the mean ± standard deviation. Differences
between the numbers of PCNA- and TUNEL-positive cells in the
metaphysis/diaphysis were assessed using Student's t-test.
Difference among the numbers of PCNA-positive cells in the
metaphysis, TUNEL-positive cells in the metaphysis, PCNA-positive
cells in the diaphysis and TUNEL-positive cell cells in the
diaphysis, was analyzed using ANOVA. Statistical analysis was
performed using GraphPad Prism, version 5.0 (GraphPad Software,
Inc.). P<0.01 was considered to indicate a statistically
significant difference.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR) analysis
The BALB/c nu/nu mice were sacrificed by overdose of
anesthesia. The tibiae were removed and separated into the
metaphysis and diaphysis. For RT-PCR, total RNA were extracted from
the MDA-MB-231 cells, metaphysis and diaphysis of the normal BALB/c
nu/nu mice using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. The first-strand complementary DNA was
synthesized using Superscript II reverse transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.). The PCR analysis was performed
using 2X Es Taq MasterMix (CWBio, Inc., Beijing, China) on a T100™
Thermal Cycler (Bio-Rad, Berkeley, CA, USA) and performed using the
following primers: Human MMP-9 (Gene ID: 4318) sense
5′-GGGACGCAGACATCGTCATC-3′ and antisense
5′-TCGTCATCGTCGAAATGGGC-3′), Human-MMP2 (Gene ID: 4313) sense
5′-GATACCCCTTTGACGGTAAGGA-3′ and antisense
5′-CCTTCTCCCAAGGTCCATAGC-3′, Mus-MMP9 (Gene ID: 17395) sense
5′-GCAGAGGCA TACTTGTACCG-3′ and antisense
5′-TGATGTTATGATGGTCCCACTTG-3′; and Mus-MMP2 (Gene ID: 17390) sense
5′-ACCTGAACACTTTCTATGGCTG-3′ and antisense
5′-CTTCCGCATGGTCTCGATG-3′. Human-β-actin (Gene ID: 60) sense
5′-CATGTACGTTGCTATCCAGGC-3′ and antisense
5′-CTCCTTAATGTCACGCACGAT-3′ and Mus-β-actin (Gene ID: 11461) sense
5′-GGCTGTATTCCCCTCCATCG-3′ and antisense
5′-CCAGTTGGTAACAATGCCATGT-3′ were used as internal controls (gene
IDs from www.ncbi.nlm.nih.gov/gene/). The conditions for RT-PCR
were similar to those previously described (21). The amplified PCR products were
separated on 2% agarose gels and digitized using the SmartGel™
Image Analysis system (Sagecreation, Beijing, China).
Results
Development of breast cancer bone
metastasis and TRAP staining
At 4 weeks post-intracardiac injection of MDA-MB-231
cells, 7/10 mice developed osteolytic lesions in the tibia,
detected on soft X-ray examination (Fig. 1A; white arrow). Breast cancer cells
positive for MGB1, which are exclusively overexpressed in primary
and metastatic human breast cancer (22), were abundant in the metastatic
lesions of the metaphysis (Fig.
1B) and diaphysis (Fig. 1C).
The TRAP staining showed that several TRAP-positive multinucleate
cells were present within the breast cancer bone metastasis nests
and on the surface of trabecular bone (Fig. 1D and E).
Immunolocalization of PCNA and in situ
detection of apoptosis
PCNA-positive cells were observed in the metaphyseal
area (Fig. 2Ab and c), but few
were observed in the diaphyseal area (Fig. 2Ad and f). In the metaphyseal tumor
nest, a few scattered TUNEL-positive apoptotic cells were observed
(Fig. 2Bb and c). However, in the
diaphyseal tumor nest, a higher number of TUNEL-positive tumor
cells were present, compared with that in metaphyseal area
(Fig. 2Be and f). ANOVA revealed
that, in the metaphysis, the number of PCNA-positive cells was
significantly higher, compared with the number of TUNEL-positive
cells (307.78±27.04, vs. 61.12±7.59 cells/mm2,
respectively; P<0.01; n=7; Fig.
