Introduction
Lung cancer is a major cause of cancer-associated
mortality worldwide and targeted therapies represent important
agents for the treatment of the disease (1). Therefore, the characterization of
molecular alterations of tumors is crucial for identifying patients
who are likely to benefit from these types of therapy. Advanced
non-small cell lung cancer (NSCLC) patients harboring sensitive
epidermal growth factor receptor (EGFR) gene
mutations or anaplastic lymphoma kinase (ALK) gene
rearrangements can be treated with specific tyrosine kinase
inhibitors (TKIs) (2). The
incidence of ALK rearrangements in NSCLC is ~3–5% and they
occur more often in never or light ex-smokers, in younger patients
and in those with lung adenocarcinoma. Patients exhibiting the
ALK gene rearrangement respond well to approved ALK
inhibitors, including crizotinib. The most common ALK
rearrangement is the echinoderm microtubule associated protein
like 4 (EML4)-ALK fusion. This results from an
inversion in the short arm of chromo-some 2, which causes the
fusion of the N-terminal domain of EML4 to the intracellular
kinase domain of ALK (3′-gene region), giving a
constitutively active ALK tyrosine kinase (3). Tumor tissue is the preferred
definitive sample type used for molecular analyses. However, for
numerous patients, this type of sample is not available. As a
result, the evaluation of surrogate sample types for the molecular
characterization of tumors has been gaining increasing
interest.
Several studies have already reported the usefulness
of circulating free tumor (ct) DNA for the analysis of somatic
mutations in NSCLC (4) and the
European Medicines Agency approved the assessment of EGFR
mutations using ct DNA when tumor tissue is unavailable.
The mRNA has been isolated from plasma, serum,
platelets and circulating tumor cells (CTCs) from patients
suffering from various types of malignancies, including lung cancer
(5–7), and has been used as a biological
marker for the early detection and diagnosis, or as a therapeutic
and prognostic indicator for the disease (8,9).
However, to the best of our knowledge, few studies have reported
the use of ct mRNA from serum or plasma to analyze the presence of
ALK rearrangements in NSCLC (10,11).
To demonstrate the feasibility of performing a
molecular characterization of lung adenocarcinoma using ct nucleic
acids, the present study planned a prospective study in which
patients with advanced NSCLC were enrolled. The present study
reported a series of 12 cases of lung adenocarcinoma tested for
aberrant ALK expression on ct mRNA purified from plasma and
analyzed using a one-step reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). Aberrant ALK expression
was detected in two patients and the results were confirmed by
fluorescence in situ hybridization (FISH) and
immunohistochemistry findings of respective solid biopsies.
Furthermore, in order to assess the reliability of the present
study, ct mRNA from healthy donors and other cancer patients was
also tested. Although this is a preliminary study, requiring
further confirmation, the present results supported the possibility
of detecting ALK aberrant expression on plasma from patients
with NSCLC.
Materials and methods
Patients
Aberrant ALK expression was firstly tested
using the mRNA from plasma and formalin-fixed paraffin-embedded
(FFPE) tissues from 12 patients with NSCLC (5 male and 7 female;
age range, 45–80 years). These patients underwent a bronchial
biopsy at the Unit of Thoracic Endoscopy, University Hospital of
Pisa (Pisa, Italy) between February and September 2015. All
patients belong to a prospective study aiming to evaluate the use
of ct nucleic acids for the molecular characterization of lung
adenocarcinoma (data not shown). The present study was headed by
the Unit of Pathological Anatomy, University Hospital of Pisa and
was approved by the local Ethics Committee. Furthermore, ALK
expression was analyzed on ct mRNA from the plasma of 4 healthy
donors (1 male and 3 female; age range, 48–62 years) and 4 patients
with thyroid and colon cancers (2 male and 2 female; age range,
42–70 years). This project required the collection of whole blood
samples into venous blood collection tubes using
ethylenediaminetetraacetic acid tripotassium as an anticoagulant.
Written informed consent was obtained from all enrolled
patients.
