Introduction
Prostate cancer is the second most frequently
diagnosed cancer in males worldwide, and is the most frequently
diagnosed cancer among males in more developed countries (1). Despite improvements in the clinical
management and treatment of prostate cancer, the five-year relative
survival rate for patients with advanced prostate cancer is ~30%.
It is estimated that 220,800 men in the USA developed prostate
cancer during 2015, and 27,540 men (12.5%) were predicted to
succumb to this malignancy (2).
Novel therapies are required for the effective treatment of
prostate cancer, and biomarkers based on the molecular biology of
the disease are required for early diagnoses.
Aberrant expression or localization of β-catenin has
been frequently observed in prostate tumor cells (3), thus suggesting that activation of the
Wnt/β-catenin signaling pathway may be involved in the development
of prostate cancer. The Wnt/β-catenin pathway regulates cell fate
during embryonic development, and its activation is involved in the
development of cancer (4–7). In the absence of Wnt signaling,
cytoplasmic β-catenin levels are reduced due to destruction
complex-induced degradation. The destruction complex consists of
axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3β
(GSK3β) and casein kinase 1. Alterations that interfere with
β-catenin degradation lead to the accumulation of β-catenin in the
cytoplasm and subsequent translocation to the nucleus. In
conjunction with the T-cell factor (TCF)/lymphoid enhancing factor
transcription factors, β-catenin then induces cellular responses
through the transcriptional activation of target genes.
Despite previous reports demonstrating that the
Wnt/β-catenin pathway is activated in prostate cancer (3), mutations in APC, β-catenin
(CTNNB1) and axin 1 (AXIN1), are rarely detected (The
Cancer Genome Atlas Network; http://cancergenome.nih.gov/). Therefore, additional
mechanisms may be responsible for the upregulation of β-catenin in
prostate cancer. Frequently rearranged in advanced T-cell
lymphomas-1 (FRAT1) overexpression activates the Wnt/β-catenin
pathway by binding to GSK3β. Expression of FRAT1 is elevated in
several types of human cancer, including ovarian cancer (8), esophageal cancer (9), glioma (10–12),
astrocytoma (13), lung (14,15)
and pancreatic cancer (16). A
recent report demonstrated that N-myc downstream-regulated gene 1
regulates β-catenin phosphorylation and nuclear trans-location
through FRAT1 in prostate cancer cells (17), which indicates a potential
association between FRAT1 and aberrant Wnt/β-catenin signaling in
prostate cancer. Therefore, the aim of the present study was to
investigate the role of FRAT1 in prostate cancer.
Materials and methods
Prostate tissue microarray
A prostate tissue microarray (reference no.
HProA100PG01), consisting of three normal prostate tissue samples
and 59 prostate adenocarcinoma tissue samples, was obtained from
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China).
Cell culture, plasmid construction and
transfection
The PC-3 and DU-145 human prostate cancer cell
lines, and the NIH3T3 mouse embryonic fibroblast cell line were
obtained from the American Type Culture Collection (Manassas, VA,
USA). Cells were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), penicillin (50 U/ml) and streptomycin (50
μg/ml). Cell lines were maintained in a humidified incubator
at 37°C and 5% CO2. Culture medium was refreshed every 3
days and cells were subcultured when confluent.
The FRAT 1 overexpression plasmid (HA-FRAT1/pCEFL),
FRAT1 short hairpin (sh) RNA knockdown plasmid
(pSilencer-FRAT1), and control shRNA plasmid
(pSilencer-control) were generated as described previously
(9). TCF reporter plasmids,
TOPFLASH and FOPFLASH were obtained from Upstate Biotechnology,
Inc. (Lake Placid, NY, USA), and the pRL-TK wild-type control
vector was purchased from Promega Corporation (Madison, WI,
USA).
Transfections were performed in 24-well or 6-well
plates. DU-145 or PC-3 cells (2×105) were seeded into
each well of a 6-well tissue culture plate and were incubated for
24 h. When the cells were 70–80% confluent, the culture media was
removed and cells were washed with pre-warmed sterile
phosphate-buffered saline. Lipofectamine 2000 Transfection Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
cells with the plasmids according to the manufacturer's protocol.
Cells were collected 24 h post-transfection and western blot
analysis and reporter assays were performed. To establish stable
HA-FRAT1/pCEFL or pCEFL empty vector control cell lines,
post-transfection, the medium was supplemented with 400
μg/ml G418 and resistant clones were pooled and maintained
in culture. Exogenous hemagglutinin (HA)-tagged FRAT1 expression
was confirmed by western blot analysis.
