Introduction
B-cell-specific Moloney murine leukemia virus
insertion site-1 (Bmi-1), a member of the polycomb family, was
originally described as interacting with c-Myc to initiate lymphoma
in mice (1). Evidence suggests
that Bmi-1 is critical for numerous physiological and pathological
processes, including axial patterning (2), hematopoiesis (3), regulation of proliferation,
senescence (4) and self-renewal of
cancer stem cells (5). In
addition, Bmi-1 is overexpressed in various malignancies (6–8),
including multiple myeloma (MM) (9), and may regulate proliferation and
carcinogenesis. Furthermore, our previous study demonstrated that
silencing of Bmi-1 sensitizes MM cells to bortezomib (10). Bmi-1 may be post-transcriptionally
regulated by microRNAs (miRNAs) (11–14);
however, whether miRNAs target Bmi-1 in MM remains unclear.
miRNAs are endogenous non-coding small RNAs that
post-transcriptionally regulate gene expression. They exert this
effect by binding to the 3′untranslated regions (UTRs) of target
mRNAs, resulting in degradation or repression of translation of
mRNAs. miRNAs may function as oncogenes or tumor suppressor genes
in human cancer (15). Studies
have suggested that numerous cancer-associated miRNAs are located
in genomic breakpoint regions (16). Certain miRNAs may be epigenetically
inactivated, particularly by hypermethylation, and re-expression of
these miRNAs may result in the downregulation of target oncogenes
(17). Hypermethylation of miR-203
in chronic myeloid leukemia (CML) (18), hepatocellular carcinoma (19) and MM (20) has been described. Restoration of
miR-203 downregulates the oncogenic BCR-ABL1 fusion protein and
cyclic AMP responsive element binding protein 1 (CREB1) in CML and
MM, respectively, thereby inhibiting cellular proliferation and
demonstrating the tumor suppressor activity of miR-203.
Computational analysis indicates that a single mRNA molecule may be
regulated by multiple miRNAs and that one miRNA may regulate
multiple mRNAs (21). Therefore,
miR-203 may target mRNAs other than CREB1 in MM.
The present study used bioinformatics databases,
TargetScan, PicTar and miRanda, to predict the potential miRNAs
that may target Bmi-1. A luciferase reporter assay was performed to
determine that miR-203 directly targets the 3′UTRs of Bmi-1.
Enforced expression of miR-203 significantly inhibits cell growth
and regulates G1/S transition in MM cells. miR-203 was
downregulated in MM and the expression of miR-203 was negatively
correlated with Bmi-1 expression. Taken together, these results
indicated that Bmi-1 is a critical target of the tumor suppressor
activity of miR-203 in MM.
Materials and methods
Patient samples
A total of 45 patients with newly diagnosed MM [24
males, 21 females; median age, 55 years (range, 45–75 years)] were
recruited from the Department of Hematology, Affiliated Union
Hospital of Fujian Medical University (Fuzhou, China). The
diagnosis of MM was based on standard criteria [International
Myeloma Working Group, 2003 (22)], and the presence of >60% plasma
cells in the bone marrow. A total of 18 healthy donors [10 males, 8
females; median age, 50 years (range, 45–68 years)] were recruited
as a control group. The study was approved by the Institutional
Review Board of Affiliated Union Hospital of Fujian Medical
University, and informed consent was obtained from all subjects
prior to sample collection.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Mononuclear cells (MNCs) were isolated from patient
bone marrow samples using Ficoll-Hypaque density gradient
centrifugation (Inno-Train Diagnostik GmbH, Kronberg im Taunus,
Germany). Total RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). To
determine miR expression, total RNA (1 µl/sample) was
reverse-transcribed using miR-specific stem-loop RT primers,
reverse transcriptase, RT buffer, dNTPs and an RNase inhibitor,
according to the manufacturer's protocol (TaqMan®
MicroRNA Reverse Transcription kit; Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR was performed using an Applied
Biosystems™ StepOnePlus™ Real-Time PCR System (Thermo Fisher
Scientific, Inc.). The 20-µl reaction mixture contained the
corresponding complementary DNA (2 µl), miRNA-specific TaqMan
primers (1 µl), TaqMan Universal PCR Master mix (10 µl; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and double-distilled
H2O (7 µl). The PCR cycling conditions were as follows:
Denaturation at 95°C for 10 min, followed by 50 cycles of
denaturation at 95°C for 15 sec and annealing at 60°C for 1 min.
