Introduction
Increasing environmental pollution and social
stressors have led to a decline in male reproductive health,
resulting in poor sperm quality and potential male infertility
(1,2). Male infertility is a worldwide health
concern that affects >20 million men. It is a multifactorial
disorder, which is triggered by environmental and physiological
factors (3). In these cases,
ejaculated spermatozoa are usually observed to have reduced
quantity or poor quality. Diagnosis of male infertility
predominantly relies on the traditional evaluation of semen
parameters, whereas genetic factors are often overlooked. Previous
studies have reported that genetic factors are frequently
associated with male fertility, and may provide novel insights into
the understanding of male infertility and the assessment of sperm
quality (4–6).
Spermatozoa are produced in the testes, and their
mRNAs maturate during the spermatogenesis process (7). Sperm mRNAs are believed to be
silenced following the release of sperm from the testes; therefore,
changes in the expression levels of sperm mRNAs may reflect
abnormal spermatogenesis (8). At
the protein level, previous studies have compared the proteomic
differences between normal and aberrant spermatozoa (9–15). A
set of proteins has been identified, which was confirmed to be
associated with sperm quality. However, the proteins investigated
were limited to the ones in high abundance, due to the
technological bias of current proteomics, thus often not fully
exploring the underpinning pathways. At the transcriptional level,
sperm mRNAs have been used as biomarkers to evaluate sperm quality,
and biomarkers associated with abnormal sperm are promising targets
for intervention, which may be developed into male contraceptive
targets (16,17). In the present study, a systematic
comparative analysis was performed in order to screen for
testis-specific and teratospermia-associated genes, aiming to
identify specific targets associated with sperm quality. The
current study provides novel insights into the clinical research of
sperm biology and identifies a potential panel of biomarkers for
the assessment of sperm quality.
Materials and methods
Data collection
Gene expression data of normal human tissues
(GSE14938) (18) and teratospermia
(GSE6967 and GSE6968) (19) were
downloaded from the Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/). The
raw data files, which contained the signal intensities and values
for every spot on the arrays, were downloaded. The annotation file
of the array platform was also downloaded using the GEO query
package. Specifically-expressed testis and epididymis genes were
identified by statistically comparing the expression of genes from
the testis/epididymis with genes from other normal tissues,
including bladder, cervix, colon, heart, kidney, liver, lung,
ovary, prostate, spleen and stomach, using one-tailed t-test with a
threshold of P<0.001. Highly expressed genes in the testis and
epididymis compared with other tissues, and highly expressed genes
in normozoospermia compared with in teratospermia were
statistically screened by Student's t-test. P<0.01 was
considered to indicate a statistically significant difference.
Statistical analysis was performed using SPSS software, version
18.0 (SPSS, Inc., Chicago, IL, USA).
Sample preparation
Testes were used for immunohistochemistry in the
present study. Young adult testes were collected from five young
fathers (27–33 years old), who had passed away in automobile
accidents, had no history of pathology that may affect reproductive
functions, and were willing to donate their bodies for medical
research while still alive. Donation of organs for medical research
was approved by their immediate family members. The elderly samples
were collected from five men (78–82 years old) undergoing
epididymal excision for prostatic cancer. All procedures were
approved by the Ethics Committee of Yu Huang Ding Hospital (Yantai,
China). One testis from each donor was processed for protein
extraction, whereas the other underwent immunohistochemistry.
In order to perform sperm protein quantification,
seminal plasma was collected from two groups consisting of 30
healthy young adults, 30 young men with teratospermia (normal
morphology <4%) and 30 young men with asthenozoospermia
(progressive motility <32%) aged between 28 and 32 years.
Seminal plasma was collected and processed according to the World
Health Organization (WHO) Laboratory Manual for the Examination and
Processing of Human Semen (5th edition, 2010). The donations were
authorized by the donors with written informed consent according to
the regulations and permission of the Ethics Committee of the Yu
Huang Ding Hospital (www.who.int/reproductivehealth/publications/infertility/9789241547789/en/).