Western blot analysis
Total protein was extracted from cultured cells
using urea buffer (8 M urea, 1 M thiourea, 0.5% CHAPS, 50 mM
dithiothreitol and 24 mM spermine). The protein concentration was
determined using the Bradford Protein Assay Dye Reagent (cat. no.
500-0006; Bio-Rad Laboratories Inc., Hercules, CA, USA). The
samples (40 μg of protein) were then separated by 12% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and were
transferred to nitrocellulose membranes. The membranes were blocked
with 5% non-fat milk in Tris-buffered saline-Tween 20 at room
temperature for 1 h, and then incubated with primary antibodies
overnight at 4°C. The membranes were subsequently incubated for 1 h
at room temperature with horseradish peroxidase (HRP)-conjugated
secondary antibodies. Signals were detected using an enhanced
chemiluminescence detection system (GE Healthcare Life Sciences,
Chalfont, UK) and evaluated using ImageJ software (version, 1.42q;
National Institutes of Health, Bethesda, MD, USA) (18). The following primary antibodies
were used: Anti-HA (cat. no. sc-805; dilution, 1:1,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-FRAT1 (cat. no.
ab108405; dilution, 1:1,000; Abcam, Cambridge, UK), anti-β-catenin
(cat. no. 610153; 1:2,000; BD Biosciences, Franklin Lakes, NJ,
USA), anti- phosphorylated (p)- β-catenin (Ser33/37/Thr41; cat. no.
9561; dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers
MA, USA), anti-β-actin (cat. no. A3854; dilution, 1:5,000;
Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and
anti-β-tubulin (cat. no. ab6046; dilution, 1:5,000, Abcam, MA,
USA). The following HRP-conjugated secondary antibodies were used:
Goat anti-rabbit immunoglobulin G (IgG) H&L (cat. no. ab97051;
dilution, 1:1,000; Abcam), goat anti-mouse IgG H&L (cat. no.
ab6789; dilution, 1:1,000; Abcam).
Cell growth assay
Cells that were stably or transiently transfected
were harvested and re-plated in triplicate in 24-well plates. Cells
were counted every 2 days with a hemocytometer, and a Trypan blue
exclusion assay was used to assess cell viability. In brief, 0.1 ml
Trypan blue stock solution was mixed with 0.1 ml cell suspension
before loading onto a hemocytometer. Cells were counted immediately
under a light microscope at low magnification.
Immunohistochemical analysis
The prostate tissue micro-array was deparaffinized,
rehydrated and incubated with 3% hydrogen peroxide for 10 min to
block endogenous peroxidase activity. The slide was subsequently
blocked with normal goat serum at room temperature for 1 h, and
then incubated at 4°C overnight with a primary antibody against
FRAT1 (cat. no. ab137391; dilution, 1:400; Abcam, Cambridge, UK).
The slide was then incubated with a biotinylated goat anti-rabbit
IgG H&L secondary antibody (dilution, 1:250; cat. no. BP-9100,
Vector Laboratories, Inc., Burlingame, CA, USA) for 40 min at room
temperature, and subsequently stained with the VECTASTAIN Elite ABC
HRP kit (Vector Laboratories, Inc.) followed by counterstaining
with hematoxylin. The localization of FRAT1 in each sample spot
within the tissue microarray slide was categorized as membranous,
cytoplasmic or nuclear. The staining intensity was graded as
follows: No staining (0), weak (1), moderate (2) and intense staining (3). Samples that could not be graded were
scored as 'not applicable'. For each sample, FRAT1 expression was
determined as the average score of the sample spot, and then
further subgrouped into low (score 0) or high (scores 1–3) FRAT1
expression.
Reporter assays
Following transfection with either TOPFLASH (100 ng)
or FOPFLASH (100 ng), the internal control plasmid pRL-TK (5 ng)
and HA-FRAT1/pCEFL, pCEFL, pSilencer-FRAT1 or
pSilencer-control-transfected cells were cultured for 36 h
at 37°C before they were lysed using Passive Lysis Buffer (cat. no.
E1941; Promega Corporation) to measure the luciferase reporter
activity using the Dual-Luciferase Reporter Assay system (Promega
Corporation) according to the manufacturer's instructions. Firefly
luciferase activity was normalized to Renilla luciferase
activity. Data are presented as the mean ± standard deviation (SD)
for independent triplicate cultures.
