Introduction
In Western industrialized nations, >25% of the
population is affected by IgE-mediated allergic disorders (1); an epidemiological study has reported
rising disease prevalence and increasing rates of allergen
sensitization worldwide (2).
Allergen-specific immunotherapy (ASIT), also referred to as
desensitization, hyposensitization or specific immunotherapy, was
introduced almost a century ago (3), and administers slowly increasing
doses of a relevant antigen until a maintenance dosage is achieved
or the patient is free of symptoms. ASIT appears to induce the
production of ‘protective substances’ that block the allergic
reaction. The thermostable protective antibodies in serum from
patients treated with ASIT are primarily IgG and are known as
‘blocking antibodies’ due to their ability to inhibit the
interaction between the allergen and IgE (4,5).
This phenomenon indicates that immune responses must be directed to
the inhibitory antibody epitopes of the allergen or antigen. Thus,
mapping of epitopes for a given allergen is useful for
understanding the immune basis of ASIT, and for designing and
developing novel allergen vaccines (5,6).
A mimotope is a macromolecule that mimics the
structure of an epitope. It may cause an immune response similar to
the one elicited by the epitope itself. A mimotope will be
recognized by an antibody against the mimicked epitope. The binding
portion of an antigen, or B-cell epitope, may be a short peptide
from the protein sequence or a patch of atoms on the protein
surface in the three-dimensional space. B-cell epitope prediction
is useful to understand the immune basis of antibody-antigen
recognition. Mimotopes that structurally mimic B-cell epitopes may
be mapped using phage-displayed random peptide libraries. Mimotopes
may then be used to develop novel diagnostics, therapeutics and
vaccines (4).
The primary source of indoor allergens worldwide is
the house dust mite, specifically Dermatophagoides
pteronyssinus and D. farinae (7). In China, a cross-sectional survey of
6,304 patients suffering from asthma and/or rhinitis in 17 cities
across mainland China revealed that 59.0 and 57.6% of participants
had positive skin prick responses for D. farinae and
D. pteronyssinus, respectively (8). Therefore, characterizing the
allergens produced by these two species is relevant to mitigating
allergic disease. Although 33 groups of house dust mite allergens
have been identified to date (www.allergen.org/), the groups 1, 2, 4, 5 and 7
constitute the known primary and mid-potency allergens (9). Allergens of groups 1 and 2 constitute
50–60% of IgE binding to house dust mite extracts; the allergens of
groups 4, 5 and 7 bind individually and together in proportion to
the primary allergens, contributing >30% of the total titer
(10). However, certain studies
have revealed that group 5 allergens from D. pteronyssinus
(Der p 5) are an important group of dust mite allergens in humans
(11–13). Furthermore, recombinant Der p 5
peptide expressed in a pGEX vector system was demonstrated to have
strong reactivity with serum IgE from >50% of asthma patients
(14). Although group 1 and 2
allergens have been well characterized, as house dust mites are
pervasive, it is important to have a good understanding of each
allergen they produce. To increase understanding of domestic mite
hypersensitivity, our laboratory cloned and expressed the dust mite
allergen Der f 5 of D. farinae (15). The present study identified
mimotopes of Der f 5 using phage-displayed random peptide libraries
against monoclonal antibodies (mAbs) specific to house dust mite
allergen Der f 5.
Materials and methods
Prokaryotic expression, purification
and renaturation of pET28a (+)-Der f 5
The plasmid pET28a (+)-Der f 5 was constructed as
described previously (15). pET28a
(+)-Der f 5 (5 ng) was used to transform BL21 (DE3) competent E.
coli cells (Agilent Technologies, Inc., Santa Clara, CA, USA).
The BL21 E. coli cells expressing pET28a (+)-Der f 5 were
cultured at 37°C overnight on lysogeny broth (LB) plates containing
50 µg/ml kanamycin. Expression was induced with isopropyl
β-D-1-thiogalactopyranoside (IPTG) as previously described
(15). Recombinant Der f 5 (rDer f
5) was isolated, purified, re-natured and verified by SDS-PAGE and
western blotting as described previously (16). The purified recombinant fusion
protein was analyzed using a 4800 matrix-assisted laser
desorption/ionization time of flight (MALDI-TOF/TOF) mass
spectrometer (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), as described previously (17). The spectra generated were
mass-calibrated using known standards and the peaks de-isotoped
(17). The masses obtained were
searched using MASCOT (SwissProt Database; Matrix Science, Ltd.,
London, UK) and a 50 ppm mass tolerance window. Significant matches
from Peptide Mass Fingerprinting were confirmed by tandem mass
spectrometry (MS/MS) using the search criteria described and an
MS/MS-tolerance window of 0.5 Da (17).
