Introduction
Lung cancer remains a leading cause of
cancer-associated mortality worldwide, and non-small cell lung
cancer (NSCLC) is a predominant type, accounting for 75–80% of lung
cancer cases (1,2). Although surgical resection,
chemotherapy and radiotherapy are available, the long-term survival
rates of patients with NSCLC remain poor. Therefore, a shift in
paradigm is required from increasing the survival rates of patients
with lung cancer patients to preventing lung cancer development.
Previous investigations on cancer have focused on natural herbs in
preventing or controlling cancer as an alternate therapy (3–5). In
addition, natural substances and plant derivatives have been used
to treat cancer patients with reduced toxicity. Previous reports
have also suggested that flavonoids in fruit can enhance anticancer
effects (6–10), indicating that these substances are
suitable for chemoprevention. Citrus platymamma Hort. ex
Tanaka (Byungkyul in Korean) has been used in Korean folk medicine
for the treatment of various diseases, including cancer.
Flavonoids, which has been reported in citrus species, including
C. platymamma, exert antiproliferative, anticancer,
antioxidant, anti-inflammatory and antidiabetic activities
(11–14). However, the molecular mechanism
underlying the anticancer effect of flavonoids from C.
platymamma (FCP) on lung cancer remains to be elucidated.
Proteomics is now an important area of investigation
in various fields, including cancer biology. Proteome analysis has
been applied in the investigation of various types of cancer
in-vitro and in vivo (15–19),
including lung cancer (20–22).
Previously, the anticancer mechanisms of therapeutic agents have
been elucidated by performing comparative proteome analysis on A549
cells (23–25). The present study was conducted to
investigate the mechanism of the anti-cancer effect of FCP-treated
A549 cells by examining the expression of proteins involved in
cancer cell survival, apoptosis, differentiation, invasion,
metastasis and metabolism. Increased understanding of the molecular
mechanisms underlying the anticancer effects of FCP may provide
novel insights in the prevention of lung carcinogenesis, which may
assist in developing novel strategies not only to prevent cancer
development, but also to improve quality of life for patients with
lung cancer.
Our previous study demonstrated that FCP induced
G2/M cell cycle arrest and apoptosis in A549 human lung cancer
cells (26). Therefore, the
present study aimed to identify the differentially expressed key
proteins, which may underlie the anticancer effects of FCP on A549
cells, using a proteomic approach. In total, eight differentially
expressed proteins were identified using two-dimensional gel
electrophoresis, coupled with matrix-assisted laser
desorption/ionization time-of-flight/time-of-flight tandem mass
spectrometry (MALDI-TOF/TOF-MS) analysis; 14-3-3ε (YWHAE) was
upregulated, and cofilin-1 (CFL1), annexin A1 (ANXA1), annexin A4
(ANXA4), endoplasmin (HSP90B1), cytoskeratin 8 (KRT8), elongation
factor Ts (tsf) and uncharacterized protein (KRT79) were
downregulated. The expression levels of YWHAE, CFL1, ANXA4 and KRT8
were also validated by immunoblotting. To date, this is first
study, to the best of our knowledge, to use proteomic techniques to
investigate the molecular mechanisms underlying the anticancer
effects of FCP on A549 cells.
Materials and methods
Chemical and reagents
The A549 human lung cancer cells and WI-38 normal
human fetal lung fibroblast cells were obtained from the Korea Cell
Line Bank (Seoul, Korea). RPMI 1640 medium was purchased from GE
Healthcare Life Sciences Hyclone Laboratories (Logan, UT, USA).
Fetal bovine serum (FBS) and antibiotics (streptomycin/penicillin)
were purchased from Gibco; Thermo Fisher Scientific, Inc. (Waltham,
MA, USA). 5-diphenyltetrazolium bromide (MTT) was obtained from
Sigma-Aldrich (St. Louis, MO, USA). Materials and chemicals used
for electrophoresis were obtained from Bio-Rad Laboratories, Inc,
(Hercules, CA, USA). FCP was provided by Animal Bio-Resources Bank
(Jinju, Korea). Antibodies targeting CFL1 (cat. no. AB3831), ANXA4
(cat. no. ABC885) and KRT8 (cat. no. MAB1611) and β-actin (cat. no.
MABT825) were purchased from EMD Millipore (Billerica, MA, USA).
Antibody targeting YWHAE (cat. no. BS6109) was obtained from
Bioworld Technology, Inc. (St. Louis Park, MN, USA). Horseradish
peroxidase (HRP)-conjugated goat anti-mouse IgG
(ALX-211-205TS-C100) and anti-rabbit IgG (ADI-SAB-301-J) were
purchased from Enzo Life Sciences, Inc. (Farmingdale, NY, USA). All
other chemicals used in the present study were purchased from
Amresco, Inc. (Solon, OH, USA) and Sigma-Aldrich. All chemicals
used were of the highest grade commercially available.
Cytotoxicity assay
The A549 cells and WI-38 cells (1×105)
were grown in RPMI-1640 medium supplemented with 10%
heat-inactivated FBS and 1% penicillin/streptomycin in a humidified
incubator with 5% CO2 in air 37°C. The cells were
cultured in 12-well plates and incubated overnight. Subsequently,
the cells were treated with 0, 100, 200, 300, 400 and 500
µg/ml of FCP for 24 h. Following incubation, 100 ml of 0.5
mg/ml MTT solution was added to each well and incubated for 3 h at
37°C in the dark. The formazan contained in the cells was
solubilized by the addition of 500 ml of DMSO, and the absorbance
was measured at 540 nm using an enzyme-linked immunosorbent assay
plate reader.
Preparation of the total cellular extract
for 2-DE
The total proteins were extracted from the A549
cells in the untreated (control) and FCP-treated groups. Following
incubation with FCP, the cells were lysed in lysis buffer,
containing 7 M urea, 2 M thiourea and 4% (w/v) CHAPS, on ice for 1
h. The lysates were then centrifuged at 13,000 × g for 15 min at
4°C, and the collected supernatant was stored at −80°C until
analysis. The total protein was used for 2-DE. The protein
concentration was determined using the Non-Interfering™ protein
assay kit (G-Biosciences, St. Louis, MO, USA), in accordance with
the manufacturer's protocol.
2-DE and image analysis
An equal quantity (150 µg) of protein per
sample was loaded onto a 18 cm linear IPG strip (pH 4–7; Amersham
Biosciences; Uppsala, Sweden) for first-dimensional
isoelectrofocusing, which was followed by 12% second dimension
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) on an Ettan DALT II system (Amersham Biosciences). The
gels were stained with silver nitrate, as described previously with
modifications (27), and three
independent gels were used in triplicate. Briefly, gels were
incubated in fixation solution (50% ethanol and 5% acetic acid) for
15 h, washed once with 30% ethanol for 15 min followed by three
times with distilled water for 5 min each. The gels were stained
with silver nitrate (0.3%) in the dark for 25 min at room
temperature. The gels were subsequently rinsed with water three
times and developed with solution containing 3% sodium carbonate,
0.02% sodium thiosulfate and 0.05% formalin. The gels were scanned
and image analysis was performed using Progenesis Samespots
software (Nonlinear Dynamics, Newcastle, UK). Using this software,
the differentially expressed spots were identified by automatic
matching of the detected protein spots. Those spots differing
significantly (P<0.05) in their intensities (fold-change ≥2), in
the FCP-treated A549 cells were used for further analysis.