Tumor growth in nude mice
NIH3T3/control or NIH3T3/FRAT1 (4×106)
cells were prepared in 0.2 ml saline, and were injected bilaterally
and subcutaneously, each into the left and right forelegs of four
female nude mice (age, 4–6 weeks; weight, 16–20 g; Beijing HFK
Bioscience Co., Ltd.). Prior to tumor cell implantation, mice were
allowed to acclimatize to laboratory conditions for 3 days. The
mice were housed in a pathogen-free environment and monitored every
2 days. Animals had free access to standard food and water, and
were maintained in 12 h light/dark cycles throughout the course of
treatment. At the end of the experiment, the mice were sacrificed
by cervical dislocation. The date when a palpable tumor first arose
and the weight of the removed tumor were recorded. The mice were
treated in accordance with the Regulations of Laboratory Animal
Quality issued by the Chinese Ministry of Science and Technology
(Beijing, China). Animal experiments were approved by the
Institutional Animal Care and Use Committee of Cancer Hospital,
Chinese Academy of Medical Sciences (reference no.
NCC2015A019).
Statistical analysis
Data are presented as the mean ± SD. A two-tailed,
unpaired Student's t-test was used to compare independent samples
from two groups. Data were analyzed using the SPSS software program
(version 16.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
FRAT1 is expressed exclusively in the
nuclei of normal prostate basal cells and is overexpressed in human
prostate cancer
FRAT1 mRNA expression in established human
cell lines was first investigated using the Human Protein Atlas
database (http://www.proteinatlas.org/ENSG00000165879-FRAT1/cell).
Notably, FRAT1 mRNA expression levels in PC-3 prostate
cancer cells were observed to be among the highest across all of
the cell lines included in the analysis (Fig. 1A).
In order to explore the clinical implications of
FRAT1 expression in prostate cancer, data from The Cancer
Genome Atlas cBioPortal database (http://www.cbioportal.org/) were analyzed (19–21).
As shown in Fig. 1B, upregulation
of FRAT1 mRNA expression levels was frequent in patients
with prostate adenocarcinoma (41/216, 19%; Memorial Sloan Kettering
Cancer Center; http://www.cbioportal.org/study?id=prad_mskcc#summary).
The protein expression of FRAT1 in normal human