2C). In the diaphysis, the number of TUNEL-positive cells was
significantly higher, compared with the number of PCNA-positive
cells (291.96±20.78, vs. 8.04±1.09 cells/mm2,
respectively; P<0.01; n=7; Fig.
2C).
 | Figure 2Immunohistochemical and statistical
analyses of PCNA and TUNEL staining for apoptosis. (A)
Immunohistochemical analysis of PCNA in the (a) metaphysis and (b)
diaphysis in the normal bone marrow of the control group.
Immunohistochemistry for PCNA in the (c) metaphyseal and (d)
diaphyseal tumor metastases. More PCNA-positive tumor cells (brown
color) were detected in the metaphyseal tumor tissue, compared with
the diaphyseal tumor tissue. (e and f) Higher magnification of c
and d, respectively. (B) TUNEL staining for apoptotic tumor cells
in the (a) metaphysis and (b) diaphysis in the normal bone marrow
tissue of the control group. TUNEL staining for apoptotic tumor
cells in the (c) metaphyseal and (d) diaphyseal tumor metastases.
More TUNEL-positive apoptotic tumor cells (blue color) were
observed in the diaphyseal tumor tissue, compared with the
metaphyseal tumor tissue. (e and f) High magnifications of c and d,
respectively. (C) Statistical analyses of the numbers of
PCNA/TUNEL-positive tumor cells in the metaphysis and diaphysis.
*P<0.01. Scale bar=50 µm in Aa-d and Ba-d; 25
µm in Ae and f, and Be and f. TUNEL, TdT-mediated dUTP
nick-end labeling; Meta, metaphysis; Dia, diaphysis; T, tumor
cells; CB, cortical bone; *, normal bone tissue. |
Immunolocalization of MMP2, MMP9 and
MMP13
In the metaphyseal area containing numerous
PCNA-positive tumor cells, MMP9-immunopositivity was significantly
more marked (Fig. 3Ab and c),
whereas staining for MMP2 was faint (Fig. 3Bb and c). In contrast, the
diaphyseal metastasis, containing more TUNEL-positive cells, showed
weak expression of MMP9 (Fig. 3Ae and
f), compared with MMP2 (Fig. 3Be
and f). No significant differences were found in the
immunolocalization and immunoreactivity of MMP13 between
PCNA-positive and negative areas (data not shown). ANOVA revealed
that, in the PCNA-positive metastatic area (metaphysis), the
staining intensity of MMP2 was significantly weaker, compared with
that of MMP9 (0.126±0.007, vs. 0.300±0.036, respectively;
P<0.01; n=7; Fig. 3C). In the
TUNEL-positive metastatic area (diaphysis), the staining intensity
of MMP2 was significantly more marked, compared with that of MMP9
(0.205±0.020, vs. 0.103±0.009; P<0.01; Fig. 3C).
 | Figure 3Immunohistochemical and statistical
analyses of MMP9 and MMP2. (A) Immunohistochemical analysis of MMP9
in the (a) metaphysis and (b) diaphysis of normal bone marrow in
the control group. Immunohistochemical analysis of MMP9 in tumor
tissue of the (c) metaphysis and (d) diaphysis (brown color). The
expression of MMP9 was significantly higher in the metaphysis,
compared with the diaphysis. (e and f) High magnification of c and
d, respectively. (B) Immunohistochemical analysis of MMP2 in the
(a) metaphysis and (b) diaphysis of normal bone marrow in the
control group. Immunohistochemical analysis of MMP2 in tumor tissue
of the (c) metaphysis and (d) diaphysis (brown color). The
expression of MMP2 was significantly higher in the diaphysis,
compared with the metaphysis. (e and f) High magnification of c and
d, respectively. (C) Statistical analyses of the immunostaining
intensity of MMP9 and MMP2 in the metaphysis and diaphysis.