MALDI-TOF/TOF MS analysis
Selected protein spots were excised manually from
the 2-DE gel, and protein digestion was performed (28) with modifications. Briefly, the
excised gel pieces were washed with 100 µl 100 mM
NH4HCO3 for 5 min and then dehydrated in 100
µl acetonitrile for 10 min. Following drying in a
lyophilizer (SFDSM06; Samwon Freezing Engineering Co., Busan, South
Korea), the gel pieces were rehydrated in 5–10 µl 50 mM
NH4HCO3 containing 20 ng/µl trypsin
(Promega Corporation, Madison, WI, USA) on ice. After 45 min, the
trypsin solution was removed and replaced with 10–20 µl 50
mM NH4HCO3 without trypsin, and digestion was
performed for a minimum of 16 h at 37°C. Subsequently, the peptide
mixtures were targeted onto a MALDI-TOF/TOF plate and analyzed
using a Voyager-DE STR mass spectrometer (Applied Biosystems;
Thermo Fisher Scientific, Inc.), equipped with delay ion
extraction.
Database search
The proteins were identified using the Mascot
program (http://www.matrixscience.com). The
Swissprot database and peptide mass fingerprinting (PMF) data were
used to identify matching proteins. The following parameters were
used for the database searches: Taxonomy, Homo sapiens
(human); cleavage specificity, trypsin with one missed cleavage
permitted; peptide tolerance of 100 ppm for the fragment ions;
permitted modifications, fixed cysteine carbamidomethylation,
variable oxidation of methionine. Protein scores >84 were
considered statistically significant (P<0.05).
Western blot analysis
The A549 cells (2×105) were cultured in
6-well plates and incubated with FCP (363 µg/ml) for 24 h.
The cell lysates was prepared, and 30 µg of proteins were
separated by 12% SDS-PAGE and transferred onto a PVDF membrane. The
blots were blocked with 5% nonfat dry milk for 1 h at room
temperature and then incubated with primary antibodies overnight
(dilution, 1:1,000), followed by incubation with HRP-conjugated
goat anti-mouse IgG (dilution, 1:1,000) for 2 h at room
temperature. The signal was visualized using ECL detection reagent
(GE Healthcare Life Sciences) and quantified by densitometry using
the Image J (http://rsb.info.nih.gov) program. The
densitometry readings of the bands were normalized to the
expression of β-actin. The experiment was repeated three times.
Gene ontology (GO) analysis
GO analysis was performed using the Agbase database
(http://www.agbase.msstate.edu/), as
previously described (29). GO
annotations were obtained from GORetriever by submitting the spot
identities. The annotation results were summarized based on the GOA
and whole proteome GOSlim set using GOSlimViewer (agbase.msstate.edu/cgi-bin/tools/goslimviewer_select.pl).
Statistical analysis
All statistical analyses were performed using SPSS
software (SPSS for Windows, ver. 10.0; SPSS, Inc. Chicago, IL,
USA). The data are presented as the mean ± standard deviation of at
least three independent experiments. The statistical significance
between the control and test groups was determined using one-way
analysis of variance followed by Student's t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
FCP specifically inhibits the
proliferation of A549 human lung cancer cells
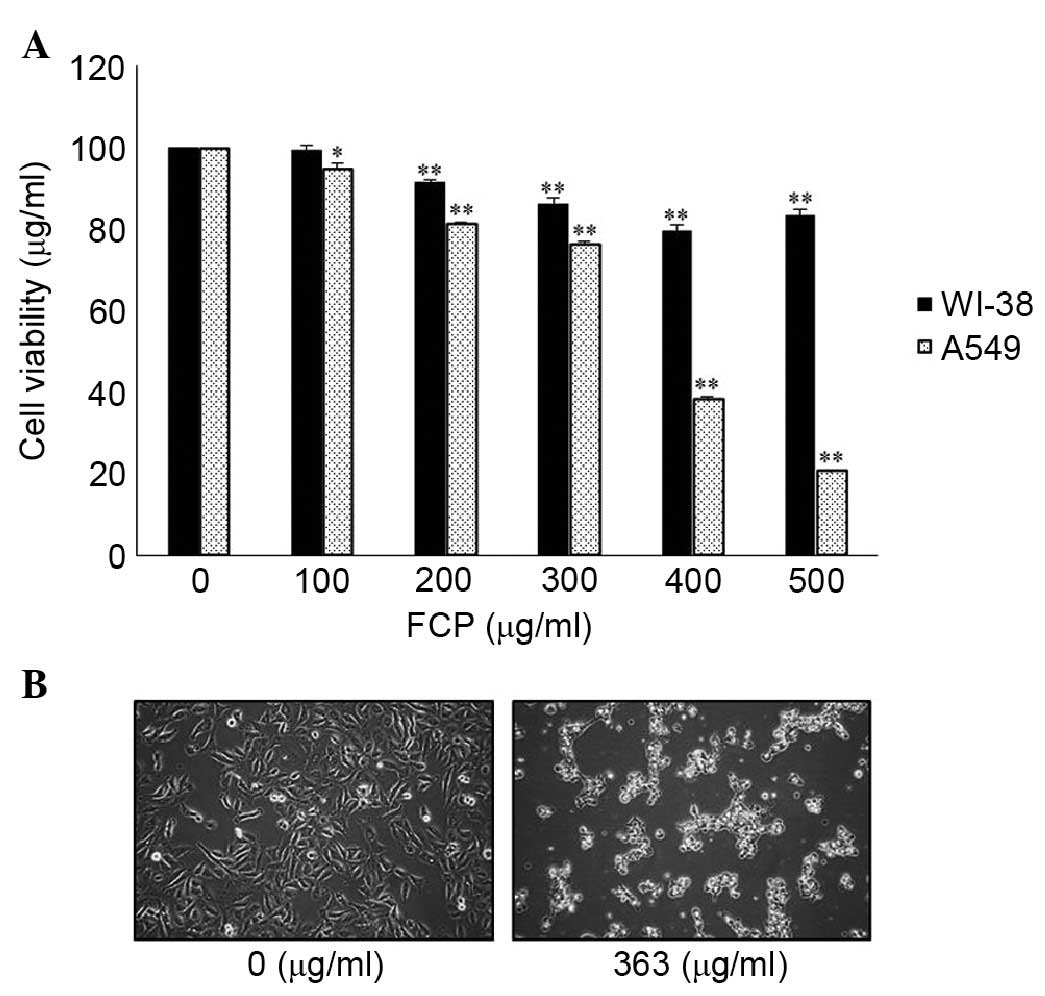
To investigate the anticancer activity of FCP, A549
human lung cancer cells and WI-38 human fetal lung fibroblasts were
treated with the indicated concentrations (0–500 µg/ml) of
FCP for 24 h. FCP inhibited the growth of the A549 cells in a
dose-dependent manner, however, no definite cytotoxic effects were
found in the WI-38 cells (Fig. 1A)
and cytotoxicity was negligible at concentrations of ≥200
µg/ml. These results showed that FCP exhibited a level of
specific cytotoxicity towards cancer cells. The 50% inhibitory
concentration for FCP treatment was found to be ~363 µg/ml.