prostate tissue and prostate adenocarcinoma tissues was analyzed by
immunohistochemical analysis using a human prostate cancer tissue
microarray. Expression of FRAT1 was observed in all three cases of
normal prostate epithelium, exclusively in the nuclei of basal
cells (Fig. 2A). These results are
consistent with the in situ hybridization results of a
previous study, demonstrating that FRAT1 protein expression was
present in all samples of normal esophageal squamous cell
epithelium and in the basal layers (9). In the present study, nuclear FRAT1
expression was detected in 68% (40/59) of prostate adenocarcinoma
samples (Fig. 2B). Since only a
small fraction of cells (basal cells) in the normal prostate tissue
samples were observed to express FRAT1, this protein was determined
to be overexpressed in prostate adenocarcinoma tissues.
FRAT1 expression status affects prostate
cancer cell growth
The next aim of the present study was to investigate
whether the expression status of FRAT1 influences the growth of
human prostate cancer cell lines. As determined using the Catalogue
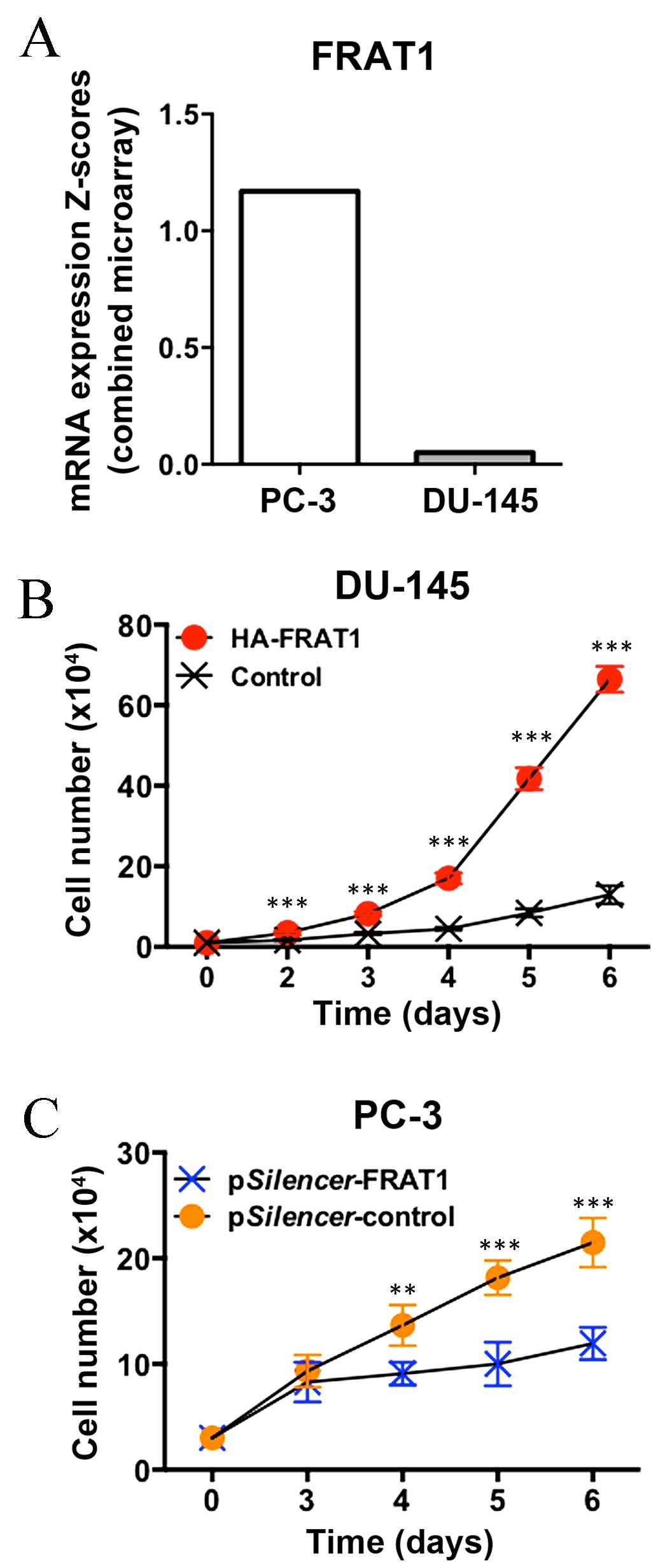
of Somatic Mutations in Cancer database (http://cancer.sanger.ac.uk/cosmic), FRAT1 mRNA
expression levels in PC-3 cells were markedly higher when compared
with DU-145 cells (Fig. 3A).
Forced overexpression of FRAT1 markedly promoted the
growth rate of DU-145 cells, with a 4-fold increase observed at day
6 (P<0.001; Fig. 3B).
Conversely, knockdown of FRAT1 expression by RNA interference
(RNAi) significantly inhibited the cell growth rate of PC-3 cells,
with a 30–50% decrease at days 4–6 (P=0.003 at day 4; P<0.001 at
days 5 and 6; Fig. 3C). These data
indicate that increased FRAT1 expression may provide a growth
advantage in prostate cancer cells.
FRAT1 activates β-catenin-dependent
transcriptional activity
It has previously been reported that FRAT1
positively regulates the activity of the Wnt/β-catenin signaling
pathway (17). In order to
investigate whether this process may also occur in prostate cancer
cells, the effect of FRAT1 expression on the transcriptional
activity of β-catenin was investigated using a TOPFLASH/FOPFLASH
reporter assay. This assay consists of wild type (TOP) or mutated
(FOP) binding sites for the β-catenin/TCF complex upstream of a
minimal thymidine kinase promoter and luciferase open reading
frame.