Bioinformatics
All proteins were functionally grouped into several
categories according to the Gene Ontology Consortium (http://www.geneontology.org/) and literature
annotation in PubMed.
Immunohistochemistry
After fixation in Bouin's solution for 12 h,
testicular tissue blocks were processed for paraffin embedding by
conventional methods and divided into 4 µm tissue sections.
Antigen retrieval was conducted in a microwave oven for 15 min.
Endogenous peroxidases in the tissue sections were inhibited by
incubation with 3% (v/v) H2O2 for 1 h.
Subsequently, 3% (w/v) bovine serum albumin (BSA; Sigma-Aldrich,
St. Louis, MO, USA) in Tris-buffered saline (TBS) was used to block
non-specific binding with antibodies at room temperature. Sections
were incubated with primary antibodies against HSPA4L (ab81221) and
PGK2 (ab186742; Abcam, Cambridge, MA, USA) diluted 1:100 in
blocking solution, overnight at 4°C. The sections were then washed
several times with TBS and were incubated with horseradish
peroxidase-conjugated anti-rabbit immunoglobulin (Ig)G (ZB-2306;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) at a final dilution of 1:400 for 1 h at 37°C. A
3,3′-diaminobenzidine kit (Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.) was used to visualize peroxidase activity
at the binding sites. Hematoxylin was used to counterstain the
sections. Subsequently, the sections were dehydrated and mounted
before undergoing bright-field microscopy (DM LB2; Leica
Microsystems GmbH, Nussloch, Germany). Pre-immune rabbit IgG was
used as a negative control. Positive immunostaining was used to
provide quantitative results. Briefly, images of immunostained
sections were analyzed using commercial Image-Pro Plus version 6.0
(Media Cybernetics, Inc., Rockville, MD, USA). The unequal
illumination was processed by shading correction, and a reference
slide was used to correct the measurement system. A total of 10
fields from each section were scored for each tissue. When the
immunostained images were converted to grayscale, a linear
combination between the average gray signal intensity and the
relative area of positive staining cells was defined as the
integrated optical density.
Quantitative assessment of protein
expression in spermatozoa
After liquefaction, human ejaculated spermatozoa
were washed in phosphate buffered saline (PBS). The sperm pellet
was placed on 1% (w/v) gelatin-coated slides, air-dried and fixed
with ice-cold methanol for 10 min. Slides were blocked for 1 h at
room temperature with 3% (w/v) BSA in PBS and incubated at 37°C for
1 h with anti-HSPA4L [ab81221; Abcam; diluted 1:50 in PBS
containing 3% (w/v) BSA]. Following three washes with PBS, the
corresponding secondary antibody was applied (fluorescein
isothiocyanate-labeled goat anti-rabbit IgG; 1:200 in PBS
containing 3% (w/v) BSA; ZF-0311; Beijing Zhingshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China). Samples were subsequently
washed in PBS and deionized water. Propidium iodide (0.01 mg/ml;
Invitrogen; Thermo Fisher Scientific, Inc.) counterstaining
visualized the nuclei. Quantitative assessment of protein
expression in spermatozoa was achieved by scanning confocal
microscopy. Slides were systematically examined at a magnification
of ×400 according to the WHO manual 2010. Inspection was performed
in sequence until a total of 200 spermatozoa had been assessed.
Using LSM 510 META software (LSM 5 version 3.2; Carl Zeiss AG,
Oberchoken, Germany) the fluorescence intensity value per stained
cell was calculated automatically by subtracting the background
fluorescence intensity, which was determined by scanning a
sperm-free area. SPSS software, version 18.0 (SPSS, Inc.) was used
to perform Pearson's correlation coefficient analysis. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of significantly expressed
genes in the testis and epididymis
By comparing gene expression data sets from 11
normal tissues with that of the testis and epididymis, a total of
1,085 significantly expressed genes were identified in the testis
and epididymis. From this total, 770 genes exhibited greater
expression in the testis, and 97 genes in the epididymis. These 867
genes were used as background data for subsequent screening
(Fig. 1).