RNU6B served as an endogenous housekeeping control for data
normalization of miR levels. mRNA was reverse-transcribed in RT
reactions containing 500 ng total RNA extracted from samples, 2 µl
5X PrimeScript™ Buffer (Takara Bio, Inc., Otsu, Japan), 0.5 µl 1X
PrimeScript™ RT Enzyme Mix I (Takara Bio, Inc.) and 0.5 µl oligo
(dT) primer. The 10-µl reactions were incubated for 42 min at 37°C,
followed by 30 sec at 85°C and subsequent exposure to 4°C. The
endogenous mRNA levels of Bmi-1 were detected using the
SYBR® Green PCR Master mix kit (Takara Bio, Inc.),
according to the manufacturer's protocol. The PCR cycling
conditions were as follows: An initial denaturation step at 95°C
for 15 min, followed by 40 cycles of denaturation at 94°C for 15
sec, annealing at 55°C for 30 sec and extension at 72°C for 30 sec.
β-actin served as an internal control. The following primers
(Invitrogen; Thermo Fisher Scientific, Inc.) were used: Forward,
5′-AAATCAGGGGGTTGAAAAATCT-3′ and reverse,
5′-GCTAACCACCAATCTTCCTTTG-3′ for Bmi-1; and forward,
5′-TTGTTACAGGAAGTCCCTTGCC-3′ and reverse,
5′-ATGCTATCACCTCCCCTGTGTG-3′ for β-actin. The comparative
quantification cycle (Cq) method was used to measure the relative
changes in expression; 2-ΔΔCq represents the fold-change in
expression (23).
Cell culture
HEK-293T cells were cultured in Dulbecco's modified
Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.) with
10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.). U266 and RPMI8226 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and cultured in RPMI 1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% FBS,
50 U/ml penicillin and 50 µg/ml streptomycin.
Lentivirus production and
infection
The pri-miR-203 sequence was amplified and cloned
into the pWPXL lentiviral vector (donated by Dr Didier Trono, École
Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). The
primers were as follows: Forward, 5′-GAATTCCGTCTAAGGCGTCCGGTA-3′
and reverse, 5′-GCGGCCGCGTTCCCACAGCACAGC-3′. The pWPXL vectors were
transfected into HEK-293T cells with the packaging plasmid psPAX2
and the VSV-G envelope plasmid pMD2.G (donated by Dr Didier Trono)
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). Cell supernatants were collected at 48 h
post-transfection and passed through a 0.22-mm filter. The virus
particles were harvested 48 h following transfection. The titer of
purified virus was 2.0×108 IU/ml. Cells (1×105) were infected with
1×106 recombinant lentivirus-transducing units and 6 µg/ml
Polybrene (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany).
Cell proliferation
Cell proliferation was measured using the Cell
Counting kit (CCK)-8 assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). Cells were seeded into 96-well plates
(4×103/well), and 10 µl CCK-8 solution was added to 90 µl culture
medium. Cells were subsequently incubated for 2 h at 37°C and the
optical density was measured at 450 nm and 650 nm. Three
independent experiments were performed.
Cell cycle and apoptosis determination
by flow cytometry
For cell cycle analysis, cells were collected and
fixed in ice-cold 70% ethanol overnight. The fixed cells were
washed with phosphate-buffered saline (PBS) and stained with
freshly-prepared PBS containing 25 µg/ml propidium iodide (Sigma),
10 µg/ml RNaseA, 0.05 mM ethylene diamine and 0.2% Triton X-100
tetra-acetic acid for 30 min in the absence of light. For each
sample, at least 20,000 cells were acquired on an EPICS Altra flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA) and analyzed using
Multicycle AV software version 5.0 (Phoenix Flow Systems, San
Diego, CA, USA).
For apoptosis detection, cells (2×105)
were collected and stained with an Annexin V-phycoerythrin
(PE)/7-aminoactinomycin D (AAD) Apoptosis Detection kit
(Sigma-Aldrich; Merck Millipore) according to the manufacturer's
protocol. Cells were acquired and analyzed as above.
Oligonucleotide transfection
The miR-203 mimic and negative control were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
sequence of the RNA duplex control was 5′-UUCUCCGAACGUGUCACGU-3′.
Oligonucleotide transfection was performed with Lipofectamine
2000.