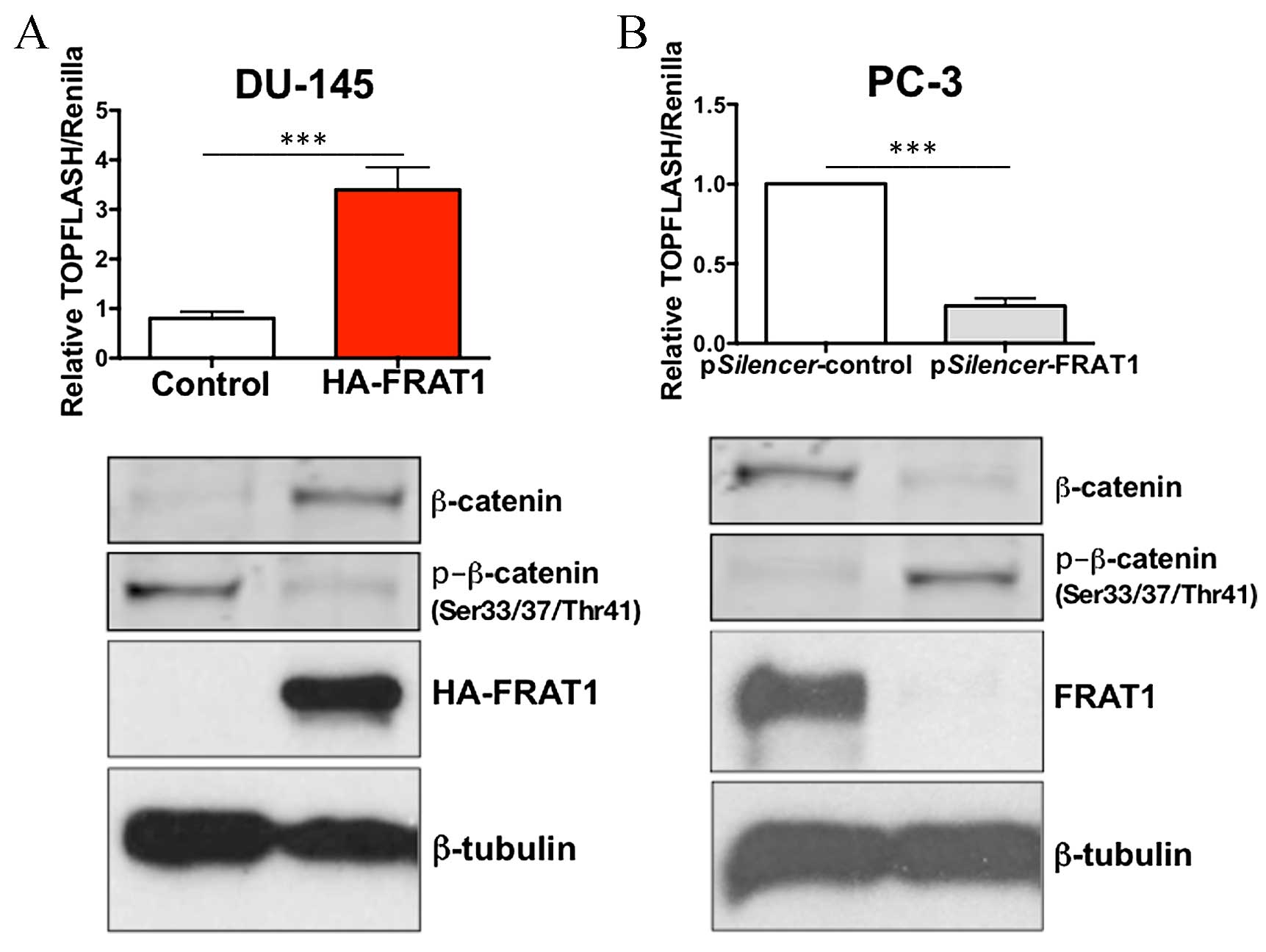
Overexpression of FRAT1 in DU-145 cells
significantly increased TOPFLASH activity, with a 3-fold increase
compared to the control cells (P<0.001; Fig. 4A). A minimal effect of FRAT1
overexpression on the FOPFLASH reporter was detected (data not
shown). In addition, knockdown of FRAT1 in PC-3 cells using RNAi,
markedly inhibited β-catenin/TCF-dependent transcriptional
activity, with a >70% reduction in TOPFLASH activity
(P<0.001; Fig. 4B).
Overexpression of FRAT1 in DU-145 cells inhibited
the expression of β-catenin phosphorylated at Ser33/37 and Thr41
residues, which provides an explanation for the observed increase
in total β-catenin levels (Fig.
4A). Conversely, depletion of FRAT1 in PC-3 cells resulted in
an increase in the protein expression levels of p-β-catenin (at
Ser33/37 and Thr41 residues), and decreased levels of total
β-catenin expression (Fig. 4B).
These data confirm that FRAT1 may function to stabilize β-catenin
in prostate cancer cells.