FISH and immunohistochemistry
The diagnosis of lung adenocarcinoma was performed
on hematoxylin and eosin stained sections from FFPE lung tissues.
FISH was performed using break-apart probes for ALK (Abbott
Molecular, Des Plaines, IL, USA). The FISH test was considered
positive if 15% or more of the tumor cells had separate 5′ (green)
and 3′ (red) probe signals or had isolated 3′ signals. Overlapping
red and green signals (resulting in yellow) indicated cells in
which ALK was not rearranged. Immunohistochemical staining of the
lesion tissue was performed using a rabbit monoclonal primary
anti-ALK antibody (clone D5F3; ready to use; Roche-Ventana Medical
Systems, Inc., Tucson, AZ, USA) in combination with an OptiView
3,3′-diaminobenzidine immu-nohistochemistry detection kit and an
OptiView Amplification kit (Ventana Medical Systems, Inc.). All the
hematoxylin and eosin staining, FISH and immunohistochemical
evaluations were performed by two independent pathologists
(Professor Gabriella Fontanini and Dr Greta Alì).
Nucleic acid extraction
Nucleic acids were extracted from FFPE tissues and
plasma samples. The DNA and RNA were purified from the FFPE tissues
using the QIAamp DNA Mini kit and the RNeasy FFPE kit (Qiagen,
Valencia, CA, USA), respectively. The plasma was isolated from
whole blood within 2–4 h of sample collection by centrifugation at
1,730 g for 10 min at 4°C. Once isolated, plasma samples were
immediately centrifuged again at 12,500 g for 10 min at 4°C and
frozen at −80°C until processing. The isolation of ct DNA and RNA
was performed separately from 3 ml plasma using the QIAmp
Circulating Nucleic Acid kit (Qiagen). The ct mRNA was then further
purified, after a DNase digestion step, using the RNeasyMinElute
Clean Up kit (Qiagen). The analyses of solid and liquid biopsy
samples were performed independently by different
investigators.
Mutational analysis
The mutational status of the tumor tissue and plasma
samples were determined by a Sequenom Mass-Array (MALDI-TOF MS)
using the Myriapod Lung Status kit (Diatech Pharmacogenetics Srl,
Jesi, Italy) together with the analysis software
MASSARRAY® TYPER 4.0 (Diatech Pharmacogenetics Srl)
(12), which allows the
simultaneous genotyping of 307 variants in the EGFR, KRAS, BRAF,
PIK3CA, NRAS, ALK, ERBB2, DDR2, MAP2K1 and RET genes.
The analysis of ct DNA was also performed using the more sensitive
Easy Real-Time PCR kits (Diatech Pharmacogenetics Srl) for the most
common variants of EGFR and KRAS.
ALK expression analysis
The aberrant expression of mRNA encoding for ALK was
analyzed using the Easy-ALK kit (Diatech Pharmacogenetics Srl),
which uses primers and probes specific for the ALK tyrosine
kinase domain. This method is a one-step procedure, during which
mRNA molecules are reverse-transcribed and directly amplified for
both ALK and a control gene, β-actin (ACTB).
In addition, each experimental run contains a positive transcript
control for both ALK and ACTB expression and a
negative template control. The analysis was performed on a
Rotor-Gene Q PCR thermocycler and analyzed by the Rotor-Gene 6000
Series software (Qiagen). Thermocycling conditions were as follows:
42°C for 5 min; 95°C for 10 sec; 40 cycles of 95°C for 5 sec and
60°C for 30 sec. According to manufacturer's protocol, after the
run is completed, sample Cq values are determined for both
ALK (target expression assay) and ACTB (control
expression assay). If ACTB amplification is detected, the
RNA sample is of good quality and ALK expression can be
assessed. If ACTB amplification is absent, the sample was
not further evaluated as the quality was not good enough. Under
normal conditions, without any rearrangements, ALK
expression should not be detectable using this method.
