Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies and the second leading cause of
cancer-associated mortality worldwide (1). The highest incidences of HCC are
found in Africa and Asia, including China (2,3), and
the majority of cases are associated with a past or concurrent
chronic hepatitis B or hepatitis C infection, alcohol abuse or
aflatoxin B1 exposure (4).
Although treatments for HCC are evolving, surgery remains the most
preferred and effective modality for treating HCC. Although liver
transplantation is also a treatment option, the shortage of donated
organs limits its wider application in clinical practice. Other
therapeutic measures, including chemotherapy and transcatheter
arterial chemoembolization have been used to treat HCC, but have
failed to produce an observable effect (5). Therefore, increased knowledge
concerning the underlying mechanisms of the tumorigenesis and/or
progression of HCC is urgently required to identify an effective
method for managing patients with HCC.
It is widely accepted that genetic and epigenetic
alterations, including gene amplifications and mutations,
chromosomal translocations and non-coding RNAs, are associated with
HCC tumorigenesis (6), and
mutations in several specific genes, including tumor protein 53,
β-catenin (7) and sirtuin 1
(8), are reported to be associated
with the development and progression of HCC. In addition, next
generation sequencing techniques have revealed possible roles for
additional mutations in genes, including low density lipoprotein
receptor-related protein 1B (9),
interferon regulatory factor 2 (10) and AT-rich interactive domain 1A
(11). However, the scientific
value of the majority of previous reports regarding genetic
involvement in HCC have been limited by small sample numbers or the
use of outdated analytical techniques. Therefore, the possible
involvement of other gene abnormities in the development of HCC
requires consideration. The GTPase of the immunity-associated
protein (GIMAP) gene family is known to contribute to the
pathogenesis of non-small cell lung cancer (NSCLC) and the immune
reaction mounted against it (12);
however, this gene family has not been investigated with regards to
its possible roles in HCC.
In the present study, the techniques of reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis, immunohistochemistry (IHC) and ELISA were used to compare
the proteins and mRNA found in samples of HCC tissue and matched
samples of non-cancerous tissue, and found that the mRNA and
protein levels of GIMAP5 and GIMAP6 were downregulated in the HCC
tumor tissue samples. Additionally, when comparing the mRNA and
protein levels of GIMAP5 and GIMAP6 in samples of blood obtained
from normal subjects and patients with HCC, similar results were
found. The identification of these downregulated levels of GIMAP
family members may contribute to an improved understanding of the
molecular mechanisms involved in the development and progression of
HCC, and may lead to future clinical applications for GIMAP5 and
GIMAP6.
Materials and methods
Patients
Between December 2011 and December 2013, samples of
blood and liver tissues were collected from 60 patients (44 males
and 36 females; mean age, 55.4 years; range, 18–76 years) who had
been diagnosed with HCC, based on criteria used by the American
Association for the Study of Liver Diseases (www.aasld.org). The protocol for the present study was
approved by the ethics committee of Longgan Hospital (Shenzhen,
China), which also waived the requirement to obtain informed
consent from the tissue donors.
Serum collection and storage
Samples of blood (10 ml) were collected from
patients with HCC and normal control subjects, placed into labeled
tubes and allowed to clot for 30 min at room temperature. The blood
was centrifuged for 5 min at 3,000 g and 4°C, following which, 0.5
ml aliquots of serum were transferred into tubes and labeled. The
serum was then frozen immediately, ideally within 10 min of
aliquoting, and stored at −80°C until further use.
RNA extraction
Tissue samples were homogenized in 50 mM Tris-HC1
(pH 7.5) and centrifuged at 2,500 × g for 5 min. TRIzol reagent
(Sigma-Aldrich, St. Louis, MO, USA) was used to isolate total RNA
from the samples of HCC cancer tissue and the serum of patients
with HCC, according to the manufacturer's protocol. Control sample
RNA was purchased from Ambion, Inc. (Austin, TX, USA) and BioChain
Institute, Inc. (Newark, CA, USA). RNA quality was assessed using
an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc. Palo Alto,
CA, USA), and RNA samples with integrity scores >5 were
subjected to RT-qPCR analysis.