In addition, morphological changes, including cell shrinkage and
density, and decreased cell numbers were observed in the
FCP-treated A549 cells (Fig. 1B).
These results suggested that the FCP specifically inhibited the
A549 lung cancer cells.
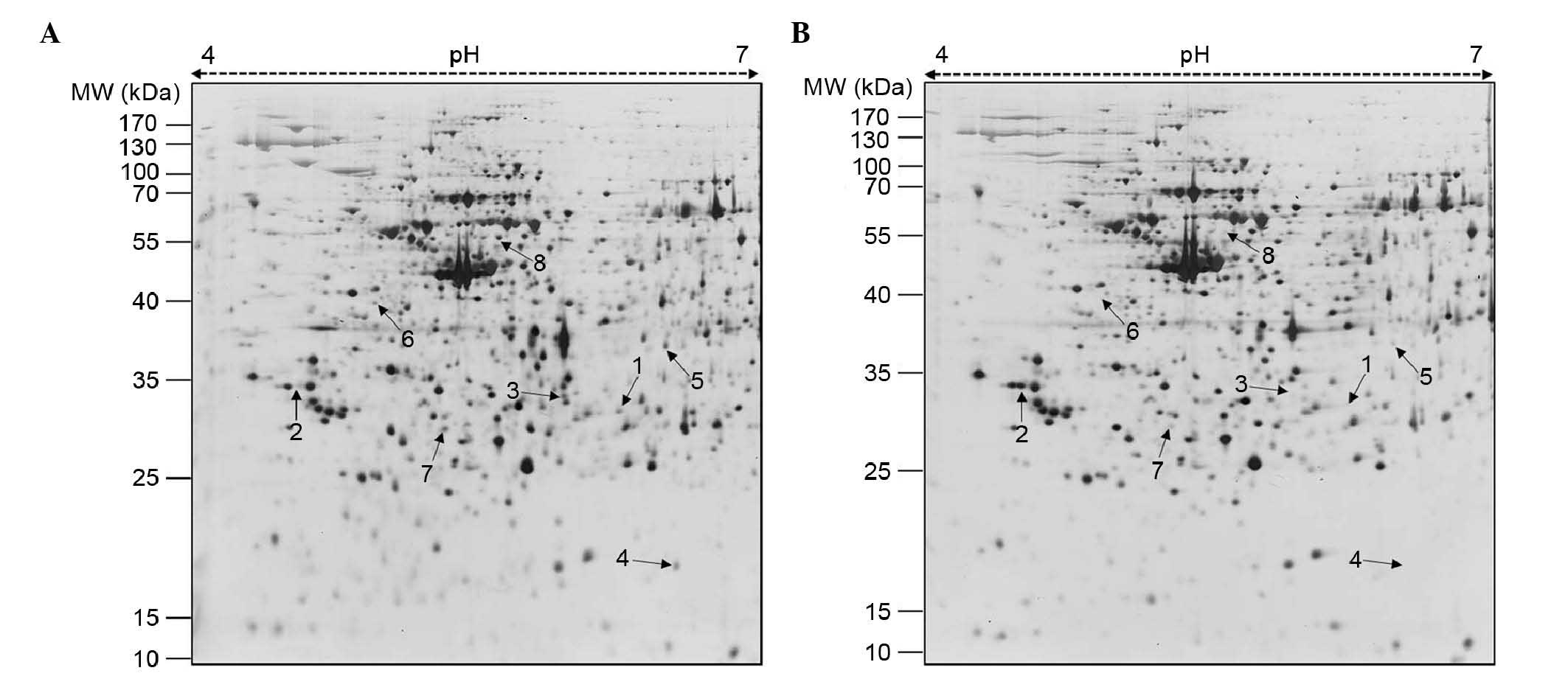
2-DE analysis and identification of
differentially expressed proteins using MALDI-TOF/TOF-MS
Subsequently, to investigate the mechanism
underlying the inhibitory effects of FCP on A549 cell
proliferation, 2-DE gel analysis was performed. The representative
2-DE patterns of the untreated (control) and FCP-treated A549 cells
were obtained by resolving 150 µg of total proteins on IPG
strips (18 cm; pH 4–7) in the first dimension and 12% SDS-PAGE in
the second dimension (Fig. 2). In
total, 15 differentially expressed protein spots were identified
(≥2-fold; P<0.05) using Progenesis Samespots image analysis
software (version 4.0). Finally, eight differentially expressed
proteins, one upregulated and seven downregulated, were detected in
the FCP-treated A549 cells using MALDI-TOF/TOF-MS analysis and
database searching (Fig. 2 and
Table I). Specifically, YWHAE was
upregulated, and ANXA4, CFL1, tsf, HSP90B1, ANXA1, KRT8 and KRT79
were downregulated in the FCP-treated A549 cells, compared with the
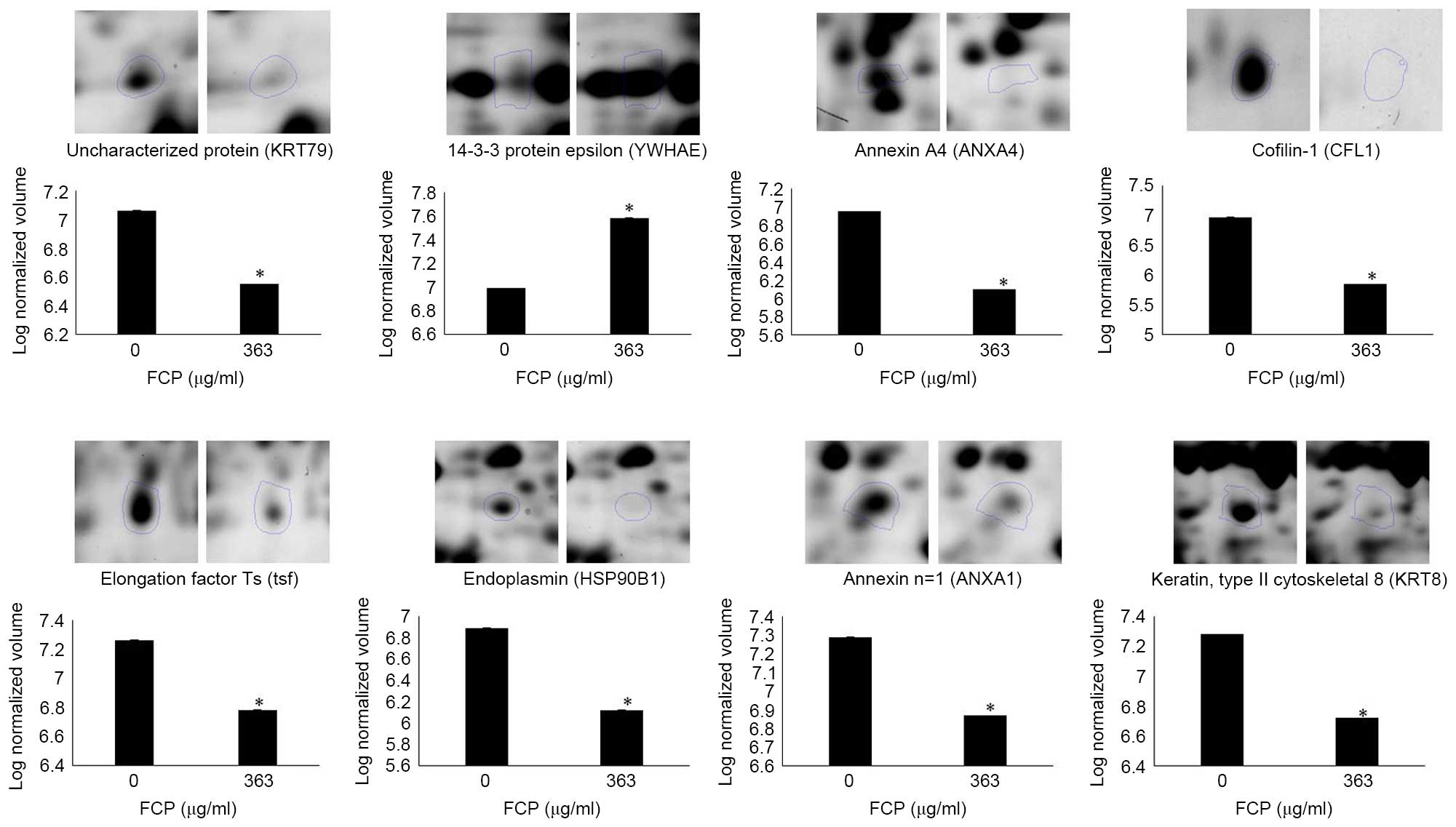
control cells (enlarged 2-DE images in Fig. 3). The annotation of all identified
proteins with their corresponding Swissprot accession number,
experimental and theoretical molecular weight, experimental and
theoretical isoelectric point, sequence coverage and number of
peptide matches, Mascot score, expression and statistical values
are shown in Table I. Based on
protein functions, the identified proteins were divided into the
following categories: Cytoskeletal proteins (CFL1, KRT8 and KRT79),
signal transduction (YWHAE, ANXA1 and ANXA4), molecular
chaperons/heat shock proteins (HSP90B1) and protein metabolism
(tsf). The identified proteins were predominantly involved in tumor
growth, cell cycle, apoptosis, migration and signal
transduction.
 | Table IList of differentially expressed
proteins in A549 cells treated with flavonoids isolated from
Citrus platymamma, identified using MALDI-TOF/TOF-MS
analysis. |
Table I
List of differentially expressed
proteins in A549 cells treated with flavonoids isolated from
Citrus platymamma, identified using MALDI-TOF/TOF-MS
analysis.
| Spot no. | Accession
numbera | Protein name (gene
name)a | Swiss-Prot entry
namea | Theoreticala/experimentalc Mr (kDa) | Theoreticala/experimentalc pI value | Sequence coverage
(%)/peptides matched | Mascot
scoreb | Fold changec | P-value