Results
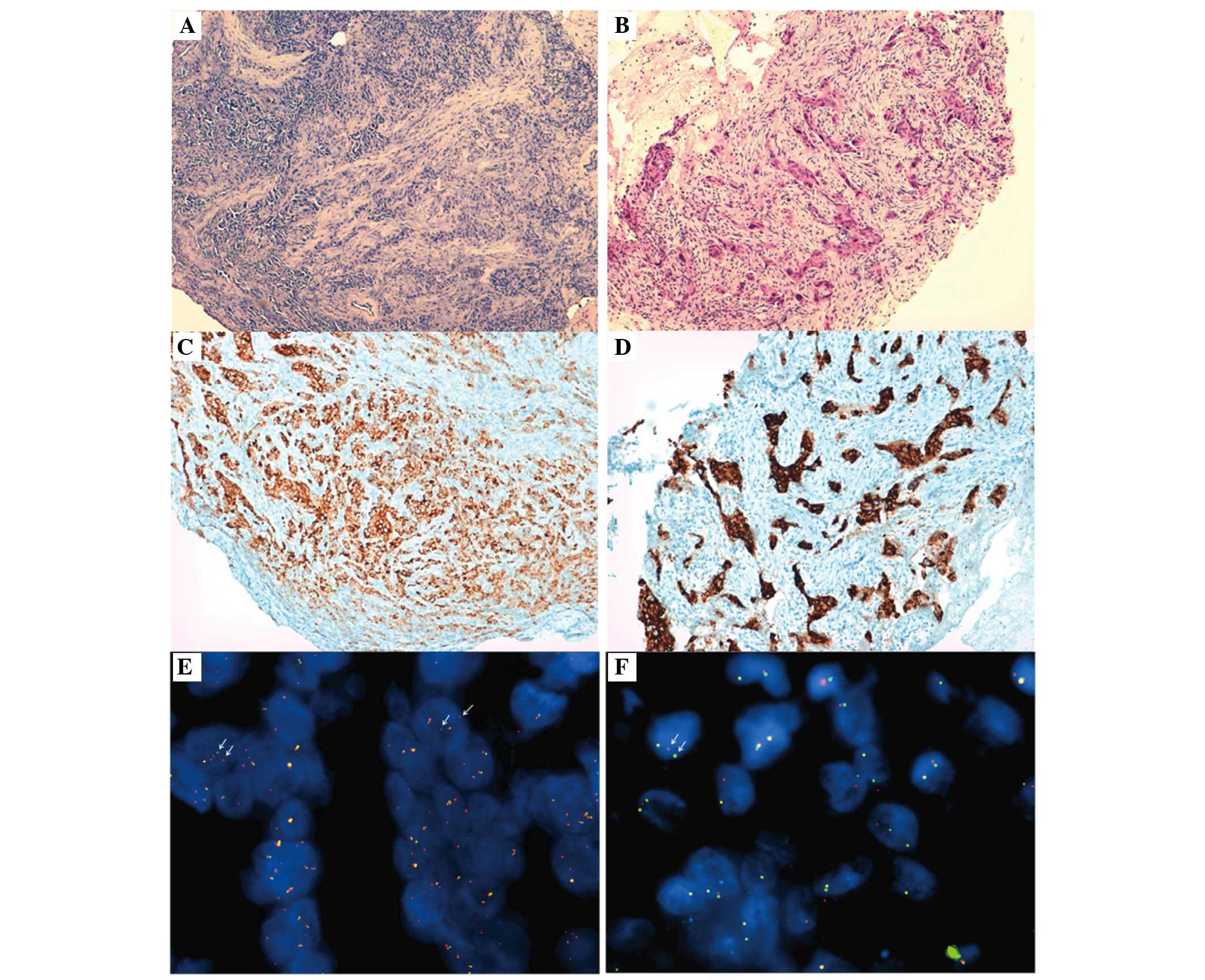
FISH and immunohistochemistry
The immunohistochemical analysis detected strong
granular cytoplasmic expression of the ALK protein only in 2/12
NSCLC enrolled patients. In the two positive cases, now referred to
as patient 1 and 2, FISH reported that 68 (34 ALK positive nuclei
from 50 examined) and 16% (8 ALK positive nuclei from 50 examined)
of the neoplastic cells, respectively, were positive for the
EML4-ALK rearrangement, according to the scoring method
proposed by Kwak et al (13). Figure
1 presents the hematoxylin and eosin staining sections,
immunohistochemistry and FISH images from the two ALK-positive
patients. In detail, patient 1 was a 44-year-old African man who
never smoked, with a clinical diagnosis of cT4N3M1a-b lung
adenocarcinoma with tumor metastases involving the liver, bones and
brain. Patient 2 was a 57-year-old European woman with a previous
history of smoking, with a clinical diagnosis of cT4N3M1a-SIVb lung
adenocarcinoma with cerebral and thoracic involvement (data not
shown).