RT-qPCR
cDNA templates of different genes were obtained by
RT of the RNA using Super M-MLV RT (BioTeke Corporation, Beijing,
China), and the final reaction mixture of volume 20 µl contained 10
µl of 2X Taq PCR Master-mix (BioTeke Corporation), 1 µl of each
primer (GIMAP5, forward 5′-CATGTTAGGGAAGCTCAGTC-3′, reverse
5′-GAAGGGTTCTACTGTGTCTCA-3′; GIMAP6, forward
5′-TGGATGCTCTGGATGTTGCA-3′, reverse 5′-TCCTGCTCATCCCCTTGTG-3′;
hypoxanthine phosphoribosyltransferase 1 (HPRT1), forward
5′-GACCAGTCAACAGGGGACAT-3′, reverse
5′-GTGTCAATTATATCTTCCACAATCAAG-3′), 1 µl cDNA template and 7 µl of
RNase free H2O. Thermal cycling parameters for the
amplification were as follows: Denaturation step, 95°C for 5 min;
followed by 36 cycles at 95°C for 20 sec, 52°C for 20 sec and 72°C
for 30 sec; and the reaction was stopped by a step at 25°C for 5
min. The relative expression level of the targeted genes was
normalized to HPRT1 according to the 2−ΔΔCq method
(13).
IHC
The samples of the HCC and matched normal tissues
were fixed in formalin and embedded in paraffin. Subsequently, 4-µm
sections of the 60 paraffin-embedded HCC tumor tissue samples and
matched non-tumor tissue samples were deparaffinized in xylene,
rehydrated in an alcohol series and washed in distilled water. The
sections were then treated by microwave for antigen retrieval for 8
min at 450 W, and incubated with serum-free blocking reagent (Dako,
Carpentaria, CA, USA) for 10 min at room temperature to inhibit
non-specific staining. Subsequently, the slides mounted with the
tissue sections were incubated with rabbit anti-GIMAP5 (1:200; cat.
no. ab74575; Abcam, Cambridge, MA, USA) or rabbit anti-GIMAP6
(1:200; cat. no. ab126067; Abcam) antibodies in a humid chamber
overnight at 4°C. After three 5 min washes with 0.01 M
phosphate-buffered saline (PBS), goat anti-rabbit secondary
antibody (1:200; cat. no. A0277; Beyotime Institute of
Biotechnology, Jiangsu, China) was added to the sections and
incubated at 37°C for 30 min prior to five PBS washes. Peroxidase
activity was detected using the enzyme substrate, 3,3
N-diaminobenzidine tertrahydrochloride. Scores of the
immunohistochemical assay was determined by scanning the slides
using an Aperio ScanScope GL (Leica Microsystems GmbH, Wetzlar,
Germany) at ×400 magnification. ImageScope software (Leica
Microsystems GmbH) was then used to assess the scanned images based
on the percentage of positively stained cells and the staining
intensity. Scores for the expression levels of GIMAP5 and GIMAP6
were based on the staining intensity and the percentage of
positively-stained cells, as follows: 0, 0–9% of cells stained
positive; 1, 10–29% of cells stained positive; 2, 30–69% of cells
stained positive; 3, 70–100% of cells stained positive.
Statistical analysis
All data were analyzed using Prism 5 software
(GraphPad Software Inc., La Jolla, CA, USA). All values are
presented as the mean ± standard error of the mean, obtained from
analyses of three independent experiments. Differences between
groups were analyzed using the non-paired Student's two-tailed
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
mRNA expression levels of GIMAP5 and
GIMAP6 in HCC tissues and matched non-tumor tissues
As shown in Fig.
1A, the mRNA expression of GIMAP5 was significantly
downregulated in the 30 HCC tissue samples, compared with the 30
samples of adjacent matched non-tumor tissues (P<0.001). Similar
results were shown for the expression of GIMAP6 in another set of
30 HCC tissues and 30 samples of adjacent non-cancerous tissue
(P<0.001; Fig. 1B). These
results indicated that GIMAP5 and GIMAP6 were dysregulated at the
mRNA level in HCC tissues, and this dysregulation may be involved
in the pathogenesis of HCC.