*P<0.01. Scale bars=50 µm in Aa-d and Ba-d; 25
µm in Ae and f, and Be and B. MMP, matrix metalloproteinase;
Meta, metaphysis; Dia, diaphysis; T, tumor cells; CB, cortical
bone; *, normal bone tissue. |
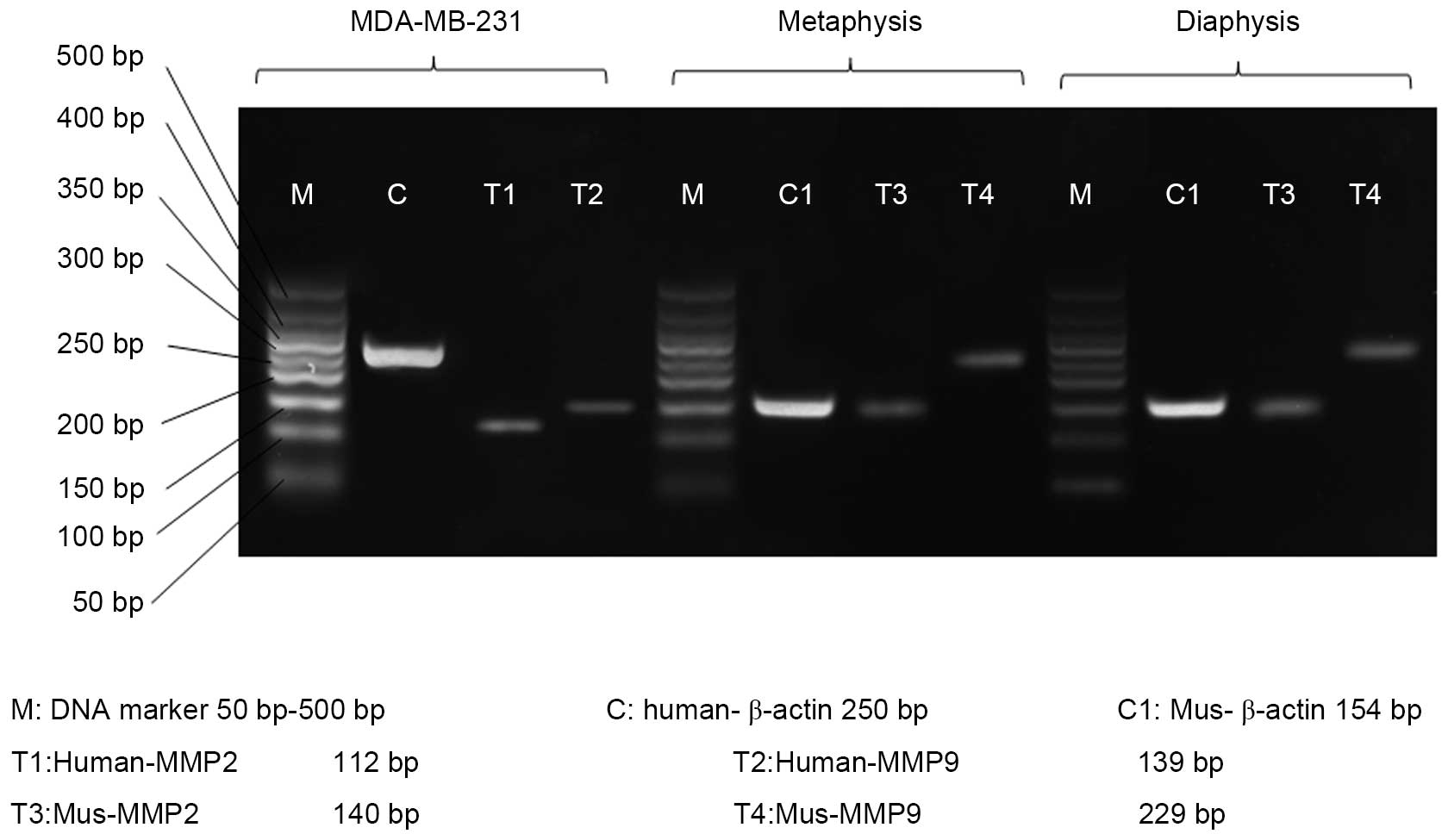
RT-PCR
The present study performed RT-PCR to investigate
the source of MMP2 and MMP9. The results revealed that MMP2 and
MMP9 mRNA were expressed in the MDA-MB-231 cells, metaphysis and
diaphysis of the tibiae of the BALB/c nu/nu mice without tumor cell
administration (Fig. 4).