Mutational analysis
The mutational analysis of EGFR, KRAS, BRAF,
PIK3CA, NRAS, ALK, ERBB2, DDR2, MAP2K1 and RET genes, on
solid and liquid biopsies, revealed that no patient with NSCLC
harbored any of the tested mutations in their tumors.
ALK expression analysis
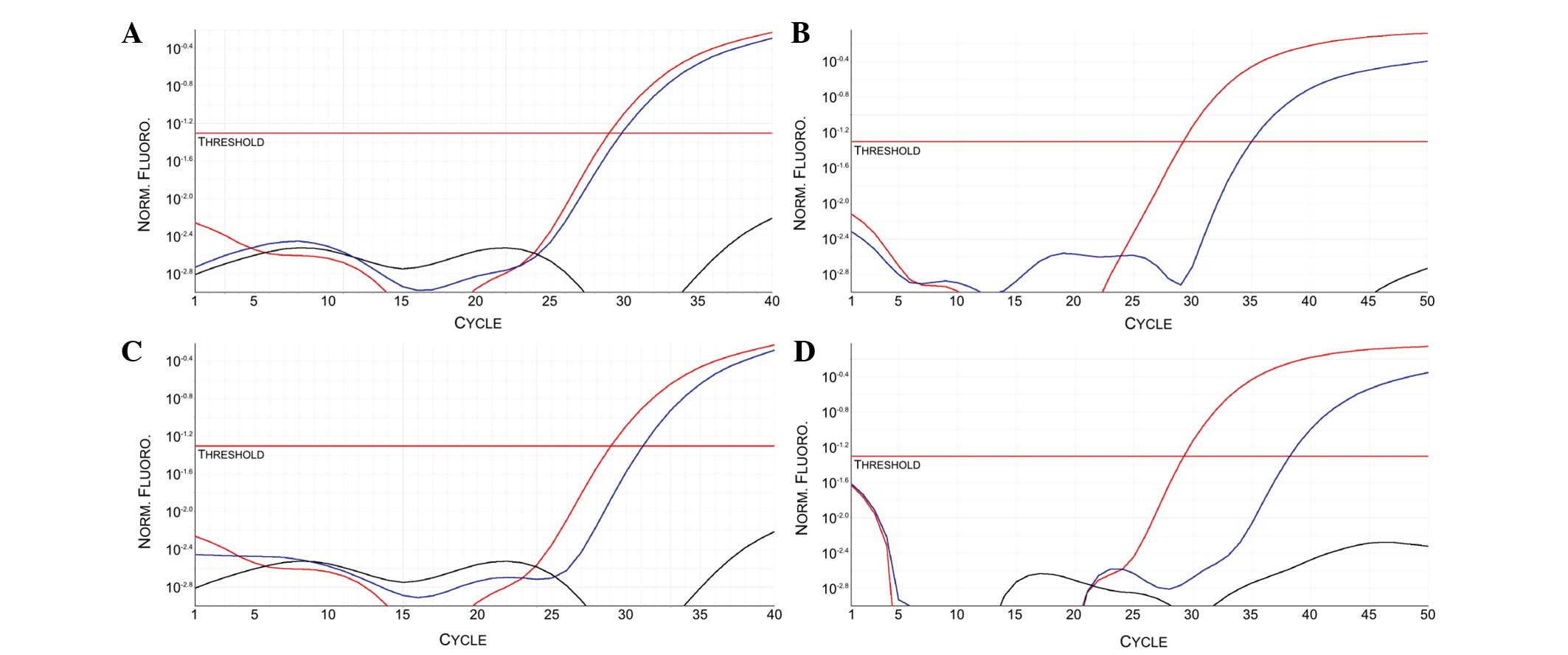
The ct mRNA samples from all the enrolled patients,
including the healthy donors and the other cancer patients,
exhibited a satisfactory ACTB amplification, confirming
their good quality for the evaluation step (data not shown).