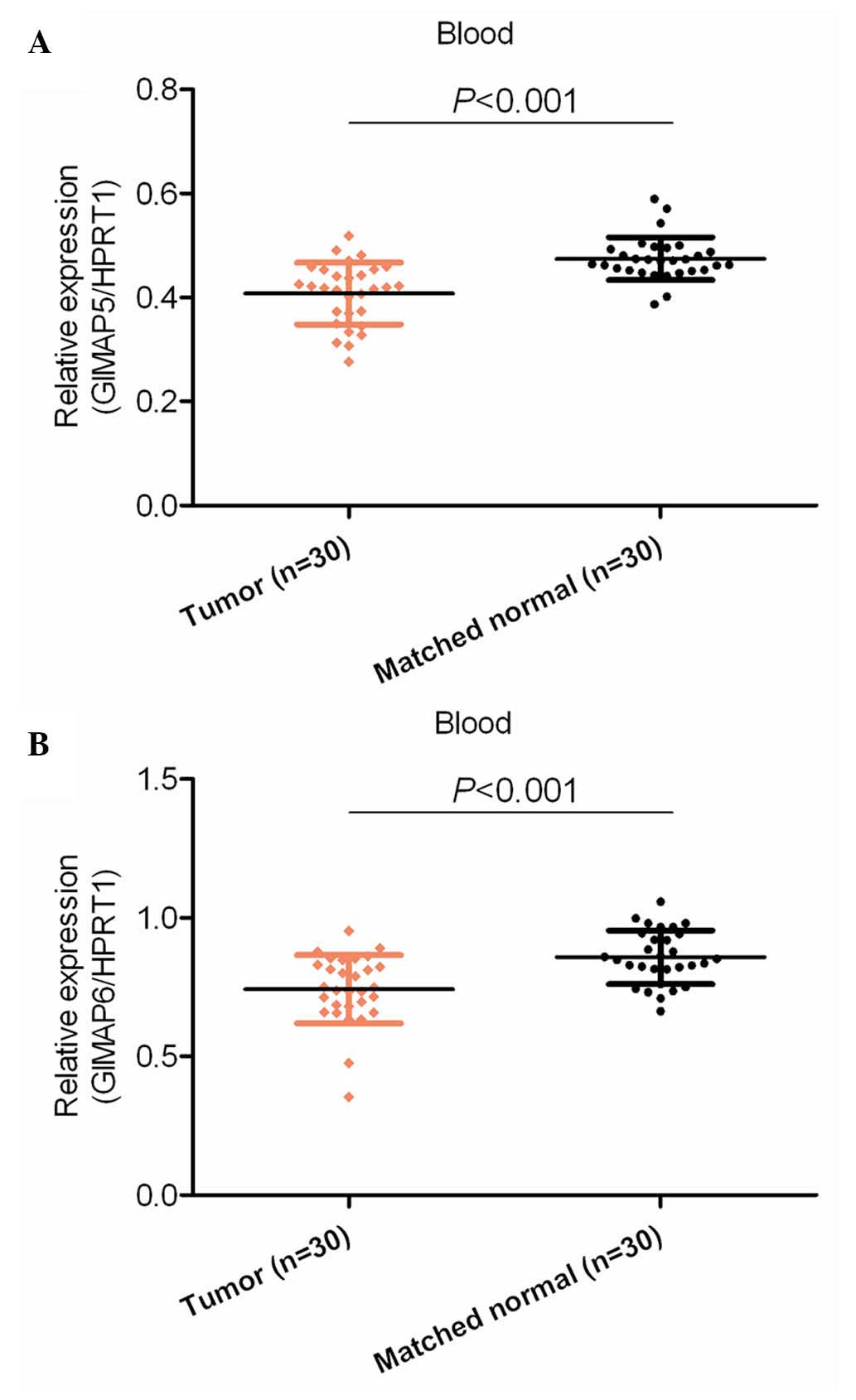
mRNA expression levels of GIMAP5 and
GIMAP6 are downregulated in the serum of patients with HCC
Dysregulation of mRNA may or may not be reflected in
the blood of patients. Therefore, the present study examined the
mRNA expression levels of GIMAP5 and GIMAP6 in the serum of blood
from patients with HCC and healthy control subjects. It was found
that the mRNA expression levels of GIMAP5 and GIMAP6 were
significantly downregulated in the blood of the patients with HCC,
compared with their expression levels in the blood of the normal
healthy subjects (P<0.001; Fig.
2).