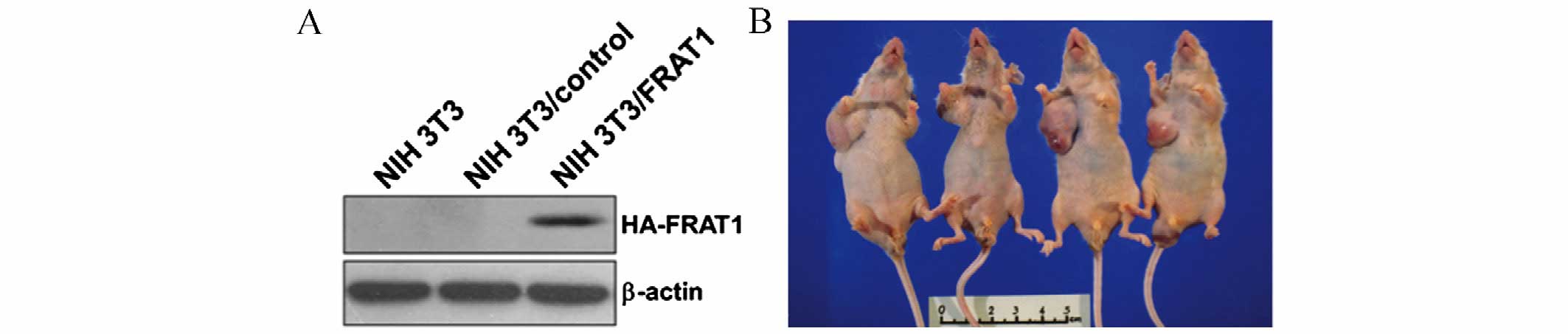
FRAT1 overexpression in NIH3T3 cells
induces tumor formation in nude mice
The in vitro results indicated that FRAT1 may
affect prostate cancer cell growth. In order to confirm the
tumorigenic potential of FRAT1 in vivo, tumor xenograft
experiments involving four nude mice were performed using NIH3T3
cells stably transfected with FRAT1 (NIH3T3/FRAT1) or control
vectors (NIH3T3/control; Fig. 5A).
At 8 weeks following injection with the transfected cells, the mice
were sacrificed and the tumors were removed and weighed. Tumors
developed in all four nude mice at the right lateral site where the
NIH3T3/FRAT1 cells were injected (Fig.
5B and Table I). Conversely,
no palpable tumors developed on the left lateral side where
NIH3T3/control cells were injected (Fig. 5B and Table I). At 3 weeks following injection
with NIH3T3/FRAT1, palpable nodular neoplasms developed and tumors
were observed at week 5. These data provide evidence to suggest
that FRAT1 serves a tumorigenic role in vivo, as its
overexpression in normal NIH3T3 mouse fibroblast cells led to the
formation of tumors in nude mice.
 | Table IFRAT1 overexpression in NIH3T3 cells
induces tumor formation in nude mice. |
Table I
FRAT1 overexpression in NIH3T3 cells
induces tumor formation in nude mice.
| No. of
cellsa | NIH3T3/FRAT1
| NIH3T3/control
|
|---|
Tumor
weight
(mg) | No.
tumors/injection | Latencyb
(days) | Tumor
weight
(mg) | No.
tumors/injection | Latencyb
(day) |
|---|
|
4×106 | 2105±409 | 4/4 | 21–35 | 0 | 0/4 | >56 |
Discussion
Prostate cancer is the fifth most common cause of
cancer-associated mortality worldwide in men, and is the eighth
most common cause of cancer-associated mortality overall (1). Molecular biomarkers and personalized
treatment strategies based on an improved understanding of the
molecular mechanisms underlying prostate cancer development and
progression would maximize therapeutic outcome (22). Despite previous reports
demonstrating that a large proportion of prostate tumor cells
exhibit aberrant expression or localization of β-catenin (3), the observation that CTNNB1,
APC and AXIN1 are rarely mutated in prostate cancer
suggests that genetic alterations may not be responsible for
activation of the Wnt/β-catenin pathway (3). The results of the present study
demonstrated that FRAT1 may contribute to the activation of the
β-catenin/TCF pathway in prostate cancer cells, due to the
observation that FRAT1 activated the β-catenin/TCF promoter
luciferase reporter gene in DU-145 prostate cancer cells. In
addition, FRAT1 was observed to affect the growth of prostate
cancer cells in vitro. Furthermore, forced expression of
FRAT1 in NIH3T3 cells was sufficient to induce cell transformation
and lead to tumor growth in vivo. These findings suggested
that FRAT1 exhibits oncogenic properties in prostate cancer.
FRAT1 is overexpressed in prostate cancer. In normal
prostate tissue, FRAT1 is expressed exclusively in the nuclei of
basal cells. At a histological level, the human prostate primarily
consists of epithelial and stromal cells. In the epithelial cell
layer, there are four differentiated cell types, including basal,
secretory luminal, neuroendocrine and transit-amplifying cells.
These cells display distinct morphologies, locations, functions and
expression markers. The basal cells, where adult prostate stem
cells are thought to reside, form a layer of flattened
cuboidal-shaped cells above the basement membrane (23). In 2014, Goksel et al
(24) reported that Wnt
pathway-associated genes, including FRAT1, were
significantly upregulated in the
CD133high/CD44high cancer stem cell (CSC)
monolayer group when compared with non-CSC counterparts. Future
studies will be required to confirm whether FRAT1 is a potential
marker for the stemness of prostate basal cells.