Luciferase reporter constructs and
luciferase assay
The wild-type and mutant 3′UTRs of Bmi-1 were cloned
downstream of a cytomegalovirus (CMV) promoter-driven firefly
luciferase cassette in a pCDNA3.0 vector (Thermo Fisher Scientific,
Inc.). The primers used were as follows: Forward,
5′-AGTGGACGTTCACCGAGTT-3′ and reverse, 5′-TCCCAAAAGCGCATTTATT-3′
for wild-type; and forward,
5′-CTAGAATGAAGATGTCCCCCATCTTATACCCCTAAC-3′ and reverse,
5′-GATGGGCGGACATCTTCTTTCTAGCAGGGAGACACTG-3′ for mutant. For the
luciferase assay, 10,000 U266 cells were cultured in 24-well plates
and cotransfected with 20 pmol RNA (negative control or miR-203
mimic), 200 ng luciferase reporter construct and 20 ng pRL-CMV
renilla luciferase reporter construct. Following a 48-h incubation,
luciferase activity was measured using the
Dual-Luciferase® Reporter assay system (Promega
Corporation, Madison, WI, USA).
Western blotting
Cells were washed with cold PBS and lysed in culture
dishes using PhosphoSafe™ extraction reagent (Merck Millipore)
containing 1% protease inhibitor cocktail (EDTA-free; Thermo Fisher
Scientific, Inc.). Protein concentrations were then determined
using Bio-Rad detergent-compatible protein assays (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Proteins (50 µg) were
loaded onto 12% SDS-PAGE gels, electrophoresed and transferred onto
polyvinylidene difluoride membranes (0.22 µm; Merck Millipore).
Membranes were blocked with 5% nonfat milk and incubated with
rabbit anti-Bmi-1 monoclonal antibody (1:1,000; catalog no. 2830S;
Cell Signaling Technology, Inc., Danvers, MA, USA) and rabbit
polyclonal anti-β-actin (1:1,000; catalog no. ab119716; Abcam,
Cambridge, MA, USA) for 1.5 h at room temperature. Following
washing, membranes were incubated for 1 h at room temperature with
a horseradish peroxidase-conjugated mouse anti-rabbit secondary
antibody (1:1,000; catalog no. bs-0295M; Bioss Inc., Woburn, MA,
USA). Protein bands were visualized using an Enhanced
Chemiluminescence reagent (Pierce Biotechnology, Inc., Rockford,
IL, USA).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software version 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA). Data are presented as the mean ± standard error. The data