Patients 1 and 2 exhibited a specific PCR amplification curve in
the ALK sample reactions (Fig. 2).
None of the other analyzed patients exhibited ALK
amplification (data not shown). The gene expression results on ct
mRNA were confirmed also on mRNA purified from NSCLC and other
cancer type FFPE tissues.
Discussion
The molecular characterization of ct nucleic acids
may be helpful for decision-making for the treatment of patients
with NSCLC whenever tumor tissue is not available. The plasma may
represent a surrogate source of tumor nucleic acids for both
genotyping and gene expression analyses.
The rearranged ALK gene acts as an oncogene
in lung adenocarcinoma and it can arise from fusions with several
partners, including EML4, HIP1 and TPR
(14–16). Patients with ALK
rearrangements can be successfully treated with ALK inhibitors,
including crizotinib (17,18).
For numerous years, CTCs have been suggested for
tumor molecular characterization (6,7,9),
however they are very difficult to detect and isolate from normal
nucleated cells in the blood, and their clinical use remains
limited.
To the best of our knowledge, little is known about
the evaluation of aberrant ALK expression in plasma and
serum, with the exception of two previous studies, one performed by
Kudo et al (10) on serum
samples using the MassArray system and one by Nilsson et al
(11) on the mRNA from plasma and
platelets by RT-PCR. Nilsson et al (11), in particular, assessed the
possibility of detecting the three most common EML4-ALK
variants on the mRNA from platelets and plasma in patients with
NSCLC. The authors used quantitative two-steps PCR TaqMan assays
and reported a sensitivity of 21 and 65% of the RT-PCR test in the
plasma and platelets RNA, respectively.
In order to demonstrate the feasibility of
evaluating aberrant expression of ALK on lung adenocarcinoma
using ct mRNA purified from plasma, the present study analyzed 12
patients with NSCLC and identified ALK rearrangements in two
cases. All patients with NSCLC included in the present study
belonged to a prospective series of 34 lung adenocarcinoma patients
(data not shown). The percentage of ALK rearranged cases in
the present study is consistent with the reported incidence of 3–5%
in NSCLC (3).
The described positive patients had 68 and 16% of
neoplastic nuclei positive for ALK rearrangement by FISH,
and both were positive for ALK aberrant mRNA expression in
plasma by the one-step RT-qPCR technique. Although this method is
unable to characterize the specific rearranged ALK variants,
the presence of aberrant ALK mRNA levels in plasma samples
may be enough to indicate that patients are candidates for
TKI-based therapy where tumor biopsies are not available. Notably,
when a solid biopsy is not available, it is challenging to
determine which of the most common EML4-ALK variants should
be analyzed on plasma, since EML4 is not the only ALK
rearrangement partner. In this context, the evaluation of aberrant
expression of ALK, directly on a small quantity of ct mRNA
extracted from few milliliters of plasma, can represent a promising
diagnostic tool. Furthermore, the absence of an ALK amplification
curve in ALK negative NSCLC patients, healthy donors and
other cancers patients, confirmed the specificity of the used
primers and probes. Further studies on a larger series of samples
are required to confirm this data.
In addition, the detection of aberrant ALK
mRNA expression both on tissue and plasma can be useful whenever
immunohistochemistry and FISH results are discordant. It has been
previously demonstrated that a single FISH or immunohistochemical
analysis may not detect all the ALK-positive cases and that certain
patients with discordant testing respond to TKIs (19). In several previous studies, the use
of RT-PCR for the detection and characterization of specific
ALK fusions has been evaluated, and the sensitivity and
specificity reported ranged between 94 and 100% (20–22).