Identification of poorly expressed genes
in teratospermia
The reduced expression of certain genes in
teratospermia may be a potential biomarker for evaluating sperm
quality. By comparing the differential gene expression in normal
spermatozoa and teratospermia, 3,406 genes exhibited significantly
lower expression in teratospermia. From this total 1,087 and 750
genes had higher expression levels in the testis and epididymis,
respectively. When specific testis and epididymis genes were
compared, it was determined that 431 testis and epididymis specific
genes were poorly expressed in teratospermia. Of these genes,
highly expressed genes in the testis were termed to be of testis
origin, and highly expressed ones in the epididymis were termed to
be of epididymis origin. Finally, 348 testis origin genes and 30
epididymis origin genes were obtained, respectively (Fig. 1).
Specific genes in the testis and
epididymis are target genes that are significantly associated with
spermatogenesis and sperm maturation
Chromosomal distribution analysis of the 1,085
identified genes highlighted that the genes were mainly expressed
on chromosomes 17 (ratio=2.19) and 19 (ratio=2.67), whereas less
were expressed on chromosomes Y (ratio=0.44) and 13 (ratio=0.55)
(Table I). The gene ontology
analysis demonstrated that proteins corresponding to these genes
were primarily involved in functions of metabolism (22%), structure
(16%), protease/protease inhibitor (16%), as well as the major
functions of transcription (11%) and signal transduction (10%)
(Fig. 2A). Over-representative
analysis indicated that these proteins were significantly involved
in the biological processes of cell cycle, mitosis and
spermatogenesis (Table II).
 | Table IChromosomal distribution of human
teratospermia-associated genes and testis/epididymis specific
genes. |
Table I
Chromosomal distribution of human
teratospermia-associated genes and testis/epididymis specific
genes.
| Chromosome | Chromosomal size
(Mbp) | Observed
| Expected
| Ratio
|
|---|
| Sp | TE | Sp | TE | Sp | TE |
|---|
| 1 | 251 | 367 | 100 | 276 | 88 | 1.33 | 1.14 |
| 2 | 243 | 262 | 71 | 267 | 85 | 0.98 | 0.84 |
| 3 | 198 | 229 | 62 | 218 | 69 | 1.05 | 0.90 |
| 4 | 191 | 140 | 38 | 210 | 67 | 0.67 | 0.57 |
| 5 | 180 | 180 | 39 | 198 | 63 | 0.91 | 0.62 |
| 6 | 171 | 187 | 58 | 188 | 60 | 0.99 | 0.97 |
| 7 | 159 | 146 | 45 | 175 | 56 | 0.83 | 0.81 |
| 8 | 146 | 139 | 32 | 161 | 51 | 0.87 | 0.63 |
| 9 | 141 | 128 | 43 | 155 | 49 | 0.82 | 0.87 |
| 10 | 136 | 152 | 41 | 150 | 48 | 1.02 | 0.86 |
| 11 | 135 | 183 | 59 | 149 | 47 | 1.23 | 1.25 |
| 12 | 134 | 197 | 45 | 147 | 47 | 1.34 | 0.96 |
| 13 | 115 | 81 | 22 | 127 | 40 | 0.64 | 0.55 |
| 14 | 107 | 117 | 37 | 118 | 37 | 0.99 | 0.99 |
| 15 | 102 | 124 | 34 | 112 | 36 | 1.10 | 0.95 |
| 16 | 92 | 133 | 55 | 101 | 32 | 1.31 | 1.71 |
| 17 | 81 | 161 | 62 | 89 | 28 | 1.81 | 2.19 |
| 18 | 78 | 60 | 26 | 86 | 27 | 0.70 | 0.95 |
| 19 | 59 | 110 | 55 | 65 | 21 | 1.69 | 2.67 |
| 20 | 63 | 94 | 36 | 69 | 22 | 1.36 | 1.63 |
| 21 | 48 | 39 | 10 | 53 | 17 | 0.74 | 0.60 |
| 22 | 51 | 54 | 29 | 56 | 18 | 0.96 | 1.63 |
| X | 155 | 117 | 74 | 171 | 54 | 0.69 | 1.37 |
| Y | 59 | 6 | 9 | 65 | 21 | 0.09 | 0.44 |
 | Table IIOver-representative functional
analysis of testis/epididymis specific genes and key teratospermia
genes. |
Table II
Over-representative functional
analysis of testis/epididymis specific genes and key teratospermia
genes.