(t-testc) |
|---|
| 1 | F6XWG3 | Uncharacterized
protein (KRT79) | F6XWG3_HORSE | 58.10/32 | 7.62/6.22 | 27/16 | 90 | 3.3↓ | 0.010 |
| 2 | P62258 | 14-3-3 protein
epsilon (YWHAE) | 1433E_HUMAN | 26.91/37 | 4.92/4.48 | 44/13 | 141 | 3.8↑ | 0.015 |
| 3 | P09525 | Annexin A4
(ANXA4) | ANXA4_HUMAN | 36.09/35 | 5.84/5.93 | 52/32 | 342 | 7.4↓ | 0.021 |
| 4 | S7MQH4 | Cofilin-1
(CFL1) | S7MQH4_MYOBR | 18.73/16 | 8.22/6.54 | 63/11 | 105 | 14.3↓ | 0.025 |
| 5 | G8XJK6 | Elongation factor
Ts (tsf) | G8XJK6_MYCHR | 33.24/44 | 5.98/6.48 | 50/17 | 122 | 3.0↓ | 0.027 |
| 6 | P08712 | Endoplasmin
(HSP90B1) | ENPL_MESAU | 46.88/54 | 4.96/4.9 | 30/16 | 118 | 6.1↓ | 0.029 |
| 7 | G1QL21 | Annexin n=1
(ANXA1) | G1QL21_NOMLE | 38.89/29 | 6.57/5.28 | 30/13 | 97 | 2.7↓ | 0.036 |
| 8 | P05787 | Keratin, type II
cytoskeletal 8 (KRT8) | K2C8_HUMAN | 53.67/79 | 5.52/5.58 | 43/29 | 430 | 3.7↓ | 0.046 |
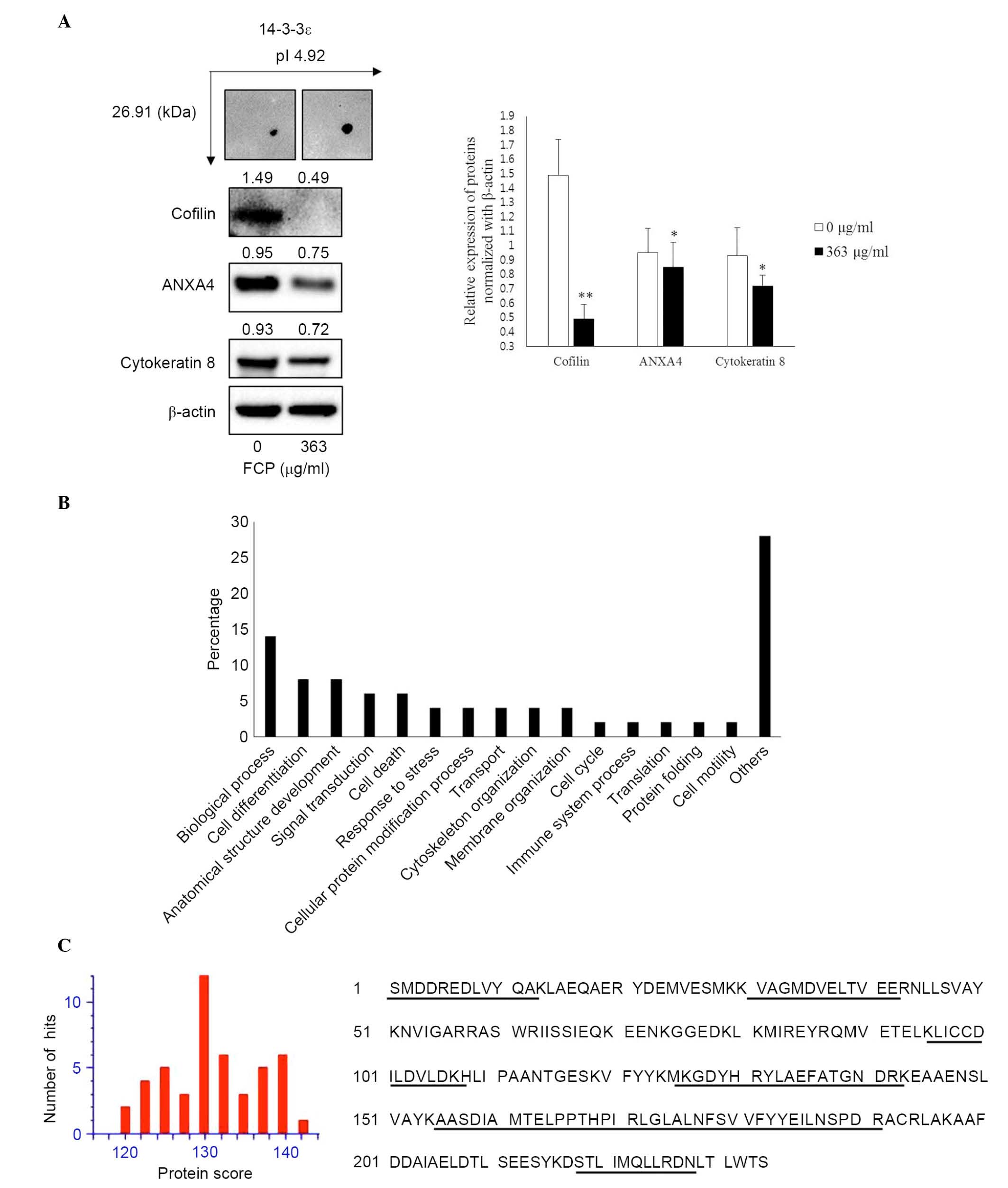
Validation of differential expressed
proteins using western blot analysis
The immunoblotting was performed to confirm the
expression of proteins, which were identified in the FCP-treated
A549 cells using proteome analysis. The results showed that YWHAE
was significantly upregulated, whereas CFL1, ANXA4 and KRT8 were
significantly downregulated in the FCP-treated A549 cells, compared
with the control (P<0.05; Fig.
4A). These findings suggested that the results of the
immunoblotting were consistent with those of the comparative
proteomic analysis.
GO analysis
The GO terms for biological processes were examined
for all eight identified proteins. The most notable functional
categories in terms of the protein expression pattern are shown in
Fig. 4B. The highest associations
were with biological processes (14%; GO:0008150). Another 8% of the
associations were with cell differentiation (GO:0030154) and
anatomical structure development (GO:0048856), whereas 6% were
associated with signal transduction (GO:0,007165) and cell death
(GO:0008219). Of the remainder, 4% were associated with response to
stress (GO:0006950), cellular protein modification process
(GO:0006464), cytoskeleton organization (GO:0007010), membrane
organization (GO:0061024) and transport (GO:0006810).
Discussion
The present study focused on the differentially
expressed proteins, which are involved in the behaviors of
FCP-treated A549 cells using proteome techniques. Of 15
differentially expressed protein spots, eight proteins were
successfully identified in the FCP-treated A549 cells using 2-DE
coupled with MALDI-TOF/TOF-MS analyses (Fig. 2 and Table I). The identified proteins were