Preparation of mAbs against rDer f
5
Conventional hybridoma technology was used to
prepare mAbs against recombinant protein rDer f 5. The animal
experiments were approved by the Institutional Animal Care and
Application Committee of the Yancheng Health Vocational &
Technical College (approval no. 20121018).
Female BALB/c mice (n=6; age, 6–8 weeks) were
purchased from the Animal Testing Center of Nanjing Medical
University (Nanjing, China). Mice were maintained at room
temperature (23±2°C) at a relative humidity of 40–70% under a 12-h
light/dark cycle, with ad libitum access to food and water.
rDer f 5 (100 µg) was mixed with Freund's Complete Adjuvant
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for the first
immunization and administered via subcutaneous injection. Further
injections of 100 µg rDer f 5 were administered once every 2–3
weeks. Following 4 injections, blood samples were collected via the
tail vein. An indirect ELISA was used to determine the titer of
antiserum with the recombinant allergen rDer f 5 as the coating
antigen and horseradish peroxidase (HRP)-conjugated rabbit
anti-mouse IgG [catalog no. ab97046; Abcam Trading (Shanghai)
Company Ltd., Shanghai, China] as the secondary antibody. When the
titer became >1:10,000, 1 mouse was selected for cell
fusion.
Cell fusion was conducted with myeloma cells and
spleen cells at a ratio of 1:20. The mixed cells were placed into a
50 ml centrifuge tube, diluted with Dulbecco's modified Eagle's
medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.), and
centrifuged at 168 × g for 5 min at 4°C. The supernatant was
discarded and the cell pellet homogenized. Polyethylene glycol
(PEG; 0.8 ml, 50%) was added slowly for 90 sec, followed by 20–30
ml DMEM. The fused cells were placed into a 37°C water bath for 10
min, centrifuged at 168 × g for 5 min at 4°C, and the
supernatant discarded. DMEM containing hypoxanthine, aminopterin
and thymidine (Sigma-Aldrich; Merck Millipore) was added to the
cell pellet. The fused cells were seeded into a 96-well plate (100
µl/well) and placed into a 5% CO2 incubator. After 4
days, the plate was assessed and the cloning efficiency of
hybridoma cells was >50% with a small quantity of cell debris;
cells were healthy. The screening and analysis was performed after
10 days.
One day prior to testing, an ELISA plate was coated
with 5 µg/ml antigen (Der f 5 prokaryotic expression product) at
100 µl/well, with PBS (pH 7.4) as the coating buffer and
HRP-conjugated rabbit anti-mouse IgG as the secondary antibody. The
following day, 100 µl supernatant from the fused cells was added to
each well. The positive wells were defined as sample well optical
density (OD) value/negative well OD value ≥2.1. A single channel
pipette was used to pick positive wells detected on the whole
plate, to perform a confirmatory assessment; cells in the confirmed
positive wells were subsequently subcloned.
For subcloning, cells in the positive wells were
spread and counted. DMEM medium (4 ml total) was placed in
centrifuge tubes. Then 100 µl cell suspension was placed into each
tube, spread evenly, with 1 ml remaining in each tube. Additional
DMEM was added to make a 4 ml total volume, spread evenly, with 100
µl remaining at the bottom of each tube. DMEM (5 ml) was added to
the centrifuge tube, mixed and dropped into the first three rows of
a 96-well plate, one drop per well, with 1.8–2 ml remaining at the
bottom of the tube. A further 5 ml DMEM was added to the tube,
mixed and dropped into the middle three rows of the 96-well plate,
one drop per well, with 1.5–1.8 ml remaining at the bottom of the
tube. A further 2.8–3 ml DMEM was added to the tube, mixed and
dropped into the last two rows of the 96-well plate, one drop per
well. Cells were observed under a light microscope 7–10 days later,
to detect the wells with growing clones. Monoclonal wells were
marked. Positive monoclonal cells were selected for subcloning.