| Biological
process | Number | P-value |
|---|
| Sperm proteins | | |
| Cell cycle | 454 |
8.04×10−12 |
| Cellular component
biogenesis | 46 |
4.38×10−2 |
| Chromosome
segregation | 66 |
3.98×10−2 |
| DNA metabolic
process | 145 |
1.08×10−3 |
| Generation of
metabolites and energy | 96 |
1.61×10−2 |
| Intracellular
protein transport | 392 |
1.59×10−5 |
| Mitosis | 184 |
4.06×10−6 |
| mRNA
processing | 191 |
1.87×10−14 |
| Oxidative
phosphorylation | 29 |
1.65×10−2 |
| Protein metabolic
process | 934 |
8.49×10−31 |
| Protein
transport | 396 |
1.77×10−5 |
| RNA
localization | 43 |
1.79×10−4 |
| Translation | 230 |
1.33×10−31 |
| Testis and
epididymis proteins | | |
| Cell cycle | 131 |
2.32×10−11 |
| Chromosome
segregation | 23 |
5.70×10−3 |
| Fertilization | 13 |
3.98×10−2 |
| Gamete
generation | 49 |
2.60×10−3 |
| Mitosis | 61 |
7.73×10−8 |
| Regulation of
catalytic activity | 81 |
2.45×10−2 |
| Reproduction | 55 |
1.67×10−3 |
| RNA
localization | 14 |
1.06×10−2 |
|
Spermatogenesis | 30 |
1.19×10−4 |
| Final target
proteins | | |
| Cell cycle | 59 |
3.01×10−6 |
| Meiosis | 9 |
3.47×10−2 |
| Mitosis | 31 |
1.28×10−5 |
Poorly expressed genes in teratospermia
may be associated with abnormal spermatogenesis, which in turn may
be involved in poor sperm quality in vitro
Poorly expressed genes in teratospermia were mainly
distributed at chromosomes 17 (ratio=1.81) and 19 (ratio=1.69), and
less so at chromosomes Y (ratio=0.09) and 13 (ratio=0.64) (Table I). All of the genes were broadly
categorized into 13 functional clusters, as presented in Fig. 2B. The proteins corresponding to
these genes were primarily involved in metabolism (26%), whereas
signal transduction (12%) and structure (10%) were also main
functions. In addition, these proteins were significantly involved
in biological processes of cell cycle, mRNA processing, protein
metabolic process and translation (Table II).
The final 336 key proteins contributed to metabolism
(19%), reproduction (16%) and protease/protease inhibitor (13%)
(Fig. 2C). These genes were mainly
involved in cell cycle, meiosis and mitosis (Table II).