were subjected to two-tailed Student's t-test. Linear
regression analysis was used to investigate the relationship
between the expression of miR-203 and Bmi-1. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-203 downregulates Bmi-1 expression
by directly targeting the Bmi-1 3′UTR
Bmi-1 is upregulated significantly in various
malignancies including MM, and may be post-transcriptionally
regulated by miRNAs. Therefore, Bmi-1 may be regulated by miRNAs in
MM. The online miRNA target-prediction tools, TargetScan
(www.targetscan.org/vert_71/), PicTar
(pictar.mdc-berlin.de/) and miRanda (www.microrna.org/microrna/home.do), were used to
predict Bmi-1-targeting miRNAs. The number of potential
Bmi-1-targeting miRNAs was 16, 12 and 82 for TargetScan, PicTar and
miRanda, respectively. Altogether, a pool of 86 miRNAs that may
potentially target Bmi-1 was selected. Of these miRNAs, miR-203 has
been reported as a tumor suppressor miRNA inhibiting cellular
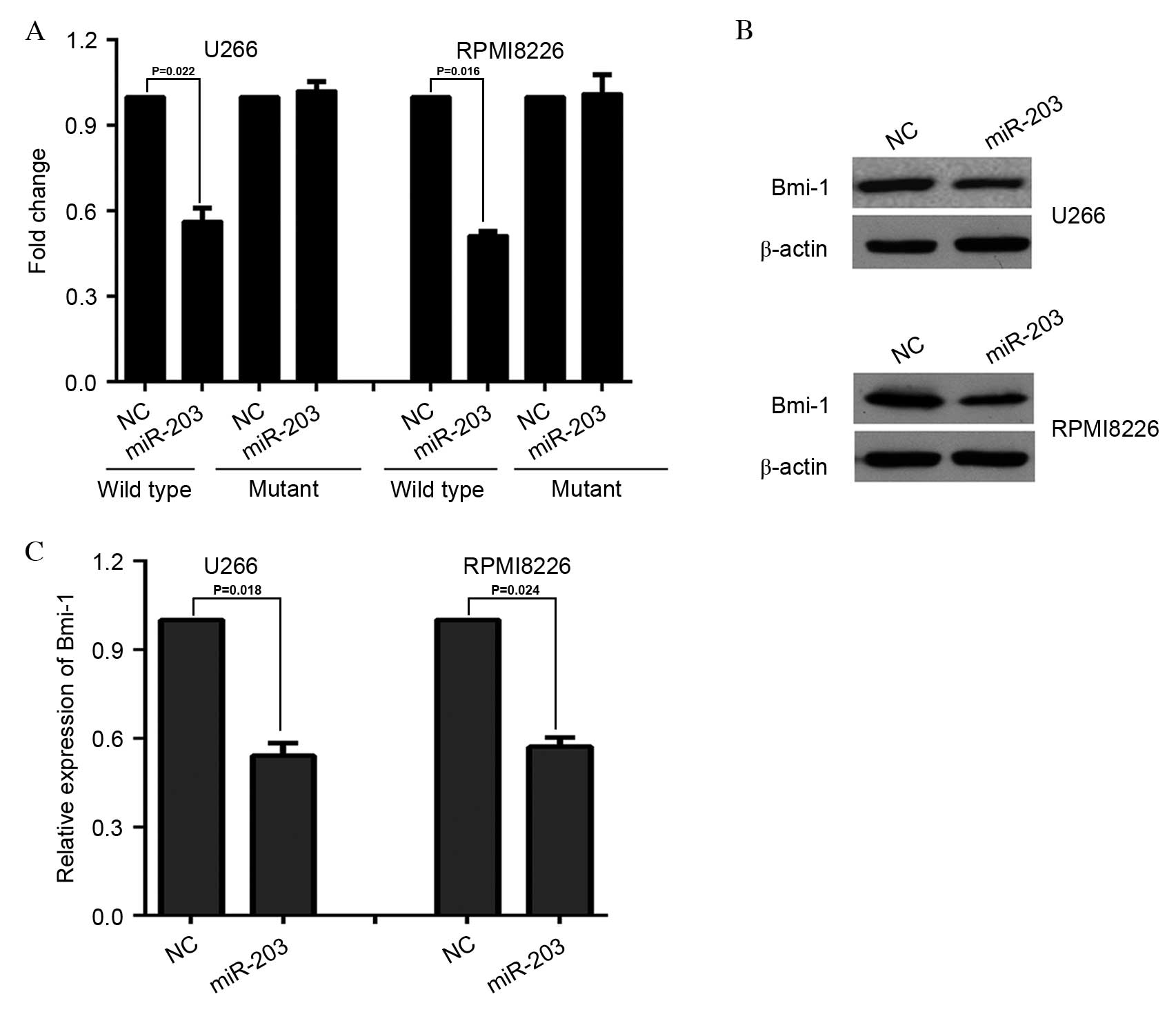
proliferation by targeting CREB1 mRNA in MM (20). Therefore, the present study focused
on miR-203. To determine whether miR-203 exerts its function by
downregulating the expression of Bmi-1 through direct binding to
its 3′UTR, full-length fragments of Bmi-1 mRNA 3′UTR (wild-type or
mutant) were constructed and inserted immediately downstream of the
luciferase reporter gene. For the luciferase assays, the miR-203
mimic was cotransfected with the luciferase 3′UTR constructs into
U266 cells and RPMI8226 cells. The results revealed that miR-203
decreased the relative luciferase activity when transfected with
wild-type, but not mutant, Bmi-1 3′UTR (P=0.022 and P=0.016 for
U266 and RPMI8226 cells, respectively; Fig. 1A). In accordance with these
results, a marked decrease was observed in endogenous Bmi-1 protein
(Fig. 1B) and mRNA (P=0.018 and
P=0.024 for U266 and RPMI8226 cells, respectively; Fig. 1C) expression levels in U266 cells
and RPMI8226 cells following infection with miR-203-expressing
lentivirus. Taken together, these results suggested that miR-203
may downregulate Bmi-1 expression post-transcriptionally by
directly targeting its 3′UTR.