predominantly involved in tumor growth and progression, and the
apoptosis of A549 cells. These results indicated that FCP inhibited
cell proliferation and induced cell death of the A549 cells by
regulating those proteins. The results of the immuneoblotting
confirmed that the expression of YWHAE was significantly
upregulated, and the expression levels of CFL8, ANXA4 and KRT8 were
significantly downregulated in the A549 cells following incubation
with FCP (363 µg/ml) for 24 h. These data suggested that FCP
inhibited the growth of A549 cells by altering the expression of
proteins, which are involved in tumor growth and progression. This
finding is consistent with those of the previous study,
demonstrating that FCP induces G2/M cell cycle arrest and apoptosis
in A549 lung cancer cells.
The 14-3-3 proteins are a highly conserved protein
family in eukaryotic cells, and comprise seven isoforms (β, ε, γ,
η, σ, τ/θ and ζ), which are crucial for regulating multiple
cellular processes, including signal transduction, cell cycle
regulation, apoptosis DNA repair, cytoskeletal regulation, cellular
metabolism, proliferation, transcription, and redox-regulation or
the stress response (30,31). Among the 14-3-3 isoforms, the
overexpression of YWHAE has been demonstrated in various types of
human malignancy, including lung cancer (32,33).
In addition, the reduced expression of YWHAE in gastric cancer is
associated with gastric carcinogenesis (34). In the present study, the expression
of YWHAE (spot no. 2) was significantly increased in the
FCP-treated A549 cells (Fig. 3).
Consistent with the 2-DE results, the expression of YWHAE was
further confirmed by immunoblotting analysis (Fig. 4A). Figure. 4C shows the protein scores for
the top hits for YWHAE when MSDB was searched with PMF and matched
peptides with 44% coverage. However, the role of the YWHAE protein
in apoptosis remains controversial; another study showed that
non-steroidal anti-inflammatory drugs induce apoptosis by the
suppression of 14-3-3ε YWHAE in colorectal cancer cells (35). Therefore, further detailed studies
are needed regarding the role of the YWHAE protein in the
anticancer effects of FCP on A549 cells.
The annexins, a family of phospholipid-binding
proteins, involved in various physiological processes, including
anticoagulation, anti-inflammatory, endocytosis and exocytosis,
signal transduction, cell proliferation, differentiation and
apoptosis (36,37). ANXA1 is a calcium-dependent
phospholipid-linked protein, differentially expressed in different
types of cancer (38). The
upregulation of ANXA1 in patients with lung cancer is associated
with a poor clinical outcome (39,40).