Discussion
MMP9 and MMP2 belong to gelatinase, which is one of
five groups of the MMP family, based on structure and substrate
specificity (23). MMP9 and MMP2
are important in cancer invasion and metastasis by degrading the
ECM and basement membrane (24).
In the present study, the immunolocalization of MMP9 and MMP2 in
osteolytic metastasis originating from human breast cancer cells
were investigated. The results showed findings consistent with
those of Ohshiba et al (15), that the expression levels of MMP9
and MMP2 were upregulated in bone metastasis nests. Notably, the
present study found that MMP9 was overexpressed in the metaphysis
with high expression levels of PCNA, whereas MMP2 was detected
predominantly in the diaphysis with marked TUNEL-positive
expression.
Metaphysis is the most common homing site for tumor
cells due to its high level of vascularization. Once tumor cells
home to metaphysis, they are stimulated to proliferate by MMP9
(25) and bone-derived growth
factors, including TGF-β (26),
for their subsequent colonization in bone. Furthermore, Nutter
et al demonstrated that the expression of MMP9 was increased
on tumor cells colonization in bone (25). These findings were verified in the
present study, which demonstrated that MMP9 was overexpressed in
the metaphysis with a high level of PCNA-positive expression in the
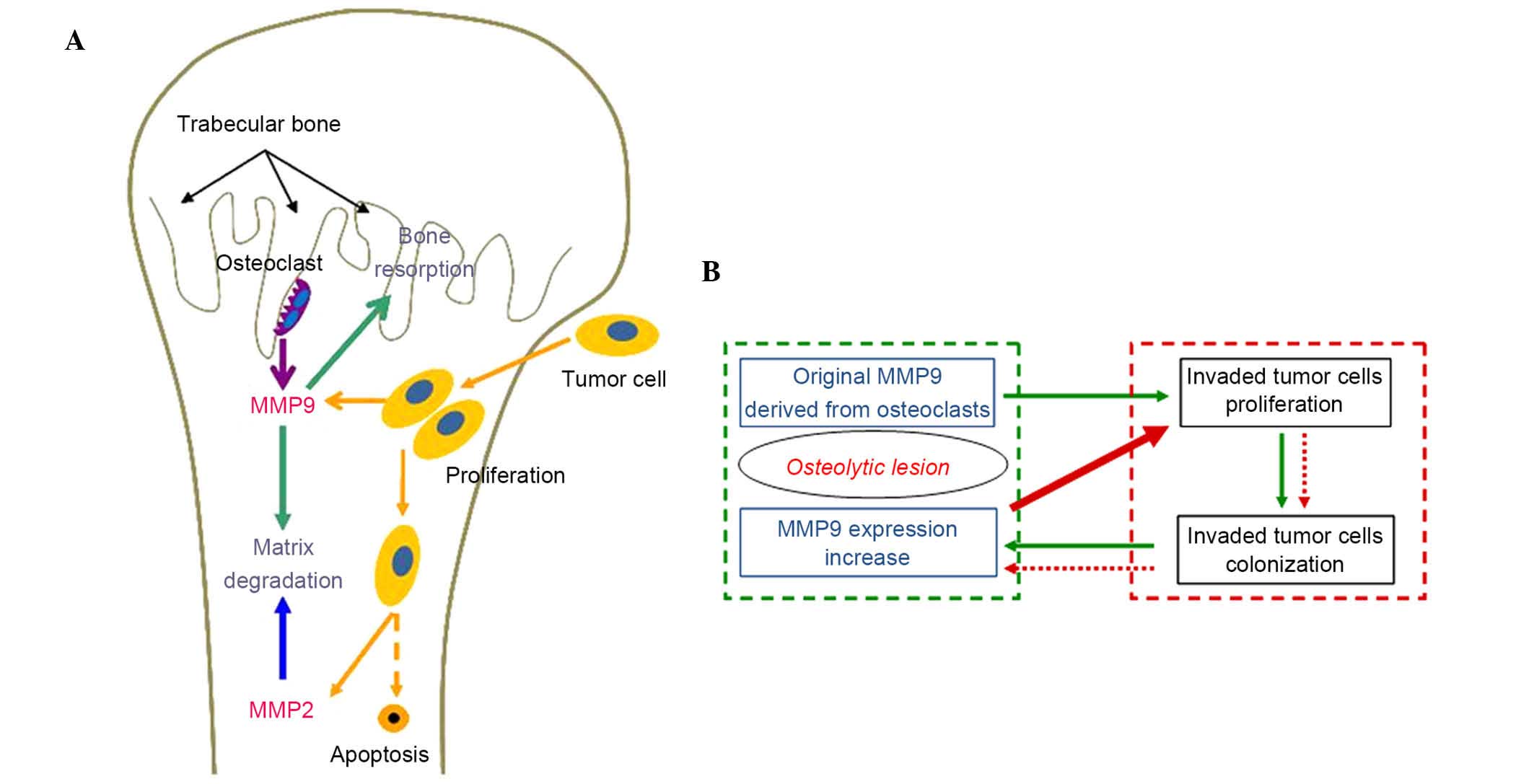
tumor cells (Fig. 5A). Tumor cells
become the predominant source of MMP9 production with the extension
of the bone metastasis nests, although MMP9 are predominantly
derived from osteoclasts and vascular endothelial cells prior to
tumor invasion. As shown in Fig.
5B, the 'vicious cycle', in which the original MMP9 derived
from osteoclasts stimulates the proliferation of invaded tumor
cells and subsequent colonization of tumor cells, accelerates the
expression of MMP9 may provide a further explanation for tumor bone
metastasis and offer a tumor prevention strategy. In addition, the
increased MMP9 is involved in the recruitment of bone-resorbing
osteoclasts, which leads to further osteolytic lesions (27,28).