Enforced expression of miR-203
inhibits MM cell growth and cell cycle transition
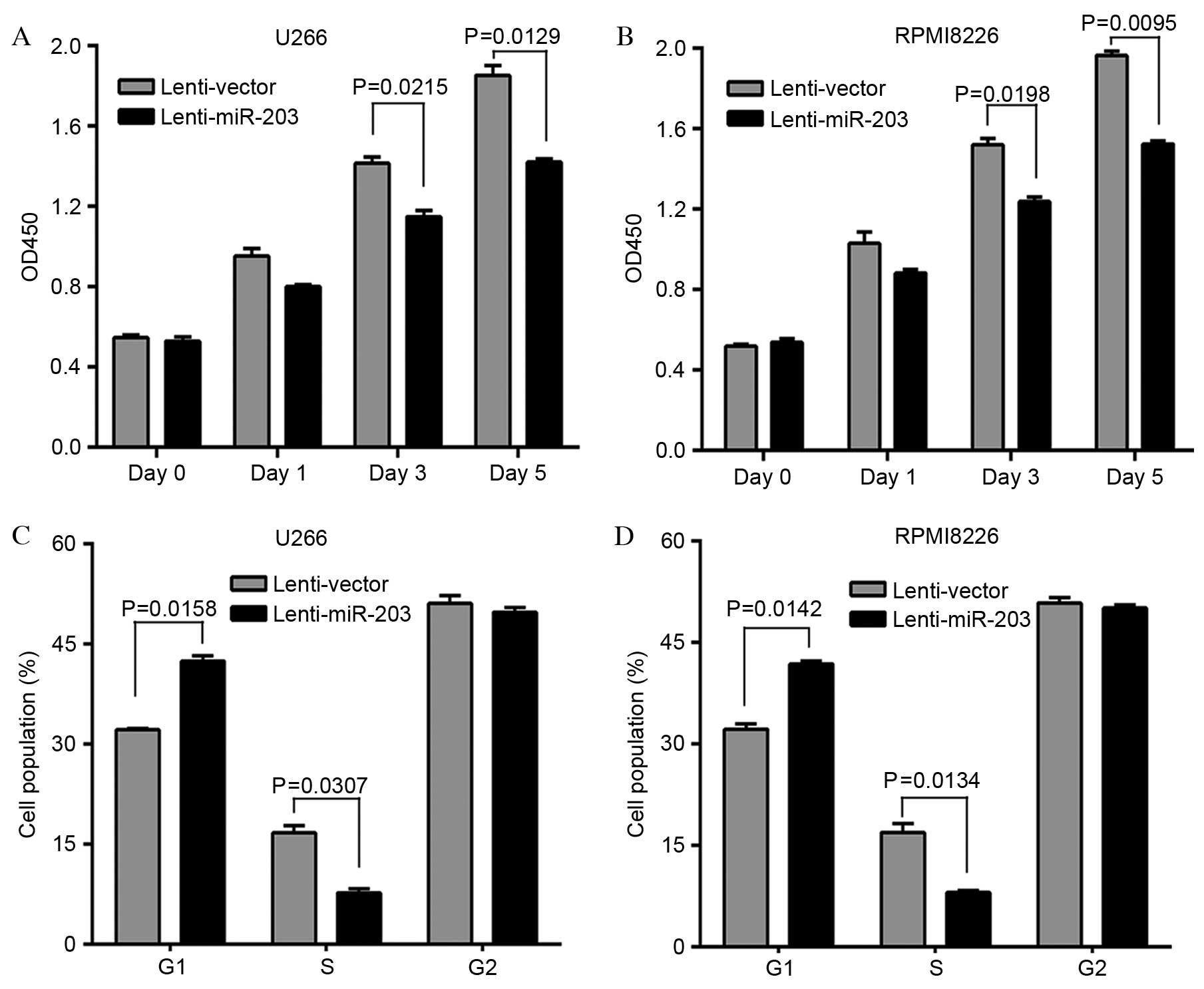
To determine the functional role of miR-203 in MM, a
lentivirus vector harboring miR-203 was constructed and used to
establish two stable MM cell lines, U266-203 and RPMI8226-203.
Analysis of proliferation (Fig. 2A and
B) revealed that ectopic expression of miR-203 resulted in a
significant decrease in the growth of U266-203 (day 3, P=0.0215;
day 5, P=0.0129) and RPMI8226-203 (day 3, P=0.0198; day 5,
P=0.0095) cells. As miR-203 inhibits MM cell proliferation, the
effect of miR-203 on MM cell cycle progression was investigated.
The cell cycle distribution demonstrated that the proportion of
cells in G1 phase was significantly increased in U266-203
(P=0.0158) and RPMI8226-203 (P=0.0142) cells compared with control
vector, whereas the cell population in S phase was significantly
reduced in U266-203 (P=0.0307) and RPMI8226-203 (P=0.0134) cells
(Fig. 2C and D). Therefore, the
growth-inhibiting effect of miR-203 on MM cells may be due to the
arrest of cell cycle progression at the G1/S transition.
Ectopic expression of miR-203 promotes
MM cell apoptosis
Annexin V-PE/7-AAD staining was performed to
evaluate the effect of miR-203 overexpression on MM cell apoptosis.