In addition, Biaoxue et al (41) demonstrated that the
co-overexpression of Hsp90-β and ANXA1 was associated with poor
survival rates and lymphatic metastasis in patients with lung
cancer patients. The increased expression of ANXA4 is associated
with drug resistance to paclitaxel, a drug commonly used for the
treatment of cancer (42). In
addition, the elevated expression of ANXA4 is associated with
advanced T stages in colorectal cancer and lymph node metastasis in
human penile squamous cell carcinoma (43,44).
In the present study, the expression levels of ANXA1, ANXA4 and
HSP90B1 (spot nos. 7, 3 and 6, respectively) were significantly
downregulated in FCP-treated A549 cells (Figs. 3 and 4A). These results indicated that FCP
exerted anticancer effects in A549 cells by suppressing the ANXA1,
ANXA4 and HSP90B1 proteins.
In the present study, the expression of CFL1 (spot
no. 4) was significantly downregulated in the FCP-treated A549
cells (Figs. 3 and 4A). CFL1, the actin regulatory protein,
is important in tumor growth and progression (45). It has been reported that CFL1 is
involved in tumor progression in ovarian carcinoma, almost 64% of
all ovarian tumors are positive for CFL1 (46). In prostate cancer, knockdown of
CFL1 was reported to increase sensitivity to docetaxel, a
chemotherapeutic agent (47). In
addition, the expression of KRT8 (spot no. 8) was also
downregulated in FCP-treated A549 cells (Figs. 3 and 4A). The increased expression of KRT8 was
significantly associated with tumor progression, and decreased
survival rates in patients with NSCLC (48). These data suggested that the
downregulation of CFL1 and KRT8 may also be involved in the
anticancer effect of FCP on A549 cells.
In conclusion, the present study demonstrated the
anticancer effects of FCP on A549 human lung cancer cells using a
proteomic approach. In the present study, eight differentially
expressed proteins (YWHAE was upregulated; CFL1, ANXA1, ANXA4,
HSP90B1, KRT8, Tsf and KRT79 were downregulated) were identified in
the FCP-treated A549 cells, which were found to be involved in
tumor growth, cell cycle, apoptosis, migration and signal
transduction (Fig. 5).
Furthermore, the expression levels of YWHAE, CLF1, ANXA4 and KTR8
were validated by immunoblotting. To the best of our knowledge, the
present study was the first to use the proteomic technique to
investigate the molecular mechanism in FCP- treated A549 cells. The
findings of the present study improve understanding of the
molecular mechanism underlying the selective growth inhibition of
FCP on A549 cells, which may offer a therapeutic potential for the
treatment of lung cancer.
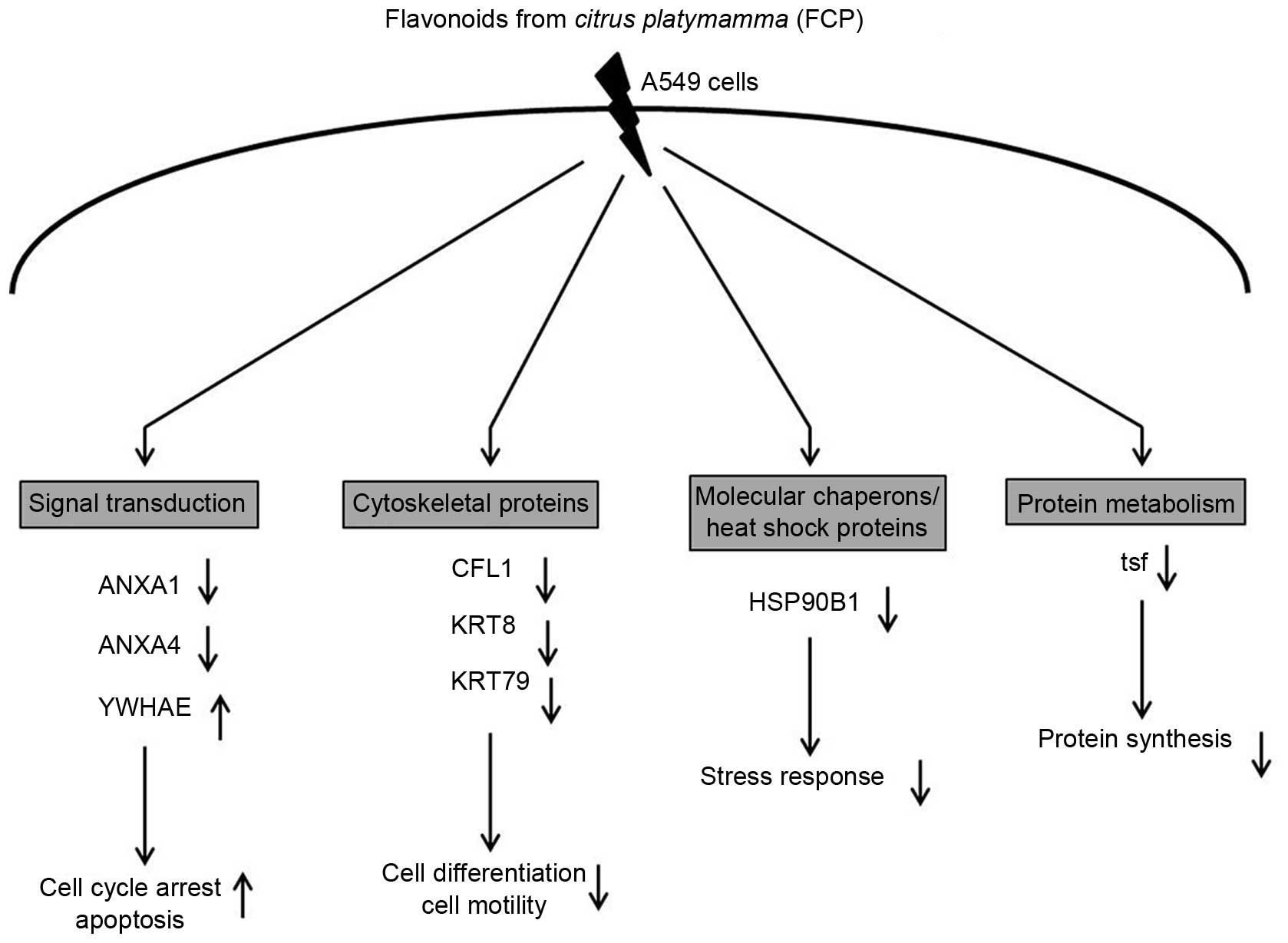 | Figure 5Biological role of eight
differentially expressed proteins identified in FCP-treated A549
cells using two-dimensional gel electrophoresis coupled with
matrix-assisted laser desorption/ionization
time-of-flight/time-of-flight tandem mass spectrometry analysis.