However, the clinical application of specific RT-PCR assays has
been limited by the number of reactions and the large quantity of
clinical samples required to investigate the different ALK
fusions. Previously, Huang et al (23) demonstrated, both on NSCLC tissue
samples and on cell-free urine samples, the efficacy of a
differential expression method, based on the presence of aberrant
high levels of the ALK kinase domain (23). Notably, the predominant
pathological consequence of ALK fusion in tumor cells is its
aberrant expression, regardless of the fusion partner. Similarly,
the one step method used in this work can be applied to evaluate
the presence of aberrant expression levels of ALK.
Furthermore, in lung adenocarcinoma,
ALK-rearrangements showed a considerable level of
intratumoral heterogeneity, which can influence the assessment and
the success of therapies (24).
The evaluation of aberrant ALK expression on ct RNA may be a
powerful tool to implement tumor characterization on solid biopsy
for primary screening and predominantly to monitor the disease
progression.
Despite the relatively unstable nature of mRNA,
particularly from the plasma, the present study confirmed that ct
mRNA is suitable for RT-qPCR, according to Nilsson et al
(11). However, to obtain good
quality mRNA for the amplification step, the optimization of
sampling phases and nucleic acid purification is urgently required,
and a one-step RT-PCR assay may be recommended to reduce the bias
associated with a distinct retrotranscription step.
Currently, no ALK RT-qPCR kits have been confirmed
on liquid biopsies; therefore, the present study used the same of
FFPE tissues and obtained satisfying results for all the analyzed
samples, since all showed a good amplification of the ACTB
control gene.
The present study represented a starting point for
further research on a larger number of patients to define the
sensitivity and specificity of this detection system, and to
delineate a specific protocol for plasma samples that can be
included in routine clinical practice for NSCLC. This would be
particularly helpful for when a solid biopsy is not available. The
analysis of ct nucleic acids may include not only characterization
of the mutational status of the EGFR, but also detection of
aberrant ALK expression.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rekhtman N, Leighl NB and Somerfield MR:
Molecular testing for selection of patients with lung cancer for
epidermal growth factor receptor and anaplastic lymphoma kinase
tyrosine kinase inhibitors: American society of clinical oncology
endorsement of the college of American pathologists/international
association for the study of lung cancer/association for molecular
pathology guideline. J Oncol Pract. 11:135–136. 2015. View Article : Google Scholar
|
|
3
|
Solomon B, Varella-Garcia M and Camidge
DR: ALK gene rearrangements: A new therapeutic target in a
molecularly defined subset of non-small cell lung cancer. J Thorac
Oncol. 4:1450–1454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Douillard JY, Ostoros G, Cobo M, Ciuleanu
T, Cole R, McWalter G, Walker J, Dearden S, Webster A, Milenkova T
and McCormack R: Gefitinib treatment in EGFR mutated caucasian
NSCLC: Circulating-free tumor DNA as a surrogate for determination
of EGFR status. J Thorac Oncol. 9:1345–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidt B, Engel E, Carstensen T,
Weickmann S, John M, Witt C and Fleischhacker M: Quantification of
free RNA in serum and bronchial lavage: A new diagnostic tool in
lung cancer detection? Lung Cancer. 48:145–147. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ross K, Pailler E, Faugeroux V, Taylor M,
Oulhen M, Auger N, Planchard D, Soria JC, Lindsay CR, Besse B, et
al: The potential diagnostic power of circulating tumor cell
analysis for non-small-cell lung cancer. Expert Rev Mol Diagn.