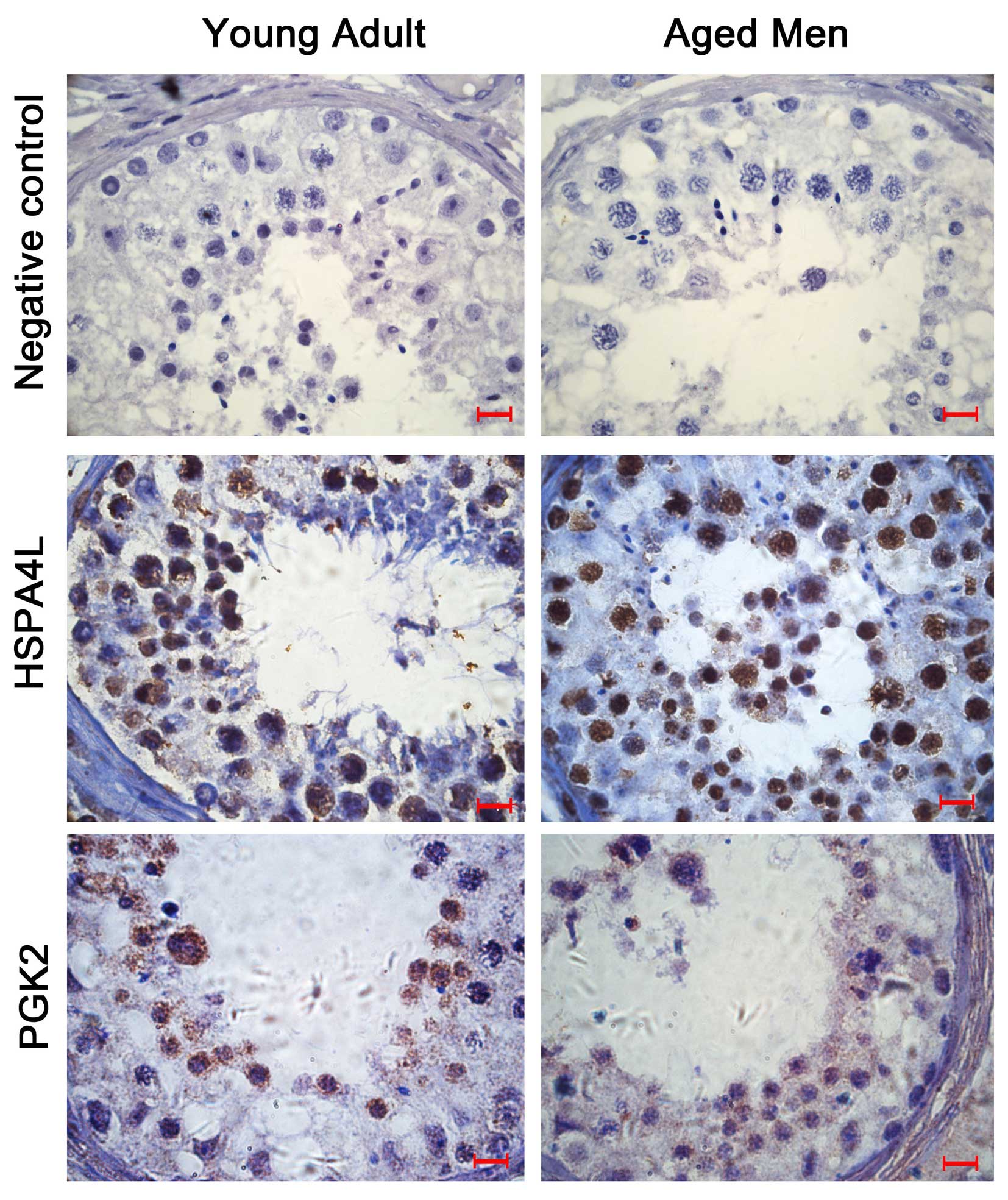
Immunohistochemical localization
From the 348 key proteins, the chaperone protein,
heat shock protein family A (Hsp70) member 4 like (HSP4AL) and
metabolism protein, phosphoglycerate kinase 2 (PGK2), were selected
for cellular localization investigation in the testis and
epididymis tissue samples obtained from young and aged men. As
presented in Fig. 3, HSPA4L was
primarily expressed in round spermatids, whereas PGK2 was primarily
expressed in spermatocytes and round spermatids. The proteins
exhibited higher expression in the testis compared with the
epididymis. In addition, the two proteins had lower expression
levels in the testis samples from the aged men, compared with the
younger men, thus suggesting there may be an age-related pattern of
expression.