Specifically, proteins involved in signal transduction were
significantly downregulated, including ANXA1 and ANXA4, whereas
YWHAE was upregulated. Cytoskeletal proteins, including CFL1, KRT8
and KRT79, and molecular chaperones/heat shock proteins, including
HSP90B1, were downregulated. Proteins involved in protein
metabolism, namely tsf, were also downregulated. The majority of
these proteins were involved in tumor growth and progression, cell
cycle, stress response and apoptosis. (↑ indicates upregulation of
protein and ↓ indicates downregulation of protein). ANXA, annexin
A; YWHAE, 14-3-3ε; CFL1, cofilin-1; KRT8, cyroskeratin 8; KRT79,
uncharacterized protein; HSP90B1; endoplasmin; tsf, elongation
factor Ts. |
Acknowledgments
This study was supported by a grant from the
National Research Foundation of Korea funded by the Ministry of
Science, ICT & Future Planning (grant nos. 2012M3A9B8019303 and
2012R1A2A2A06045015) and the National R&D Program for Cancer
Control, Ministry for Health, Welfare and Family Affairs, Republic
of Korea (grant no. 0820050).
References
|
1
|
Meoni G, Cecere FL, Lucherini E and Di
Costanzo F: Medical treatment of advanced non-small cell lung
cancer in elderly patients: A review of the role of chemotherapy
and targeted agents. J Geriatr Oncol. 4:282–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG
and Lee JS: Prediction of cancer incidence and mortality in Korea,
2013. Cancer Res Treat. 45:15–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jeong JW, Lee WS, Go SI, Nagappan A, Baek
JY, Lee JD, Lee SJ, Park C, Kim GY, Kim HJ, et al: Pachymic acid
induces apoptosis of EJ bladder cancer cells by DR5 up-regulation,
ROS generation, modulation of Bcl-2 and IAP family members.
Phytother Res. 29:1516–1524. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park KI, Park HS, Nagappan A, Hong GE, Lee
do H, Kang SR, Kim JA, Zhang J, Kim EH, Lee WS, et al: Induction of
the cell cycle arrest and apoptosis by flavonoids isolated from
Korean Citrus aurantium L. in non-small-cell lung cancer cells.
Food Chem. 135:2728–2735. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hong GE, Kim JA, Nagappan A, Yumnam S, Lee
HJ, Kim EH, Lee WS, Shin SC, Park HS and Kim GS: Flavonoids
identified from Korean Scutellaria baicalensis Georgi inhibit
inflammatory signaling by suppressing activation of NF-κB and MAPK
in RAW 264.7 cells. Evid Based Complement Alternat Med.
2013:9120312013. View Article : Google Scholar
|
|
6
|
Hatcher H, Planalp R, Cho J, Torti FM and
Torti SV: Curcumin: From ancient medicine to current clinical
trials. Cell Mol Life Sci. 65:1631–1652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee DH, Park KI, Park HS, Kang SR,
Nagappan A, Kim JA, Kim EH, Lee WS, Hah YS, Chung HJ, et al:
Flavonoids isolated from Korea Citrus aurantium L. induce G2/M
phase arrest and apoptosis in human gastric cancer AGS cells. Evid
Based Complement Alternat Med. 2012:5159012012. View Article : Google Scholar
|
|
8
|
Yumnam S, Park HS, Kim MK, Nagappan A,
Hong GE, Lee HJ, Lee WS, Kim EH, Cho JH, Shin SC and Kim GS:
Hesperidin induces paraptosis like cell death in hepatoblastoma,
HepG2 Cells: involvement of ERK1/2 MAPK [corrected]. PLoS One.
9:e1013212014. View Article : Google Scholar
|
|
9
|
Han MH, Lee WS, Lu JN, Lee WS, Lu JN, Kim
G, Jung JM, Ryu CH, Kim GY, Hwang HJ, Kwon TK and Choi YH: Citrus
aurantium L. exhibits apoptotic effects on U937 human leukemia
cells partly through inhibition of Akt. Int J Oncol. 40:2090–2096.
2012.PubMed/NCBI
|
|
10
|
Lee HJ, Nagappan A, Park HS, Hong GE,
Yumnam S, Raha S, Saralamma VV, Lee WS, Kim EH and Kim GS:
Flavonoids isolated from Citrus platymamma induce
mitochondrial-dependent apoptosis in AGS cells by modulation of the
PI3K/AKT and MAPK pathways. Oncol Rep. 34:1517–1525.
2015.PubMed/NCBI
|
|
11
|
Nogata Y, Sakamoto K, Shiratsuchi H, Ishii
T, Yano M and Ohta H: Flavonoid composition of fruit tissues of
citrus species. Biosci Biotechnol Biochem. 70:178–192. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benavente-García O and Castillo J: Update
on uses and properties of citrus flavonoids: New findings in
anticancer, cardiovascular and anti-inflammatory activity. J Agric
Food Chem. 56:6185–6205. 2008. View Article : Google Scholar
|
|
13
|
Luo G, Guan X and Zhou L: Apoptotic effect
of citrus fruit extract nobiletin on lung cancer cell line A549 in
vitro and in vivo. Cancer Biol Ther. 7:966–973. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai CS, Li S, Miyauchi Y, Suzawa M, Ho CT
and Pan MH: Potent anti-cancer effects of citrus peel flavonoids in
human prostate xenograft tumors. Food Funct. 4:944–949. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan Y, Li W, Li L, Yang X, Gu R, Liu H,
Huang K and Yu Y: Effects of tetrazanbigen on the protein
expression in human hepatocellular carcinoma cell line QGY-7701. J
Huazhong Univ Sci Technolog Med Sci. 29:304–308. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu CI, Chen CC, Chen JC, Su JH, Huang HH,
Chen JY and Wu YJ: Proteomic analysis of anti-tumor effects of
11-dehydrosinulariolide on CAL-27 cells. Mar Drugs. 9:1254–1272.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng YL, Zhang GY, Li C and Lin J:
Screening for novel protein targets of indomethacin in HCT116 human
colon cancer cells using proteomics. Oncol Lett. 6:1222–1228.
2013.PubMed/NCBI
|
|
18
|
Li X, Wang Z, Liu J, Tang C, Duan C and Li
C: Proteomic analysis of differentially expressed proteins in
normal human thyroid cells transfected with PPFP. Endocr Relat
Cancer. 19:681–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagappan A, Park HS, Park KI, Hong GE,
Yumnam S, Lee HJ, Kim MK, Kim EH, Lee WS, Lee WJ, et al:
Helicobacter pylori infection combined with DENA revealed altered
expression of p53 and 14-3-3 isoforms in Gulo−/−mice. Chem Biol
Interact. 206:143–152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen ZJ, Wang SY, Chen JA and Peng XX: The
influence of temperature on the protein expression of human lung
cancer cell line A549. Shi Yan Sheng Wu Xue Bao. 35:179–183.