When the positive rate had reached 100%, monoclonal wells were
selected for large-scale culturing.
An intraperitoneal injection of 0.5 ml liquid
paraffin was administered to each mouse (n=6; age, 6–8 weeks).
Between days 7 and 30 following injection of liquid paraffin, the
pretreated mice were intraperitoneally injected with
1×106 hybridoma cells. Between days 7 and 10 following
injection of hybridoma cells, a syringe needle was used to remove
as much liquid as possible. The mice were sacrificed by cervical
dislocation.
The collected ascitic fluid was centrifuged, and the
supernatant collected and purified. Protein A Sepharose (GE
Healthcare Life Sciences, Chalfont, UK) was used to pack the
column. The ascitic fluid was diluted 1:10 with PBS and slowly
loaded onto the column. Phosphate buffer was used to wash the
column to the minimum value that could be detected with an
ultraviolet detector. Glycine elution buffer was used to elute the
purified antibody, which was dialyzed immediately at 4°C overnight.
The purity, concentration and titer of the antibody were determined
the following day.
Western blotting was used to verify the specificity
of the mAbs. Recombinant protein (1 and 10 ng) was loaded onto 12%
SDS-PAGE gels and transferred to nitrocellulose membranes (Tiangen
Biotech Co., Ltd., Beijing, China) following electrophoresis.
Membranes were incubated with 5% skim milk powder (50 g/l) for 1 h
at room temperature. mAbs (1:1,000) were applied to the membranes
overnight at 4°C. Membranes were washed three times, 10 min each
with PBS containing 0.1% Tween-20 (PBST). The rabbit anti-mouse
IgG-HRP (1:1,000) was applied and incubated at room temperature for
1 h. Membranes were washed with PBST three times for 15 min each
and mAb binding was visualized using 1 ml TrueBlue peroxidase
substrate (Kirkegaard & Perry Laboratories, Inc., Gaithersburg,
MD, USA) for 1 min.
Using bacteriophage library to screen
Der f 5 mimic epitope
A Ph.D.™-12 Phage Display Peptide Library (New
England BioLabs, Inc., Ipswich, MA, USA) of random dodecapeptides
fused to a minor coat protein (pIII) of M13 phage was used. The
kit-28 gIII sequencing primer was 5′-HOGTATGGGATTTTGCTAAACAAC-3′,
100 pmol, 1 pmol/µl; the −96 gIII sequencing primer was
5′-HOCCCTCATAGTTAGCGTAACG-3′, 100 pmol, 1 pmol/µl.
The immunotube (MaxiSorp™; Nalge Nunc International;
Thermo Fisher Scientific, Inc.) was coated overnight at 4°C with 10
µg mAb diluted to 1 ml with TBS. The immunotube was blocked with 5
ml 1% bovine serum albumin (BSA; Sigma-Aldrich; Merck Millipore) at
37°C for 2 h and washed three times with TBS containing 0.1%
Tween-20 (TBST). Subsequently, 3 ml phage sample (containing 0.5%
BSA and 0.1% Tween-20) was added at 37°C for 2 h, with mixing every
30 min. The immunotube was again washed with TBST 10 times to
remove the unbound phage. Bound phage was washed with 1 ml
glycine-HCl (pH 2.1) for 5 min and 160 µl Tris neutralization
solution was added. Elution was repeated and eluents combined. The
eluted phage sample (10 µl) was used for gradient dilution. ER2738
competent E. coli cells provided with the Ph.D.-12 Phage
Display Peptide Library were infected during mid-log phase, coated
onto an LB/IPTG/Xgal Top-Agar plate and inverted in a 37°C
incubator. Cells were counted the following day. The single phage
plaque was used for subsequent experiments. The remaining 2.3 ml
eluted phage samples were used to infect 50 ml ER2738 during
early-log phase. Cells were oscillated and cultured at 37°C for 4.5
h to collect phage supernatants. These samples were used for the
next round of screening following precipitation and
concentration.
The phage was diluted with LB medium at a 10:1
ratio. The amplified phage monoclones were diluted to
1.0×108-1.0×1011, and the unamplified were
diluted to 1.0×101-1.0×104. Phage monoclones
(10 µl) at various dilutions were selected and added to 190 µl
ER2738 bacterial solution of mid-log phase (A600=1.0). Samples were
mixed and poured into a preheated culture tube at 45°C, with an
agar top layer. Culture tubes were gently shaken, and the solution
was poured onto an LB plate containing IPTG and Xgal. The plate was
inverted at 37°C overnight.