In the present study, the tumor cells appeared to
expand towards the diaphysis following the initial invasion taking
place in the metaphysis. Compared with the immunolocalization of
MMP9, MMP2 was expressed at high levels in the diaphysis, which
exhibits weak proliferation/increased apoptosis of tumor cells. Ni
et al reported that the upregulation of MMP2 is important in
breast cancer bone metastasis through the
microRNA-106b/MMP2/extracellular signal-regulated kinase pathway,
which affects the balance of receptor activator of nuclear
factor-κB ligand and osteoprotegerin production (29). MMP2 is secreted predominantly by
fibroblasts and osteoblasts (14,15),
however, the results of the present study showed negative
expression in the fibroblasts and osteoblasts adjacent to the
metastatic tumor cells in the diaphysis. Further investigation is
required for understanding the intricate interactions among tumor
cells and host bone marrow cells. In addition, based on existing
data, it is difficult to explain why a higher number of
TUNEL-positive cells were found in the diaphysis occupied by the
invaded breast cancer cells. In view of a previous study, which
demonstrated that breast cancer cells may induce osteoblast
apoptosis (30), the present study
hypothesized that the higher number of TUNEL-positive cells in the
diaphysis may be composed predominantly of apoptotic stromal cells
and fibroblasts induced by the invaded breast cancer cells.
Although, certain apoptotic tumor cells may be contained due to
decreased blood supply in the diaphysis.
In conclusion, the results of the present study
showed that the invaded tumor cells exhibited different
proliferation activity and apoptosis status between metaphysis and
diaphysis. MMP9 was predominantly expressed in the PCNA-positive
metaphysis, whereas MMP2 was predominantly expressed in the
diaphysis, which contained more TUNEL-positive cells. As a
consequence, it was suggested that MMP9 and MMP2 may have their own
importance in ECM degradation and trabecular bone damage in
different zones of bone metastasis. Further investigations are
required to determine the exact mechanisms.