The percentage of apoptotic U266-control and U266-203 cells was
1.5±0.008 and 36±2.04% (P<0.001), respectively (Fig. 3A); the percentage of apoptotic
RPMI8226-control and RPMI8226-203 cells was 13.66±0.98 and
30.06±1.65% (P=0.014), respectively (Fig. 3B). Therefore, ectopic expression of
miR-203 may promote MM cell apoptosis by targeting Bmi-1.
miR-203 is downregulated in MM and
negatively correlates with Bmi-1 expression
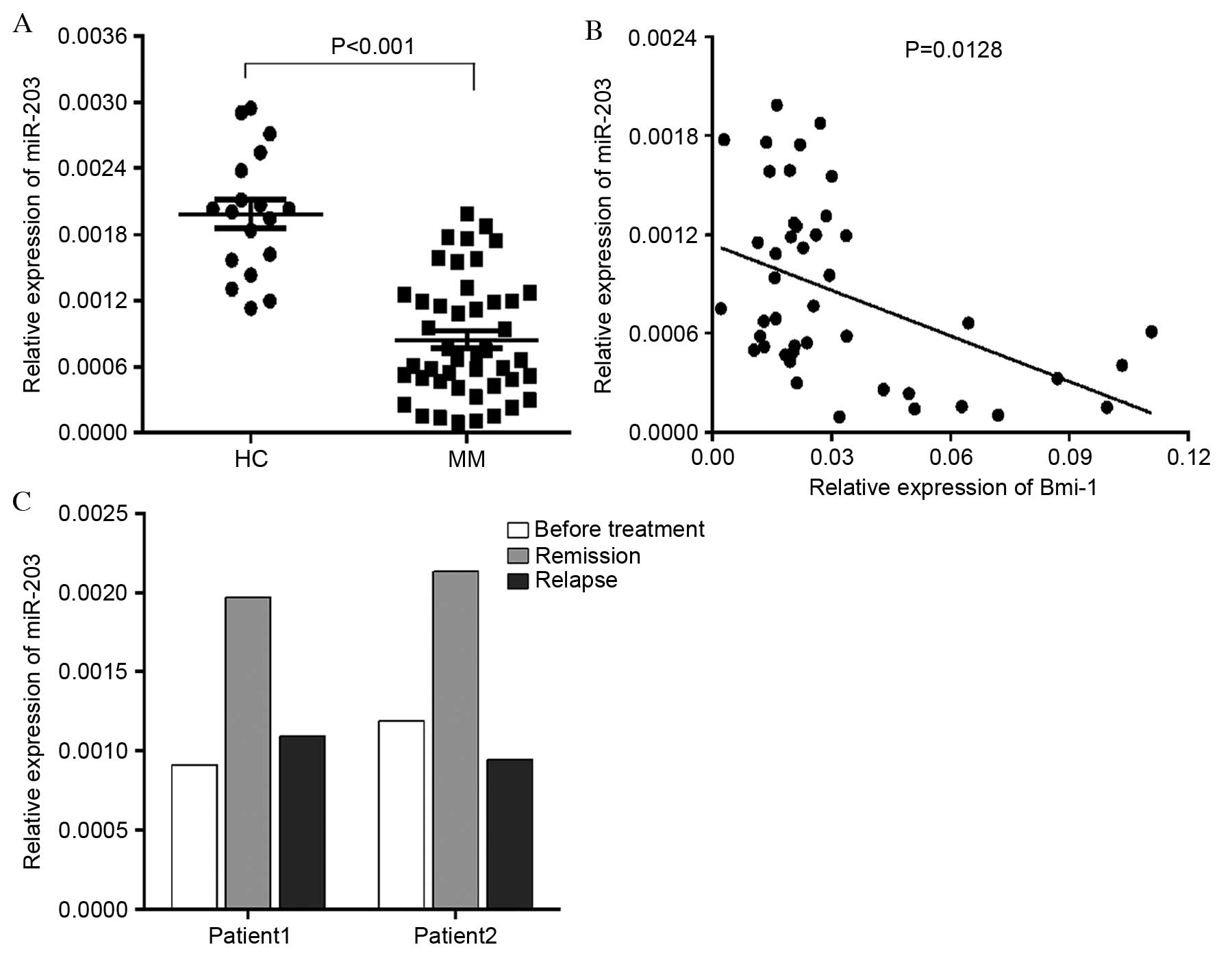
To determine miR-203 expression levels in bone
marrow MNCs from newly diagnosed MM patients and healthy donors,
RT-qPCR was performed. miR-203 was significantly downregulated in
MM patients compared with healthy controls (P<0.001; Fig. 4A). Bmi-1 was relatively highly
expressed in MM and negatively correlated with miR-203 expression
(P=0.0128; Fig. 4B). Furthermore,
miR-203 expression levels were examined in certain patients
following treatment remission and relapse. miR-203 expression
increased following remission and declined again following relapse
(Fig. 4C). Taken together, these
results demonstrated that the expression levels of miR-203 were
downregulated in MM patients, and may be associated with the
current disease state.