Monoclones were selected from LB/IPTG/Xgal plates
following the third round of screening and seeded onto a 96-well
deep well plate, with 500 µl LB (diluted at 1:100 with E.
coli ER2738) in each well. Samples were cultured at 37°C for
4.5 h and centrifuged at 42 × g for 5 min at 4°C.
Supernatant was collected to perform ELISA. Briefly, plates were
coated with 200 ng/well mAb and incubated overnight at 4°C.
Following blocking with 5% skim milk at 37°C for 1 h, the amplified
enriched phage clones were added and incubated at 37°C for 1 h.
Plates were washed and HRP-labeled anti-M13 phage IgG, provided
with the Ph.D.-12 Phage Display Peptide Library, diluted 1:5,000
was added at 100 µl/well, and incubated at 37°C for 30 min. Color
was developed with a tetramethylbenzidine (Kirkegaard & Perry
Laboratories, Inc.) chromogenic substrate system. The OD450 value
was determined following termination of the reaction.
Phages with greater OD values were selected and
single-stranded DNA was extracted using odium iodide according to
the kit manufacturer's protocol. Sequencing with primer-96 gIII,
5′-CCCTCATAGTTAGCGTAACG-3′ was performed. The obtained DNA
sequences were inverted, converted and translated into amino acid
sequences, which were used for analysis.
Results
Expression and purification of pET28a
(+)-Der f 5
BL21 E. coli cells expressing the pET28a
(+)-Der f 5 plasmid were used to isolate recombinant Der f 5.
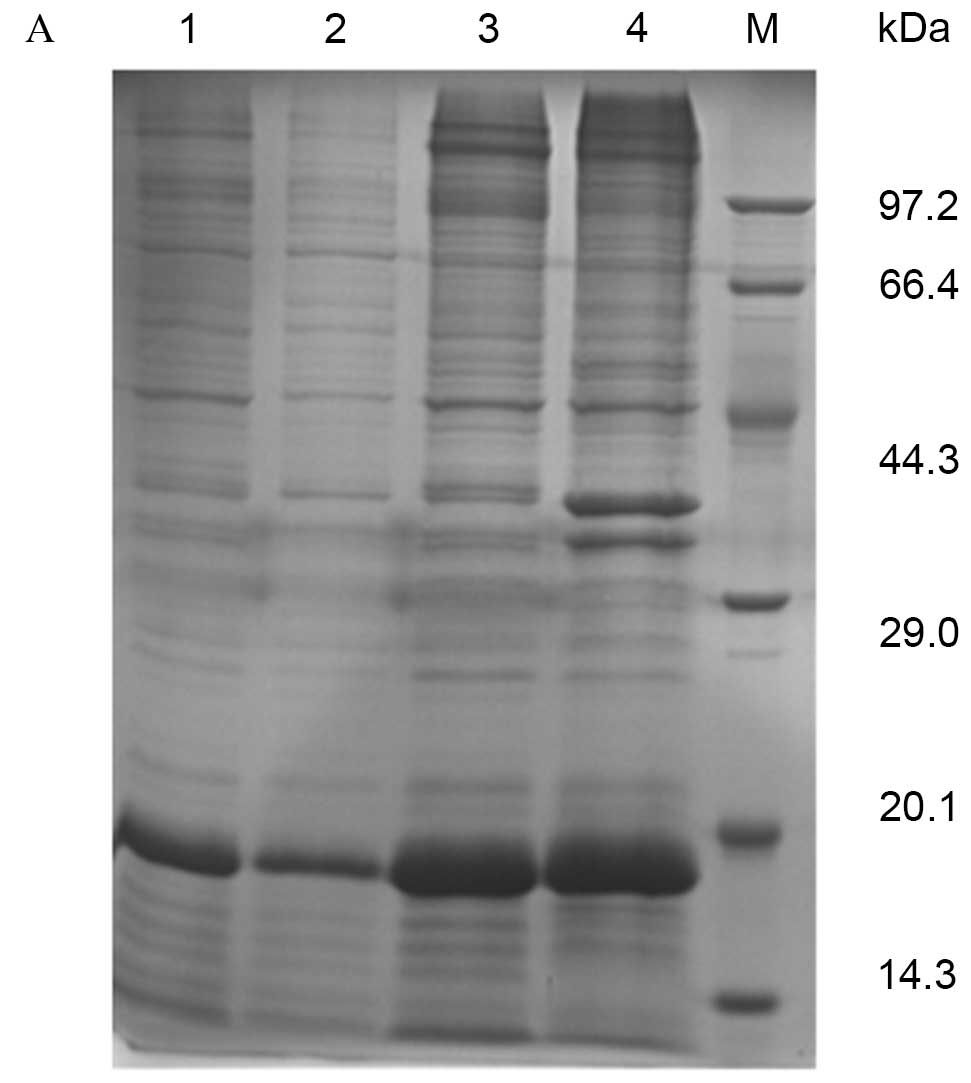
SDS-PAGE revealed specific bands at 15–20 kDa, estimated to be the
fusion protein of Der f 5 and the vector pET28a (+). Recombinant
protein was purified by affinity chromotagraphy, resulting in a
purity of 90% and a concentration of 1 mg/ml (Fig. 1A). MALDI-TOF/TOF revealed a peptide
mass fingerprint consistent with the structure of Der f 5 (Fig. 1B).