The human FRAT1 gene is a homologue of the
mouse proto-oncogene Frat1, which was demonstrated to convey
a selective advantage to cells at the later stages of murine T-cell
lymphomagenesis (25,26). FRAT1 functions as a GSK3-binding
protein, which is similar to the function of its Xenopus
homolog GSK3-binding protein (27). Ectopic expression of Frat1 in
Xenopus embryos induces secondary axis formation by
stabilizing β-catenin levels (28). Overexpression of Frat1 in
mouse cells induces β-catenin/TCF-dependent reporter gene activity
(29). In addition, Frat1
interacts with Dishevelled (Dvl), which may enable signaling from
Dvl to GSK3 (30). Finally,
low-density lipoprotein receptor related protein 5 recruits axin
and Frat1 to the cell membrane, which induces axin degradation and
Frat1-mediated inhibition of GSK-3. As a consequence, β-catenin is
not targeted for degradation, which leads to the activation of the
Wnt/β-catenin signaling pathway (31–33).
In the present study, the nuclear expression
patterns of FRAT1 in normal prostate basal cells and prostate tumor
cells were reported. Although GSK3β generally functions as part of
the β-catenin-degradation complex in the cytosol, there is evidence
to suggest that GSK3β may also reduce Wnt signaling in the nucleus
(34). Caspi et al
(34) demonstrated that GSK3β is
able to translocate to the nucleus, where it forms a complex with
β-catenin and reduces the level of β-catenin/TCF-dependent
transcriptional activity. The results of the present study suggest
that nuclear FRAT1 activates the Wnt/β-catenin signaling pathway
and confers an increase in prostate cancer cell growth, potentially
by preventing nuclear GSK3β-mediated inhibition of β-catenin/TCF
activity.
Acknowledgments
The present study was supported by grants from the
National Natural Science Youth Foundation (grant nos. 81201779 and
81502118), the Natural Science Foundation of Hubei Province (grant
no. 2014CFB250) and the National Natural Science Foundation of
China (grant nos. 81452761 and 81321091). Dr Yihua Wang was
supported by Biological Sciences, Faculty of Natural and
Environmental Sciences, University of Southampton.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kypta RM and Waxman J: Wnt/beta-catenin
signalling in prostate cancer. Nat Rev Urol. 9:418–428. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barker N and Clevers H: Mining the Wnt
pathway for cancer therapeutics. Nat Rev Drug Discov. 5:997–1014.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kahn M: Can we safely target the WNT
pathway? Nat Rev Drug Discov. 13:513–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Hewitt SM, Liu S, Zhou X, Zhu H,
Zhou C, Zhang G, Quan L, Bai J and Xu N: Tissue microarray analysis
of human FRAT1 expression and its correlation with the subcellular
localisation of beta-catenin in ovarian tumours. Br J Cancer.
94:686–691. 2006.PubMed/NCBI
|
|
9
|
Wang Y, Liu S, Zhu H, Zhang W, Zhang G,
Zhou X, Zhou C, Quan L, Bai J, Xue L, et al: FRAT1 overexpression
leads to aberrant activation of beta-catenin/TCF pathway in
esophageal squamous cell carcinoma. Int J Cancer. 123:561–568.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo G, Mao X, Wang P, Liu B, Zhang X,
Jiang X, Zhong C, Huo J, Jin J and Zhuo Y: The expression profile
of FRAT1 in human gliomas. Brain Res. 1320:152–158. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo G, Kuai D, Cai S, Xue N, Liu Y, Hao J,
Fan Y, Jin J, Mao X, Liu B, et al: Knockdown of FRAT1 expression by
RNA interference inhibits human glioblastoma cell growth, migration
and invasion. PloS One. 8:e612062013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo G, Zhong CL, Liu Y, Xue N, Liu Y, Hao
J, Fan Y, Jin J, Mao X and Liu B: Overexpression of FRAT1 is
associated with malignant phenotype and poor prognosis in human
gliomas. Dis Markers. 2015:2897502015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo G, Liu B, Zhong C, Zhang X, Mao X,
Wang P, Jiang X, Huo J, Jin J, Liu X and Chen X: FRAT1 expression
and its correlation with pathologic grade, proliferation, and
apoptosis in human astrocytomas. Med Oncol. 28:1–6. 2011.