Discussion
Bmi-1 was originally described as interacting with
c-Myc to initiate lymphoma in mice (1). Evidence has demonstrated that Bmi-1
is critical for numerous physiological and pathological processes,
including hematopoiesis (3),
senescence (4), axial patterning
(2), regulation of proliferation
and maintenance of cancer stem cell self-renewal (5). In addition, evidence has suggested
that Bmi-1 is upregulated in various malignancies, including
colorectal cancer (6), non-small
cell lung cancer (8), pancreatic
cancer (24), breast cancer
(25), prostate cancer (26) and leukemia (7). Bmi-1 is overexpressed in MM and may
regulate cell proliferation and carcinogenesis in vitro and
in vivo. Our previous study demonstrated that silencing of
Bmi-1 sensitizes MM cells to bortezomib (10). However, the reasons behind the
upregulation of Bmi-1 in MM remain unclear.
miRNAs are post-transcriptional regulators of
downstream gene expression, and may function as oncogenes or tumor
suppressor genes during tumor development and progression (27). Previous studies have suggested that
Bmi-1 may be post-transcriptionally regulated by miRNAs (11–14);
however, whether miRNAs target Bmi-1 in MM remains unclear. In the
present study, three miRNA target-prediction tools, TargetScan,
PicTar and miRanda, were used to predict all the miRNAs that may
target Bmi-1. A total of 86 miRNAs were identified that may target
Bmi-1. Of these miRNAs, miR-203 was selected for further
investigation. Previous studies have reported that miR-203 is
downregulated in MM and CML due to epigenetic silencing, and that
CREB1 is a novel target of miR-203 in MM (19,20).
However, computational analysis indicates that one miRNA may
regulate multiple mRNAs. Therefore, more mRNAs than CREB1 may be
regulated by miR-203 in MM. The present study identified Bmi-1 as a
direct downstream target of miR-203 in MM cells. A potential
miR-203 target site was identified in the Bmi-1 3′UTR. Following
cotransfection with wild-type 3′UTR, miR-203 decreased the relative
luciferase activity. However, when the potential target site was
mutated, luciferase activity was unaffected. In accordance with
these results, endogenous Bmi-1 protein and mRNA expression levels
were demonstrated to be downregulated by miR-203 in MM cells.
If miR-203 functions as a tumor suppressor in MM,
restoration of miR-203 expression in MM cells may inhibit
proliferation and/or promote apoptosis. The present study
demonstrated that ectopic expression of miR-203 in MM cells led to
a significant inhibition of proliferation and the arrest of cell
cycle progression at the G1/S transition. In addition,
the present study suggested that miR-203 re-expression may promote
MM cell apoptosis by targeting Bmi-1. Taken together, these results
confirm miR-203 as a tumor suppressor in MM. Furthermore, the
expression levels of miR-203 and Bmi-1 in MM patient and healthy
control bone marrow samples were detected. miR-203 was
significantly downregulated in MM; Bmi-1 was relatively abundant in
MM and negatively correlated with miR-203 expression. In certain
clinical circumstances, miRNA profiling may be superior to mRNA
profiling to classify tumor subtypes and predict prognosis. The
present study demonstrated that miR-203 expression may indicate
current disease status.
In conclusion, the results of the present study
demonstrated, to the best of our knowledge for the first time, that
miR-203 may target Bmi-1 in MM, inhibit MM cell growth and regulate
G1/S transition. These results suggested that miR-203
has tumor suppressor properties and therefore potential therapeutic
application in MM.
Acknowledgements
The authors thank Dr Didier Trono (School of Life
Sciences, École Polytechnique Fédérale de Lausanne, Lausanne,
Switzerland) for donating the pWPXL, psPAX2 and pMD2.G lentivirus
plasmids. The present study was supported by the National Natural
Science Foundation of China (grant no. 81201872), the Natural
Science Foundation of Fujian Province (grant no. 2013J01308) and
the Foundation of Fujian Key Laboratory of Hematology (grant no.
2009J1004).
References
|
1
|
Jacobs JJ, Kieboom K, Marino S, DePinho RA
and van Lohuizen M: The oncogene and Polycomb-group gene bmi-1
regulates cell proliferation and senescence through the ink4a
locus. Nature. 397:164–168. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong KY, Yim RL, Kwong YL, Leung CY, Hui
PK, Cheung F, Liang R, Jin DY and Chim CS: Epigenetic inactivation
of the MIR129-2 in hematological malignancies. J Hematol Oncol.