Protein expression of GIMAP5 and
GIMAP6 in tissue samples
The present study subsequently examined whether the
dysregulation observed in the mRNA expression of GIMAP5 and GIMAP6
were also observed in the respective proteins. The IHC examinations
of the protein expression levels of GIMAP5 and GIMAP6 revealed
results that were consistent with the results for mRNA expression,
with the protein levels of GIMAP5 and GIMAP6 being downregulated in
the HCC tissue samples, compared with their levels in the samples
of adjacent normal tissue (P<0.001; Fig. 3). These results suggested that the
GIMAP5 and GIMAP6 proteins may offer potential as diagnostic
biomarkers for HCC.
Protein expression of GIMAP5 and
GIMAP6 in the serum of patients with HCC and normal control
subjects
The present study also examined the protein
expression of GIMAP5 and GIMAP6 in the blood of patients with HCC
using ELISA. It was found that the GIMAP5 and GIMAP6 proteins were
expressed at lower levels in the blood of the patients with HCC,
compared with the expression levels in the blood of healthy control
subjects (P<0.001; Fig. 4),
providing further confirmation of the lower expression levels of
GIMAP5 and GIMAP6 in the tissues and blood of patients with
HCC.
Discussion
GIMAPs are a family of putative small GTPase
proteins, which are conserved in vertebrates and plants (14). This family comprises seven
proteins, which have their coding genes residing on human
chromosome 7 (12). These GIMAP
family members have unique primary structures, and thus represent a
novel family of G proteins. Previous studies have shown that GIMAPs
are expressed at the highest levels in cells of the immune system
(15), therefore, the majority of
reports have associated them with immunological functions and
immune disorders (16,17). In addition, certain studies have
demonstrated GIMAP dysregulation in a number of tumors, suggesting
their potential functions in tumorigenesis (12,18).
To extend current knowledge of the GIMAP family members associated
with HCC, the present study investigated the expression of GIMAP5
and GIMAP6 in samples of HCC tissue and in serum obtained from
patients with HCC.
GIMAP5 is the member of the GIMAP family, which has
received the most attention. Dalberg et al (19) found that reduced expression of
GIMAP5 in Jurkat cells did not affect the number of apoptotic
cells, whereas transient overexpression of GIMAP5 resulted in a
significant increase in the number of apoptotic cells; suggesting
that the overexpression of GIMAP5 is important for inducing T cell
apoptosis (19). In a study by
Hellquist et al (20),
genetic variation in GIMAP5 was found to be involved in the
susceptibility to systemic lupus erythematosus. In addition, Shiao
et al (12) compared NSCLC
tissues with adjacent non-tumor tissues, and found that all GIMAP
members, including GIMAP5, were expressed at lower levels in the
NSCLC tissues, suggesting their potential function in NSCLC
tumorigenesis (12). In the
present study, it was found that the expression of GIMAP5 was
downregulated in the tumor tissues and blood samples obtained from
patients with HCC, suggesting its involvement in the pathogenesis
of HCC, as well as its potential as a diagnostic biomarker for
HCC.
Pascall et al (21) showed that the interaction between
GIMAP6 and autophagy-related protein 8 is crucial for autophagy
(21). Additionally, GIMAP6 was
found to be downregulated in samples of NSCLC tissue, compared with
normal tissue (12). Knowledge
regarding the function of GIMAP6 in cells remains limited. In the
present study, it was found that the expression levels of GIMAP6
and GIMAP5 were downregulated in HCC tissues, compared with
adjacent non-tumor tissues, which suggested their potential
functions in the development and progression of HCC. Consistent
with these findings, the expression of GIMAP6 was also
downregulated in the serum of patients with HCC, suggesting its
potential future diagnostic application. These results were
confirmed at the mRNA and protein expression levels.
In conclusion, the present study found that the mRNA
and protein expression levels of GIMAP5 and GIMAP6 were
downregulated in samples of HCC tissue and in serum obtained from
patients with HCC, suggesting the potential application of these
two biomarkers in diagnostic procedures. Further investigations
into the feasibility of using GIMAP5 and GIMAP6 as biomarkers for
HCC are required.
Acknowledgements
This study was supported by the National Natural
Science Foundation of PR. China (grant nos. 81201831 and 81301882),
the Medical Science and Technology Research Project of Guangdong
Province (grant no. B2012266) and the Fundamental Research Funds
for the Central Universities (grant no. 20620140118).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hao C, Zhu PX, Yang X, Han ZP, Jiang JH,
Zong C, Zhang XG, Liu WT, Zhao QD, Fan TT, et al: Overexpression of
SIRT1 promotes metastasis through epithelial-mesenchymal transition
in hepatocellular carcinoma. BMC Cancer. 14:9782014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goh LY, Leow AH and Goh KL: Observations
on the epidemiology of gastrointestinal and liver cancers in the
Asia-Pacific region. J Dig Dis. 15:463–468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cauchy F, Fuks D and Belghiti J: HCC:
Current surgical treatment concepts. Langenbecks Arch Surg.