Association of HSPA4L expression with
sperm functions
Immunofluorescent analysis indicated that HSPA4L was
located on the sperm acrosome. Quantification of protein
localization was performed using confocal scanning micros-copy,
which indicated that the percentage of sperm with positive staining
for HSPA4L and fluorescence intensity was lower in patients with
asthenozoospermia compared with normal young adults (Fig. 4).
There was a significant correlation between
quantification of HSPA4L in ejaculated spermatozoa and progressive
sperm motility. Higher correlation coefficients were observed
between staining percentage and intensity and progressive sperm
motility and sperm number in the sperm samples from patients with
asthenozoospermia (Table
III).
 | Table IIIRelationship between heat shock
protein family A (Hsp70) member 4 like quantification in ejaculated
sperm and progressive sperm motility and total sperm counts in
semen samples from young adults and patients with
asthenozoospermia. |
Table III
Relationship between heat shock
protein family A (Hsp70) member 4 like quantification in ejaculated
sperm and progressive sperm motility and total sperm counts in
semen samples from young adults and patients with
asthenozoospermia.
| Parameter | Stained (%)
| Intensity
|
|---|
| r | P | r | P |
|---|
| Progressive sperm
motility | | | | |
| Young adults | 0.522 | P<0.05 | 0.397 | NS |
|
Asthenozoospermia | 0.546 | P<0.05 | 0.504 | P<0.05 |
| Total sperm
number | | | | |
| Young adults | 0.431 | NS | 0.501 | P<0.05 |
|
Asthenozoospermia | 0.524 | P<0.05 | 0.638 | P<0.05 |
Discussion
Spermatozoa are produced in the testes and mature in
the epididymis; therefore, the testis and epididymis are the two
critical organs responsible for the formation of functional
ejaculated sperm (20). This
process includes two major biological processes: Spermatogenesis
and sperm maturation. Complex internal pathways are involved in
these processes via the interactions of various functional
proteins. Notably, various testis or epididymis-specific proteins
are important for the maintenance or modification of sperm
functions (21,22). Suboptimal expression levels of
these proteins may alter sperm functions; therefore, these proteins
may serve as potential biomarkers to assess sperm quality.
The present study used a novel comparative
bioinformatics analysis to identify tissue-specific genes and genes
associated with abnormal sperm. It was hypothesized that genes with
lower expression levels, particularly testis/epididymis-specific
genes in teratospermia may be considered promising biomarkers for
the assessment of sperm quality, which may provide novel insights
for male infertility research. When normal tissues were compared
with testis/epididymis tissues, 1,085 genes were determined to be
specifically expressed in the testis and epididymis. These genes
were used as background data to screen for sperm quality-associated
genes that were associated with spermatogenesis or sperm
maturation. These identified genes were mainly sourced from
chromosomes 17 and 19. Ontological analysis indicated that gene
products corresponding to these genes functioned predominantly in
metabolism, structure and protease/protease inhibition.
Over-representative analysis determined the significant biological
processes. It was demonstrated that chromosome segregation,
fertilization, gamete generation, mitosis and spermatogenesis were
the established spermatogenesis-associated functions. Proteins
involved in chromosome segregation may be responsible for human
infertility (23). Proteins
involved in functions of mitosis may be important for the
proliferation of germ cells and the differentiation of haploid
spermatids (24).
A large number of teratospermia-associated genes
were identified in the present study, which primarily contributed
to the regulation of metabolism, signal transduction and structure.
Corresponding to functions of testis/epididymis-specific genes, a
greater proportion of the genes were involved in the functions of
transferase, transporter, cell adhesion, chaperone,
defense/immunity and receptors. These genes were mainly involved in
intercellular biological processes and were ascribed to testicular
gene expression. Further screening detected the genes that
overlapped with testis/epididymis-specific genes, and 348
testis-specific genes were identified, which were considered
promising targets for future clinical research. Notably, the
majority of these genes were associated with reproductive
activities, and were involved in the biological processes of cell
cycle, meiosis and mitosis.