6:162013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park IK, Qian D, Kiel M, Becker MW,
Pihalja M, Weissman IL, Morrison SJ and Clarke MF: Bmi-1 is
required for maintenance of adult self-renewing haematopoietic stem
cells. Nature. 423:302–305. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Molofsky AV, He S, Bydon M, Morrison SJ
and Pardal R: Bmi-1 promotes neural stem cell self-renewal and
neural development but not mouse growth and survival by repressing
the p16Ink4a and p19Arf senescence pathways. Genes Dev.
19:1432–1437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J, Chai L, Liu F, Fink LM, Lin P,
Silberstein LE, Amin HM, Ward DC and Ma Y: Bmi-1 is a target gene
for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci
USA. 104:10494–10499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK,
Choe IS, Choe YK and Kim JW: The Bmi-1 oncoprotein is overexpressed
in human colorectal cancer and correlates with the reduced
p16INK4a/p14ARF proteins. Cancer Lett. 203:217–224. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Larmonie NS, Dik WA, Beverloo HB, van
Wering ER, van Dongen JJ and Langerak AW: BMI1 as oncogenic
candidate in a novel TCRB-associated chromosomal aberration in a
patient with TCRgammadelta+ T-cell acute lymphoblastic leukemia.
Leukemia. 22:1266–1267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vonlanthen S, Heighway J, Altermatt HJ,
Gugger M, Kappeler A, Borner MM, van Lohuizen M and Betticher DC:
The bmi-1 oncoprotein is differentially expressed in non-small cell
lung cancer and correlates with INK4A-ARF locus expression. Br J
Cancer. 84:1372–1376. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jagani Z, Wiederschain D, Loo A, He D,
Mosher R, Fordjour P, Monahan J, Morrissey M, Yao YM, Lengauer C,
et al: The Polycomb group protein Bmi-1 is essential for the growth
of multiple myeloma cells. Cancer Res. 70:5528–5538. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu SQ, Xu ZZ, Niu WY, Huang HB and Zhan R:
ShRNA-mediated Bmi-1 silencing sensitizes multiple myeloma cells to
bortezomib. Int J Mol Med. 34:616–623. 2014.PubMed/NCBI
|
|
11
|
Bhattacharya R, Nicoloso M, Arvizo R, Wang
E, Cortez A, Rossi S, Calin GA and Mukherjee P: MiR-15a and MiR-16
control Bmi-1 expression in ovarian cancer. Cancer Res.
69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan
FK, Sung JJ and Yu J: MicroRNA-218 inhibits cell cycle progression
and promotes apoptosis in colon cancer by downregulating BMI1
polycomb ring finger oncogene. Mol Med. 18:1491–1498.
2013.PubMed/NCBI
|
|
14
|
Yu X, Jiang X, Li H, Guo L, Jiang W and Lu
SH: miR-203 inhibits the proliferation and self-renewal of
esophageal cancer stem-like cells by suppressing stem renewal
factor Bmi-1. Stem Cells Dev. 23:576–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lujambio A and Esteller M: CpG island
hypermethylation of tumor suppressor microRNAs in human cancer.
Cell Cycle. 6:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM,
Fernández-Piqueras J and Malumbres M: Genetic and epigenetic
silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene
expression. Cancer Cell. 13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong KY, Liang R, So CC, Jin DY, Costello
JF and Chim CS: Epigenetic silencing of MIR203 in multiple myeloma.
Br J Haematol. 154:569–578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
International Myeloma Working Group, .
Criteria for the classification of monoclonal gammopathies,
multiple myeloma and related disorders: A report of the
International Myeloma Working Group. Br J Haematol. 121:749–757.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song W, Tao K, Li H, Jin C, Song Z, Li J,
Shi H, Li X, Dang Z and Dou K: Bmi-1 is related to proliferation,
survival and poor prognosis in pancreatic cancer. Cancer Sci.
101:1754–1760. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi YJ, Choi YL, Cho EY, Shin YK, Sung
KW, Hwang YK, Lee SJ, Kong G, Lee JE, Kim JS, et al: Expression of
Bmi-1 protein in tumor tissues is associated with favorable
prognosis in breast cancer patients. Breast Cancer Res Treat.
113:83–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Leenders GJ, Dukers D, Hessels D, van
den Kieboom SW, Hulsbergen CA, Witjes JA, Otte AP, Meijer CJ and
Raaphorst FM: Polycomb-group oncogenes EZH2, BMI1, and RING1 are
overexpressed in prostate cancer with adverse pathologic and
clinical features. Eur Urol. 52:455–463. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|