Acknowledgments
This study was partially supported by the National
Nature Science Foundation of China (grant nos. 81271965, 81470719
and 81311140173) and the Specialized Research Fund for the Doctoral
Program of Higher Education (grant no. 20120131110073) to Professor
Minqi Li, and the Shandong Province Science and Technique
Foundation, China (grant no. 2014GSF118093) to Dr Jie Guo.
References
|
1
|
Coleman RE: Metastatic bone disease:
Clinical features, pathophysiology and treatment strategies. Cancer
Treat Rev. 27:165–176. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamaoka T, Madewell JE, Podoloff DA,
Hortobagyi GN and Ueno NT: Bone imaging in metastatic breast
cancer. J Clin Oncol. 22:2942–2953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kozlow W and Guise TA: Breast cancer
metastasis to bone: Mechanisms of osteolysis and implications for
therapy. J Mammary Gland Biol Neoplasia. 10:169–180. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kingsley LA, Fournier PG, Chirgwin JM and
Guise TA: Molecular biology of bone metastasis. Mol Cancer Ther.
6:2609–2617. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li M, Amizuka N, Takeuchi K, Freitas PH,
Kawano Y, Hoshino M, Oda K, Nozawa-Inoue K and Maeda T:
Histochemical evidence of osteoclastic degradation of extracellular
matrix in osteolytic metastasis originating from human lung small
carcinoma (SBC-5) cells. Microsc Res Tech. 69:73–83. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li M, Sasaki T, Ono K, de Freitas PH,
Sobhan U, Kojima T, Shimomura J, Oda K and Amizuka N: Distribution
of macrophages, osteoclasts and the B-lymphocyte lineage in
osteolytic metastasis of mouse mammary carcinoma. Biomed Res.
28:127–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jacob K, Webber M, Benayahu D and Kleinman
HK: Osteonectin promotes prostate cancer cell migration and
invasion: A possible mechanism for metastasis to bone. Cancer Res.
59:4453–4457. 1999.PubMed/NCBI
|
|
8
|
Lynch CC and Matrisian LM: Matrix
metalloproteinases in tumor-host cell communication.
Differentiation. 70:561–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nannuru KC, Futakuchi M, Varney ML,
Vincent TM, Marcusson EG and Singh RK: Matrix metalloproteinase
(MMP)-13 regulates mammary tumor-induced osteolysis by activating
MMP9 and transforming growth factor-beta signaling at the
tumor-bone interface. Cancer Res. 70:3494–3504. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scherer RL, Mclntyre JO and Matrisian LM:
Imaging matrix metalloproteinases in cancer. Cancer Metastasis Rev.
27:679–690. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee J, Weber M, Mejia S, Bone E, Watson P
and Orr W: A matrix metalloproteinase inhibitor, batimastat,
retards the development of osteolytic bone metastases by MDA-MB-231
human breast cancer cells in Balb C nu/nu mice. Eur J Cancer.
37:106–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weber MH, Lee J and Orr FW: The effect of
Neovastat (AE-941) on an experimental metastatic bone tumor model.
Int J Oncol. 20:299–303. 2002.PubMed/NCBI
|
|
14
|
Monteiro-Amado F, Castro-Silva II, Lima
CJ, Soares FA, Kowalski LP and Granjeiro JM: Immunohistochemical
evaluation of MMP-2, MMP-9 and CD31/microvascular density in
squamous cell carcinomas of the floor of the mouth. Braz Dent J.
24:3–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ohshiba T, Miyaura C, Inada M and Ito A:
Role of RANKL-induced osteoclast formation and MMP-dependent matrix
degradation in bone destruction by breast cancer metastasis. Br J
Cancer. 88:1318–1326. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morrison C, Mancini S, Cipollone J,
Kappelhoff R, Roskelley C and Overall C: Microarray and proteomic
analysis of breast cancer cell and osteoblast co-cultures: Role of
osteoblast matrix metalloproteinase (MMP)-13 in bone metastasis. J
Biol Chem. 286:34271–34285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lafleur MA, Drew AF, de Sousa EL, Blick T,
Bills M, Walker EC, Williams ED, Waltham M and Thompson EW:
Upregulation of matrix metalloproteinases (MMPs) in breast cancer
xenografts: A major induction of stromal MMP-13. Int J Cancer.