A recent study reported that various key genes are
responsible for teratospermia (4).
These genes were included in the dataset of the present study. Due
to abnormal meiotic segregation, sperm heads have aneuploid
composition leading to macrozoospermia. Aurora kinase C is involved
in meiosis and spermatogenesis, and mutation of this gene is
responsible for macrozoospermia (25). In addition, genetic mutations in
spermatogenesis associated 16, protein interacting with C kinase 1
and dpy-19 like 2 are associated with globozoospermia (26–28).
Other genes were initially reported to be associated with the
formation of teratospermia, which require further investigation.
Specifically, testis-specific genes may be considered potential
biomarkers for the assessment of ejaculated sperm quality. In the
present study, the gene products of HSPA4L and PGK2 were
characterized in human testes from elderly men, which may reflect
age or androgen-associated alterations in their expression. Further
quantification of HSPA4L in normal and poor quality sperm was
performed by immunofluorescence, and the results indicated that the
expression of HSPA4L in the sperm may be affected by testicular
alteration and was closely associated with sperm quality. It is
possible that these genes and their products may be potential
targets for the assessment of sperm quality, and warrant further
investigation.
In conclusion, 336 testis-specific genes were
determined to be associated with teratospermia, which were also
potentially associated with sperm quality and may be used as
promising biomarkers for male infertility research. HSPA4L was
determined to be associated with aberrant sperm quality. The
biomarkers identified in the present study warrant further
investigation and may be used as a panel of potential biomarkers
for the assessment of sperm quality.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81300533),
the Shandong Provincial Natural Science Foundation, China (grant
no. ZR2013HQ002), Yantai Science and Technology Program (grant no.
2015WS002) and Youth Science Foundation of Yantai Yu Huang Ding
Hospital, China (grant no. 201401).
References
|
1
|
Olea N and Fernandez MF: Chemicals in the
environment and human male fertility. Occup Environ Med.
64:430–431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rockliff HE, Lightman SL, Rhidian E,
Buchanan H, Gordon U and Vedhara K: A systematic review of
psychosocial factors associated with emotional adjustment in in
vitro fertilization patients. Hum Reprod Update. 20:594–613. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Manfo FP, Nantia EA and Mathur PP: Effect
of environmental contaminants on mammalian testis. Curr Mol
Pharmacol. 7:119–135. 2014. View Article : Google Scholar
|
|
4
|
Coutton C, Escoffier J, Martinez G,
Arnoult C and Ray PF: Teratozoospermia: Spotlight on the main
genetic actors in the human. Hum Reprod Update. 21:455–485. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Braekeleer M, Nguyen MH, Morel F and
Perrin A: Genetic aspects of monomorphic teratozoospermia: A
review. J Assist Reprod Genet. 32:615–623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brahem S, Elghezal H, Ghédir H, Landolsi
H, Amara A, Ibala S, Gribaa M, Saad A and Mehdi M: Cytogenetic and
molecular aspects of absolute teratozoospermia: Comparison between
polymorphic and monomorphic forms. Urology. 78:1313–1319. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamatani T: Human spermatozoal RNAs.
Fertil Steril. 97:275–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dadoune JP: Spermatozoal RNAs: What about
their functions? Microsc Res Tech. 72:536–551. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM and
Sha JH: Identification of several proteins involved in regulation
of sperm motility by proteomic analysis. Fertil Steril. 87:436–438.