View Article : Google Scholar
|
|
14
|
Zhang Y, Yu JH, Lin XY, Miao Y, Han Y, Fan
CF, Dong XJ, Dai SD and Wang EH: Overexpression of Frat1 correlates
with malignant phenotype and advanced stage in human non-small cell
lung cancer. Virchows Arch. 459:255–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Han Y, Zheng R, Yu JH, Miao Y,
Wang L and Wang EH: Expression of Frat1 correlates with expression
of β-catenin and is associated with a poor clinical outcome in
human SCC and AC. Tumour Biol. 33:1437–1444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan Y, Yang Z, Miao X, Li D, Liu Z and
Zou Q: The clinical significance of FRAT1 and ABCG2 expression in
pancreatic ductal adenocarcinoma. Tumour Biol. 36:9961–9968. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin R, Liu W, Menezes S, Yue F, Zheng M,
Kovacevic Z and Richardson DR: The metastasis suppressor NDRG1
modulates the phosphorylation and nuclear translocation of
β-catenin through mechanisms involving FRAT1 and PAK4. J Cell Sci.
127:3116–3130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:pl12013. View Article : Google Scholar
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chesire DR and Isaacs WB: Beta-catenin
signaling in prostate cancer: An early perspective. Endocr Relat
Cancer. 10:537–560. 2003. View Article : Google Scholar
|
|
23
|
Prajapati A and Gupta S, Mistry B and
Gupta S: Prostate stem cells in the development of benign prostate
hyperplasia and prostate cancer: Emerging role and concepts. Biomed
Res Int. 2013:1079542013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goksel G, Bilir A, Uslu R, Akbulut H,
Guven U and Oktem G: WNT1 gene expression alters in heterogeneous
population of prostate cancer cells; decreased expression pattern
observed in CD133+/CD44+ prostate cancer stem cell spheroids. J
BUON. 19:207–214. 2014.PubMed/NCBI
|
|
25
|
Jonkers J, Korswagen HC, Acton D, Breuer M
and Berns A: Activation of a novel proto-oncogene, Frat1,
contributes to progression of mouse T-cell lymphomas. EMBO J.
16:441–450. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jonkers J, Weening JJ, van der Valk M,
Bobeldijk R and Berns A: Overexpression of Frat1 in transgenic mice
leads to glomerulosclerosis and nephrotic syndrome, and provides
direct evidence for the involvement of Frat1 in lymphoma
progression. Oncogene. 18:5982–5990. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yost C, Farr GH III, Pierce SB, Ferkey DM,
Chen MM and Kimelman D: GBP, an inhibitor of GSK-3, is implicated
in Xenopus development and oncogenesis. Cell. 93:1031–1041. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jonkers J, van Amerongen R, van der Valk
M, Robanus-Maandag E, Molenaar M, Destrée O and Berns A: In vivo
analysis of Frat1 deficiency suggests compensatory activity of
Frat3. Mech Dev. 88:183–194. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van Amerongen R, van der Gulden H, Bleeker
F, Jonkers J and Berns A: Characterization and functional analysis
of the murine Frat2 gene. J Biol Chem. 279:26967–26974. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Yuan H, Weaver CD, Mao J, Farr GH
III, Sussman DJ, Jonkers J, Kimelman D and Wu D: Axin and Frat1
interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated
regulation of LEF-1. EMBO J. 18:4233–4240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hay E, Faucheu C, Suc-Royer I, Touitou R,
Stiot V, Vayssière B, Baron R, Roman-Roman S and Rawadi G:
Interaction between LRP5 and Frat1 mediates the activation of the
Wnt canonical pathway. J Biol Chem. 280:13616–13623. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thomas GM, Frame S, Goedert M, Nathke I,
Polakis P and Cohen P: A GSK3-binding peptide from FRAT1
selectively inhibits the GSK3-catalysed phosphorylation of axin and
beta-catenin. FEBS Lett. 458:247–251. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hagen T, Cross DA, Culbert AA, West A,
Frame S, Morrice N and Reith AD: FRAT1, a substrate-specific
regulator of glycogen synthase kinase-3 activity, is a cellular
substrate of protein kinase A. J Biol Chem. 281:35021–35029. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caspi M, Zilberberg A, Eldar-Finkelman H
and Rosin-Arbesfeld R: Nuclear GSK-3beta inhibits the canonical Wnt
signalling pathway in a beta-catenin phosphorylation-independent
manner. Oncogene. 27:3546–3555. 2008. View Article : Google Scholar : PubMed/NCBI
|