Preparation and identification of
mAbs
Mice were immunized with the pET-28a (+)-Der f 5
prokaryotic expression product. The titers of antisera for the
recombinant protein were determined via indirect ELISA and the
mouse with the greatest titer was selected for cell fusion. The
spleen cells and Sp2/0 myeloma cells of the immunized mice were
used to perform PEG fusion. Indirect ELISA determined the antibody
secretion status in the cell culture supernatant. The positive
clones were cultured according to the limiting dilution method, to
obtain 3 continuously secreting specific anti-Der f 5 hybridoma
cell lines, named 3G9, 6B8 and 10D6. The cell lines were prepared
into ascitic fluid, and ELISA was used to determine the titer
(Fig. 2A).
Following purification, the ascitic fluid (3 ml) was
collected with concentrations of 0.7 mg/ml 3G9, 0.4 mg/ml 6B8 and
0.67 mg/ml 10D6. SDS-PAGE revealed that the purity of the visible
antibody following purification was >85% (Fig. 2B). The recombinant protein (rDer f
5) was probed with mAbs 3G9, 6B8 and 10D6 by western blotting; all
three mAbs bound to the recombinant protein (Fig. 2C).
Screening Der f 5 mimic epitopes in
the phage library
The mAbs against recombinant Der f 5 were used to
screen the random peptide library. Following three rounds of strict
screening, the recovery rate of 6B8 had increased from
2.1×10−7 to 3.7×10−5. Following four rounds
of screening, the recovery rate of 3G9 had increased from
1.2×10−6 to 3.5×10−4. Following four rounds
of screening, the recovery rate of 10D6 had increased from
8.3×10−7 to 3.1×10−6. The results indicated
that the specific phage clones had been enriched to varying
degrees.
Following three cycles of screening, the supernatant
of antibody 6B8 was used for ELISA; 15 positive clones were
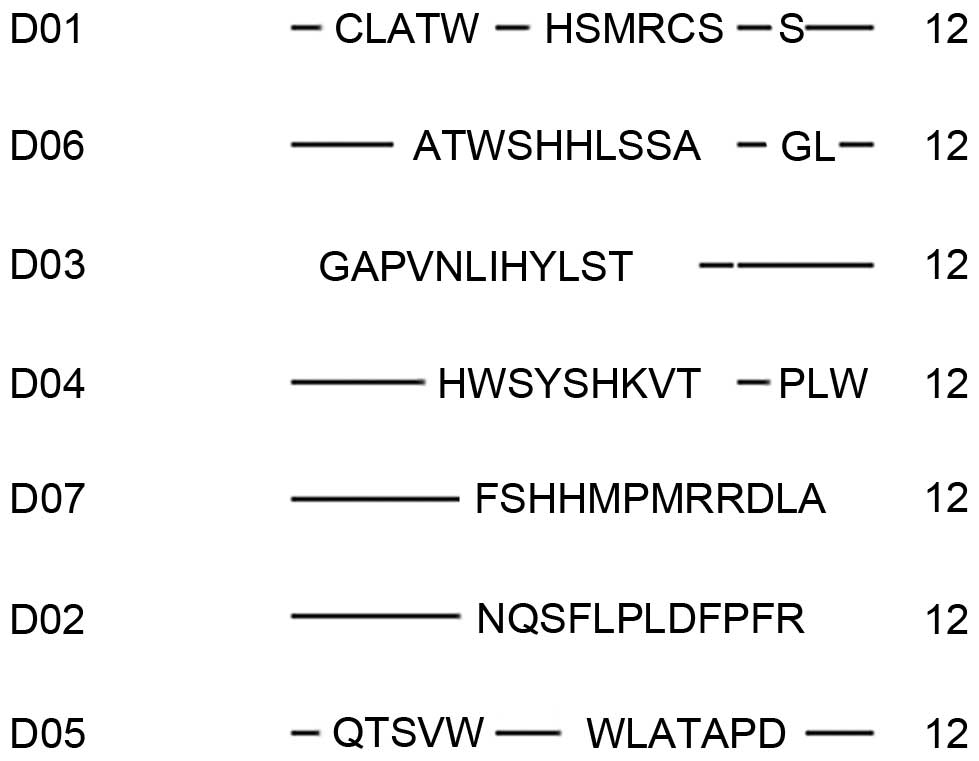
obtained. The sequencing results for these clones were consistent,
all ‘CLATWHSMRCSS’ (Table I). The
polypeptide screening of mAbs 3G9 and 10D6 performed following the
third screening cycle revealed relatively weaker positive clone
signals or a reduced positive rate. Therefore, a fourth cycle of
screening was performed on these two clones. Following three cycles
of screening, antibody 3G9 produced 7 positive clones, all