13:1605–1629. 2015. View Article : Google Scholar
|
|
7
|
Pailler E, Adam J, Barthélémy A, Oulhen M,
Auger N, Valent A, Borget I, Planchard D, Taylor M, André F, et al:
Detection of circulating tumor cells harboring a unique ALK
rearrangement in ALK-positive non-small-cell lung cancer. J
ClinOncol. 31:2273–2281. 2013. View Article : Google Scholar
|
|
8
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Faugeroux V, Pailler E, Auger N, Taylor M
and Farace F: Clinical utility of circulating tumor cells in
ALK-positive non-small-cell lung cancer. Front Oncol. 4:2812014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kudo K, Nishio M, Sakai K, Tanimoto A,
Sakatani T and Saito R: Detection of EML4-ALK in serum RNA from
lung cancer patients using MassARRAY platform. J Clin Oncol.
30:suppl; abstr 10569. 2012.
|
|
11
|
Nilsson RJ, Karachaliou N, Berenguer J,
Gimenez-Capitan A, Schellen P, Teixido C, Tannous J, Kuiper JL,
Drees E, Grabowska M, et al: Rearranged EML4-ALK fusion transcripts
sequester in circulating blood platelets and enable blood-based
crizotinib response monitoring in non-small-cell lung cancer.
Oncotarget. 7:1066–1075. 2016.
|
|
12
|
Gabriel S, Ziaugra L and Tabbaa D: SNP
genotyping using the Sequenom MassARRAY iPLEX platform. Curr
Protocs Human Genet. Chapter 2: Unit 2.12. 2009. View Article : Google Scholar
|
|
13
|
Kwak EL, Bang YJ, Camidge DR, Shaw AT,
Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al:
Anaplastic lymphoma kinase inhibition in non-small-cell lung
cancer. N Engl J Med. 363:1693–1703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hong M, Kim RN, Song JY, Choi SJ, Oh E,
Lira ME, Mao M, Takeuchi K, Han J, Kim J and Choi YL: HIP1-ALK, a
novel fusion protein identified in lung adenocarcinoma. J Thorac
Oncol. 9:419–422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi YL, Lira ME, Hong M, Kim RN, Choi SJ,
Song JY, Pandy K, Mann DL, Stahl JA, Peckham HE, et al: A novel
fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol.
9:563–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cameron L and Solomon B: Treatment of
ALK-rearranged non-small cell lung cancer: Recent progress and
future directions. Drugs. 75:1059–1070. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabillic F, Gros A, Dugay F, Begueret H,
Mesturoux L, Chiforeanu DC, Dufrenot L, Jauffret V, Dachary D,
Corre R, et al: Parallel FISH and immunohistochemical studies of
ALK status in 3244 non-small-cell lung cancers reveal major
discordances. J Thorac Oncol. 9:295–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan Y, Zhang Y, Li Y, Hu H, Wang L, Li H,
Wang R, Ye T, Luo X, Zhang Y, et al: ALK, ROS1 and RET fusions in
1139 lung adenocarcinomas: A comprehensive study of common and
fusion pattern-specific clinicopathologic, histologic and cytologic
features. Lung Cancer. 84:121–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takeuchi K, Choi YL, Soda M, Inamura K,
Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y, et
al: Multiplex reverse transcription-PCR screening for EML4-ALK
fusion transcripts. Clin Cancer Res. 14:6618–6624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Soda M, Isobe K, Inoue A, Maemondo M,
Oizumi S, Fujita Y, Gemma A, Yamashita Y, Ueno T, Takeuchi K, et
al: A prospective PCR-based screening for the EML4-ALK oncogene in
non-small cell lung cancer. Clin Cancer Res. 18:5682–5689. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Q, Deng Q, Jiang L, Fang R, Qiu Y,
Wang P, Zhou JX and Yang H: Assessment of ALK gene fusions in lung
cancer using the differential expression and exon integrity
methods. Oncol Lett. 11:1651–1656. 2016.PubMed/NCBI
|
|
24
|
Zito Marino F, Liguori G, Aquino G, La
Mantia E, Bosari S, Ferrero S, Rosso L, Gaudioso G, De Rosa N,
Scrima M, et al: Intratumor heterogeneity of ALK-rearrangements and
homogeneity of EGFR-mutations in mixed lung adenocarcinoma. PLoS
One. 10:e01415212015. View Article : Google Scholar
|