2002.In Chinese.
|
|
21
|
Zhan XQ, Guan YJ, Li C, Chen ZC, Xie JY,
Chen P and Liang SP: Differential proteomic analysis of human lung
adenocarcinoma cell line A-549 and of normal cell line HBE. Sheng
Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 34:50–56. 2002.In
Chinese.
|
|
22
|
Li MY, Xiao ZQ, Li C, Wu XY, Feng XP, Yi
H, Li JL, Chen ZC, Chen P and Liang SP: Establishment of protein
profile of human small cell lung cancer cell line NCI-H446. Ai
Zheng. 23:1116–1121. 2004.In Chinese. PubMed/NCBI
|
|
23
|
Wu H, Pan CL, Yao YC, Chang SS, Li SL and
Wu TF: Proteomic analysis of the effect of Antrodia camphorata
extract on human lung cancer A549 cell. Proteomics. 6:826–835.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Q, Xu W, Cao L, Li X, He T, Wu Z and Li
W: SAHA treatment reveals the link between histone lysine
acetylation and proteome in nonsmall cell lung cancer A549 cells. J
Proteome Res. 12:4064–4073. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu Z, Song Q, Yang J, Zhao X, Zhang X,
Yang P and Kang J: Comparative proteomic analysis of anti-cancer
mechanism by periplocin treatment in lung cancer cells. Cell
Physiol Biochem. 33:859–868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagappan A, Lee HJ, Saralamma VV, Park HS,
Hong GE, Yumnam S, Raha S, Charles SH, Shin SC, Kim EH, et al:
Flavonoids isolated from Citrus platymamma induced G2/M cell cycle
arrest and apoptosis of A549 human lung cancer cells. Oncol Lett.
In press.
|
|
27
|
Swain M and Ross NW: A silver stain
protocol for proteins yielding high resolution and transparent
background in sodium dodecyl sulfate-polyacrylamide gels.
Electrophoresis. 16:948–951. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McCarthy FM, Wang N, Magee GB, Nanduri B,
Lawrence ML, Camon EB, Barrell DG, Hill DP, Dolan ME, Williams WP,
et al: AgBase: A functional genomics resource for agriculture. BMC
Genomics. 7:2292006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aitken A: Post-translational modification
of 14-3-3 isoforms and regulation of cellular function. Semin Cell
Dev Biol. 22:673–680. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang X, Lee WH, Sobott F, Papagrigoriou E,
Robinson CV, Grossmann JG, Sundström M, Doyle DA and Elkins JM:
Structural basis for protein-protein interactions in the 14-3-3
protein family. Proc Natl Acad Sci USA. 103:17237–17242. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu TA, Jan YJ, Ko BS, Liang SM, Chen SC,
Wang J, Hsu C, Wu YM and Liou JY: 14-3-3ε overexpression
contributes to epithelial-mesenchymal transition of hepatocellular
carcinoma. PLoS One. 8:e579682013. View Article : Google Scholar
|
|
33
|
Qi W, Liu X, Qiao D and Martinez JD:
Isoform-specific expression of 14-3-3 proteins in human lung cancer
tissues. Int J Cancer. 113:359–363. 2005. View Article : Google Scholar
|
|
34
|
Leal MF, Calcagno DQ, Demachki S,
Assumpção PP, Chammas R, Burbano RR and Smith Mde A: Clinical
implication of 14-3-3 epsilon expression in gastric cancer. World J
Gastroenterol. 18:1531–1537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liou JY, Ghelani D, Yeh S and Wu KK:
Nonsteroidal anti-inflammatory drugs induce colorectal cancer cell
apoptosis by suppressing 14-3-3epsilon. Cancer Res. 67:3185–3191.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Raynal P and Pollard HB: Annexins: The
problem of assessing the biological role for a gene family of
multifunctional calcium- and phospholipid-binding proteins. Biochim
Biophys Acta. 1197:63–93. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang X, Liu S, Guo C, Zong J and Sun MZ:
The association of annexin A2 and cancers. Clin Transl Oncol.
14:634–640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lim LH and Pervaiz S: Annexin 1: The new
face of an old molecule. FASEB J. 21:968–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rong B, Zhao C, Liu H, Ming Z, Cai X, Gao
W and Yang S: Elevated serum annexin A1 as potential diagnostic
marker for lung cancer: A retrospective case-control study. Am J
Transl Res. 6:558–569. 2014.PubMed/NCBI
|
|
40
|
Biaoxue R, Xiguang C and Shuanying Y:
Annexin A1 in malignant tumors: Current opinions and controversies.
Int J Biol Markers. 29:e8–20. 2014. View Article : Google Scholar
|
|
41
|
Biaoxue R, Shuanying Y, Wei L, Zongjuan M,
Xiguang C and Qiuhong Z: Co-overexpression of Hsp90-β and annexin
A1 with a significantly positive correlation contributes to the
diagnosis of lung cancer. Expert Rev Mol Diagn. 14:1067–1079. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han EK, Tahir SK, Cherian SP, Collins N
and Ng SC: Modulation of paclitaxel resistance by annexin IV in
human cancer cell lines. Br J Cancer. 83:83–88. 2000.PubMed/NCBI
|
|
43
|
Duncan R, Carpenter B, Main LC, Telfer C
and Murray GI: Characterisation and protein expression profiling of
annexins in colorectal cancer. Br J Cancer. 98:426–433. 2008.
View Article : Google Scholar
|
|
44
|
Zimmermann U, Balabanov S, Giebel J,
Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B and
Walther R: Increased expression and altered location of annexin IV
in renal clear cell carcinoma: A possible role in tumour
dissemination. Cancer Lett. 209:111–118. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kapoor S: Cofilin-1 overexpression and its
role in tumor growth and progression in systemic malignancies. Int
J Radiat Biol. 90:1132014. View Article : Google Scholar
|
|
46
|
Zhou J, Wang Y, Fei J and Zhang W:
Expression of cofilin 1 is positively correlated with the
differentiation of human epithelial ovarian cancer. Oncol Lett.
4:1187–1190. 2012.PubMed/NCBI
|
|
47
|
Pérez-Martínez FC, Carrión B, Lucío MI,
Rubio N, Herrero MA, Vázquez E and Ceña V: Enhanced
docetaxel-mediated cytotoxicity in human prostate cancer cells
through knockdown of cofilin-1 by carbon nanohorn delivered siRNA.
Biomaterials. 33:8152–8159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fukunaga Y, Bandoh S, Fujita J, Yang Y,
Ueda Y, Hojo S, Dohmoto K, Tojo Y, Takahara J and Ishida T:
Expression of cytokeratin 8 in lung cancer cell lines and
measurement of serum cytokeratin 8 in lung cancer patients. Lung
Cancer. 38:31–38. 2002. View Article : Google Scholar : PubMed/NCBI
|