114:544–554. 2005. View Article : Google Scholar
|
|
18
|
Huang S, Van Arsdall M, Tedjarati S,
McCarty M, Wu W, Langley R and Fidler IJ: Contributions of stromal
metalloproteinase-9 to angiogenesis and growth of human ovarian
carcinoma in mice. J Natl Cancer Inst. 94:1134–1142. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Björklund M and Koivunen E:
Gelatinase-mediated migration and invasion of cancer cells. Biochim
Biophys Acta. 1755:37–69. 2005.PubMed/NCBI
|
|
20
|
Li M, Hasegawa T, Hogo H, Tatsumi S, Liu
Z, Guo Y, Sasaki M, Tabata C, Yamamoto T, Ikeda K and Amizuka N:
Histological examination on osteoblastic activities in the alveolar
bone of transgenic mice with induced ablation of osteocytes. Histol
Histopathol. 28:327–335. 2013.PubMed/NCBI
|
|
21
|
Tsunoda M and Sharma RP: Modulation of
tumor necrosis factor alpha expression in mouse brain after
exposure to aluminum in drinking water. Arch Toxicol. 73:419–426.
1999. View Article : Google Scholar
|
|
22
|
Sasaki E, Tsunoda N, Hatanaka Y, Mori N,
Iwata H and Yatabe Y: Breast-specific expression of
MGB1/mammaglobin: An examination of 480 tumors from various organs
and clinicopathological analysis of MGB1-positive breast cancers.
Mod Pathol. 20:208–214. 2007. View Article : Google Scholar
|
|
23
|
Slattery ML, John E, Torres-Mejia G, Stern
M, Lundgreen A, Hines L, Giuliano A, Baumgartner K, Herrick J and
Wolff RK: Matrix metalloproteinase genes are associated with breast
cancer risk and survival: The Breast Cancer Health Disparities
Study. PLoS One. 8:e631652013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McCawley LJ and Matrisian LM: Matrix
metalloproteinases: They're not just for matrix anymore! Curr Opin
Cell Biol. 13:534–540. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nutter F, Holen I, Brown HK, Cross SS,
Evans CA, Walker M, Coleman RE, Westbrook JA, Selby PJ, Brown JE
and Ottewell PD: Different molecular profiles are associated with
breast cancer cell homing compared with colonisation of bone:
Evidence using a novel bone-seeking cell line. Endocr Relat Cancer.
21:327–341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yin JJ, Selander K, Chirgwin JM, Dallas M,
Grubbs BG, Wieser R, Massagué J, Mundy GR and Guise TA: TGF-beta
signaling blockade inhibits PTHrP secretion by breast cancer cells
and bone metastases development. J Clin Invest. 103:197–206. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Woodward JK, Holen I, Coleman RE and
Buttle DJ: The roles of proteolytic enzymes in the development of
tumour-induced bone disease in breast and prostate cancer. Bone.
41:912–927. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Balbín M, Pendás AM, Uría JA, Jiménez MG,
Freije JP and López-Otín C: Expression and regulation of
collagenase-3 (MMP-13) in human malignant tumors. APMIS. 107:45–53.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X,
Ding Q, Zha X, Sha J and Wang S: Downregulation of miR-106b induced
breast cancer cell invasion and motility in association with
overexpression of matrix metalloproteinase 2. Cancer Sci.
105:18–25. 2014. View Article : Google Scholar
|
|
30
|
Mastro AM, Gay CV, Welch DR, Donahue HJ,
Jewell J, Mercer R, DiGirolamo D, Chislock EM and Guttridge K:
Breast cancer cells induce osteoblast apoptosis: A possible
contributor to bone degradation. J Cell Biochem. 91:265–276. 2004.
View Article : Google Scholar : PubMed/NCBI
|