2007. View Article : Google Scholar
|
|
10
|
Martinez-Heredia J, de Mateo S,
Vidal-Taboada JM, Ballescà JL and Oliva R: Identification of
proteomic differences in asthenozoospermic sperm samples. Hum
Reprod. 23:783–791. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan CC, Shui HA, Wu CH, Wang CY, Sun GH,
Chen HM and Wu GJ: Motility and protein phosphorylation in healthy
and asthenozoospermic sperm. J Proteome Res. 8:5382–5386. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siva AB, Kameshwari DB, Singh V, Pavani K,
Sundaram CS, Rangaraj N, Deenadayal M and Shivaji S:
Proteomics-based study on asthenozoospermia: Differential
expression of proteasome alpha complex. Mol Hum Reprod. 16:452–462.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen S, Wang J, Liang J and He D:
Comparative proteomic study between human normal motility sperm and
idiopathic asthenozoospermia. World J Urol. 31:1395–1401. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Parte PP, Rao P, Redij S, Lobo V, D'Souza
SJ, Gajbhiye R and Kulkarni V: Sperm phosphoproteome profiling by
ultra performance liquid chromatography followed by data
independent analysis (LC-MS (E)) reveals altered proteomic
signatures in asthenozoospermia. J Proteomics. 75:5861–5871. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amaral A, Paiva C, Attardo Parrinello C,
Estanyol JM, Ballescà JL, Ramalho-Santos J and Oliva R:
Identification of proteins involved in human sperm motility using
high-throughput differential proteomics. J Proteome Res.
13:5670–5684. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Garrido N, García-Herrero S and Meseguer
M: Assessment of sperm using mRNA microarray technology. Fertil
Steril. 99:1008–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feugang JM, Rodriguez-Osorio N, Kaya A,
Wang H, Page G, Ostermeier GC, Topper EK and Memili E:
Transcriptome analysis of bull spermatozoa: Implications for male
fertility. Reprod Biomed Online. 21:312–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
She X, Rohl CA, Castle JC, Kulkarni AV,
Johnson JM and Chen R: Definition, conservation and epigenetics of
housekeeping and tissue-enriched genes. BMC Genomics. 10:2692009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Platts AE, Dix DJ, Chemes HE, Thompson KE,
Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF
and Krawetz SA: Success and failure in human spermatogenesis as
revealed by teratozoospermic RNAs. Hum Mol Genet. 16:763–773. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hinton BT and Cooper TG: The epididymis as
a target for male contraceptive development. Handb Exp Pharmacol.
198:117–137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gupta GS: LDH-C4: A target with
therapeutic potential for cancer and contraception. Mol Cell
Biochem. 371:115–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dacheux JL and Dacheux F: New insights
into epididymal function in relation to sperm maturation.
Reproduction. 147:R27–R42. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kogo H, Kowa Sugiyama H, Yamada K, Bolor
H, Tsutsumi M, Ohye T, Inagaki H, Taniguchi M, Toda T and Kurahashi
H: Screening of genes involved in chromosome segregation during
meiosis I: Toward the identification of genes responsible for
infertility in humans. J Hum Genet. 55:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grimes SR: Testis-specific transcriptional
control. Gene. 343:11–22. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ounis L, Zoghmar A, Coutton C, Rouabah L,
Hachemi M, Martinez D, Martinez G, Bellil I, Khelifi D, Arnoult C,
et al: Mutations of the aurora kinase C gene causing
macrozoospermia are the most frequent genetic cause of male
infertility in Algerian men. Asian J Androl. 17:68–73. 2015.
View Article : Google Scholar :
|
|
26
|
Karaca N, Yilmaz R, Kanten GE,
Kervancioglu E, Solakoglu S and Kervancioglu ME: First successful
pregnancy in a globozoospermic patient having homozygous mutation
in SPATA16. Fertil Steril. 102:103–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koscinski I, Elinati E, Fossard C, Redin
C, Muller J, Velez de la Calle J, Schmitt F, Ben Khelifa M, Ray PF,
Kilani Z, et al: DPY19L2 deletion as a major cause of
globozoospermia. Am J Hum Genet. 88:344–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Mao Z, Wu M and Xia J: Rescuing
infertility of Pick1 knockout mice by generating testis-specific
transgenic mice via testicular infection. Sci Rep.
3:28422013.PubMed/NCBI
|