397:681–695. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Woo HG, Kim SS, Cho H, Kwon SM, Cho HJ,
Ahn SJ, Park ES, Lee JS, Cho SW and Cheong JY: Profiling of exome
mutations associated with progression of HBV-related hepatocellular
carcinoma. PLoS One. 9:e1151522014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laurent-Puig P, Legoix P, Bluteau O,
Belghiti J, Franco D, Binot F, Monges G, Thomas G, Bioulac-Sage P
and Zucman-Rossi J: Genetic alterations associated with
hepatocellular carcinomas define distinct pathways of
hepatocarcinogenesis. Gastroenterology. 120:1763–1773. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen J, Zhang B, Wong N, Lo AW, To KF,
Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, et al: Sirtuin 1 is
upregulated in a subset of hepatocellular carcinomas where it is
essential for telomere maintenance and tumor cell growth. Cancer
Res. 71:4138–4149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding D, Lou X, Hua D, Yu W, Li L, Wang J,
Gao F, Zhao N, Ren G, Li L and Lin B: Recurrent targeted genes of
hepatitis B virus in the liver cancer genomes identified by a
next-generation sequencing-based approach. PLoS Genet.
8:e10030652012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guichard C, Amaddeo G, Imbeaud S, Ladeiro
Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M,
Degos F, et al: Integrated analysis of somatic mutations and focal
copy-number changes identifies key genes and pathways in
hepatocellular carcinoma. Nat Genet. 44:694–698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fujimoto A, Totoki Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shiao YM, Chang YH, Liu YM, Li JC, Su JS,
Liu KJ, Liu YF, Lin MW and Tsai SF: Dysregulation of GIMAP genes in
non-small cell lung cancer. Lung Cancer. 62:287–294. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nitta T and Takahama Y: The lymphocyte
guard-IANs: Regulation of lymphocyte survival by IAN/GIMAP family
proteins. Trends Immunol. 28:58–65. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Filen S and Lahesmaa R: GIMAP proteins in
T-lymphocytes. J Signal Transduct. 2010:2685892010.PubMed/NCBI
|
|
16
|
Yano K, Carter C, Yoshida N, Abe T, Yamada
A, Nitta T, Ishimaru N, Takada K, Butcher GW and Takahama Y: Gimap3
and Gimap5 cooperate to maintain T-cell numbers in the mouse. Eur J
Immunol. 44:561–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saunders A, Webb LM, Janas ML, Hutchings
A, Pascall J, Carter C, Pugh N, Morgan G, Turner M and Butcher GW:
Putative GTPase GIMAP1 is critical for the development of mature B
and T lymphocytes. Blood. 115:3249–3257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zenz T, Roessner A, Thomas A, Fröhling S,
Döhner H, Calabretta B and Dahéron L: HIan5: The human ortholog to
the rat Ian4/Iddm1/lyp is a new member of the Ian family that is
overexpressed in B-cell lymphoid malignancies. Genes Immun.
5:109–116. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dalberg U, Markholst H and Hornum L: Both
Gimap5 and the diabetogenic BBDP allele of Gimap5 induce apoptosis
in T cells. Int Immunol. 19:447–453. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hellquist A, Zucchelli M, Kivinen K,
Saarialho-Kere U, Koskenmies S, Widen E, Julkunen H, Wong A,
Karjalainen-Lindsberg ML, Skoog T, et al: The human GIMAP5 gene has
a common polyadenylation polymorphism increasing risk to systemic
lupus erythematosus. J Med Genet. 44:314–321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pascall JC, Rotondo S, Mukadam AS, Oxley
D, Webster J, Walker SA, Piron J, Carter C, Ktistakis NT and
Butcher GW: The immune system GTPase GIMAP6 interacts with the Atg8
homologue GABARAPL2 and is recruited to autophagosomes. PLoS One.
8:e777822013. View Article : Google Scholar : PubMed/NCBI
|