‘NQSFLPLDFPFR’ and antibody 10D6 produced 13 positive clones, with
sequencing results as follows: ‘HWSYSHKVTPLW’ (1 positive clone),
‘QTSVWWLATAPD’ (7 positive clones) and ‘ATWSHHLSSAGL’ (5 positive
clones; Table I).
 | Table I.Amino acid sequences of positive
clones. |
Table I.
Amino acid sequences of positive
clones.
| Monoclonal
antibody | Random peptide
sequences | Frequency | OD values in
ELISA | Formula | Theoretical pI | GRAVY |
|---|
| 6B8 | CLATWHSMRCSS | 15 | 0.895~1.140 |
C56H88N18O17S3 | 8.08 | 0.067 |
| 3G9 | NQSFLPLDFPFR | 7 | 0.194–0.258 |
C71H101N17O18 | 5.84 | −0.250 |
|
| CLATWHSMRCSS | 4 | 0.450–0.709 |
C56H88N18O17S3 | 8.08 | 0.067 |
|
| GAPVNLIHYLST | 1 | 0.469 |
C59H93N15O17 | 6.74 | 0.550 |
| 10D6 | HWSYSHKVTPLW | 4 | 0.171–0.406 |
C75H101N19O17 | 8.61 | −0.775 |
|
| QTSVWWLATAPD | 7 | 0.405–0.799 |
C64H91N15O19 | 3.80 | −0.083 |
|
| ATWSHHLSSAGL | 5 | 0.348–0.524 |
C56H83N17O17 | 6.96 | 0.033 |
|
| FSHHMPMRRDLA | 1 | 1.147 |
C64H100N22O16S2 | 9.61 | −0.758 |
Following four cycles of screening, there were 2
sequences of positive clones from antibody 3G9: ‘CLATWHSMRCSS’ (4
positive clones) and ‘GAPVNLIHYLST’ (1 positive clone). In
addition, there were 2 sequences from antibody 10D6: ‘HWSYSHKVTPLW’
(3 positive clones) and ‘FSHHMPMRRDLA’ (1 positive clone). The 7
mimic epitopes obtained in total from all 3 mAbs were entered into
MIMOX (www.immunet.cn/mimox) for comparison;
results are presented in Fig. 3.
‘WH’ had the greatest frequency, with its derived common
subsequence ‘---[-A][-T]W[-S]H[HSFW][LM][PSKR][TLV][AST]-[DP]
[-L]-’. Specifically, the frequency of amino acid W at the sixth
position was 57%; the possibility of amino acid S at the seventh
position was 43%; the possibility of amino acid H at the eighth
position was 57%; the possibility of amino acid L at the tenth
position was 57%; and the possibility of amino acid L at the
sixteenth position was 43%. The Der f 5 amino acid sequence was
entered into ElliPro (tools.immuneepitope.org/tools/ElliPro/iedb_input). This
predicted that it may include 3 discontinous epitopes, located at
1–14, 68–84 and 38–47. These results suggested that P2, K3, K4, H5,
F11, F13, L14, R72, T77, L79, R84, T39, F40, P44, T45 and K46 were
the key amino acids.
Discussion
With the increasing prevalence of allergic disorders
resulting from house dust mite allergens, investigations are
focused on characterizing allergens and their epitopes, for the
development of novel and more effective specific immunotherapies. A
relatively novel technique, phage display, creates a mimic of a
natural epitope, referred to as a mimotope, to permit its
characterization (18). Phage
display libraries are based on random peptides that may be used to
mimic the binding site for an antibody. In the present study, three
mAbs were successfully raised against recombinant Der f 5: 6B8, 3G9
and 10D6. To locate the binding site of these mAbs, a phage surface
capsid protein displayed 12-mer peptide library was used to search
for sequences. Results revealed that the mAb 6B8 recognizes a
sequence ‘CLATWHSMRCSS’, the mAb 3G9 recognizes the three sequences
‘NQSFLPLDFPFR’, ‘CLATWHSMRCSS’ and ‘GAPVNLIHYLST’, and the mAb 10D6
recognizes the four sequences ‘HWSYSHKVTPLW’, ‘QTSVWWLATAPD’,
‘ATWSHHLSSAGL’ and ‘FSHHMPMRRDLA’. A phage surface capsid protein
display 12-mer peptide library consists of ~109
electroporated sequences amplified once to yield ~100 copies of
each sequence in 10 µl of the supplied phage. The peptides
recognized by these three mAbs have certain residues in common with
the sequences in Der f 5. Following alignment of the seven
sequences recognized by these three mAbs, the amino acids ‘WH’ were
revealed to have the greatest frequency. The common subsequence was
deduced to be ‘---[-A][-T]W[-S]H[HSFW][LM][PSKR][TLV][AST]-[DP]
[-L]- ’. These seven mimotopes may serve as a treatment vaccine to
be developed for immunotherapy.
Mimotope analysis-based methods may predict linear
and conformational epitopes and have therefore become more widely
used. Although algorithms have been suggested, identifying the
exact localization of the interaction site mimicked by mimotopes
remains an obstacle. The present study predicted the epitopes for
Der f 5 allergen using ElliPro, an online tool that implements
Thornton's method, together with a residue clustering algorithm,
the MODELLER program and the Jmol viewer (19). Three discontinuous epitopes,
located at residues 1–14, 68–84 and 38–47, were identified. The
common subsequence deduced from the seven mimotopes, combined with
the three discontinous epitopes predicted by ElliPro, resulted in
the prediction of key residues at P2, K3, K4, H5, F11, F13, L14,
R72, T77, L79, R84, T39, F40, P44, T45 and K46. Therefore,
modifying these key amino acids may be beneficial for
epitope-specific immunotherapy.
Acknowledgements
The present study was supported by the National
Sciences Foundation of China (grant nos. NSFC 30060166,
NSFC81001330 and NSFC31272369).
References
|
1
|
Valenta R: The future of antigen-specific
immunotherapy of allergy. Nat Rev Immunol. 2:446–453.
2002.PubMed/NCBI
|
|
2
|
Leung TF and Wong GW: The Asian side of
asthma and allergy. Curr Opin Allergy Clin Immunol. 8:384–390.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Freeman J: Further observation on the
treatment of hay fever by hypodermic inoculations of pollen
vaccine. Historical document. Ann Allergy. 18:427–434.
1960.PubMed/NCBI
|
|
4
|
van Neerven RJ, Knol EF, Ejrnaes A and
Würtzen PA: IgE-mediated allergen presentation and blocking
antibodies: Regulation of T-cell activation in allergy. Int Arch
Allergy Immunol. 141:119–129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Knittelfelder R, Riemer AB and
Jensen-Jarolim E: Mimotope vaccination-from allergy to cancer.
Expert Opin Biol Ther. 9:493–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tanabe S: Epitope peptides and
immunotherapy. Curr Protein Pept Sci. 8:109–118. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cui Y: Immunoglobulin E-binding epitopes
of mite allergens: From characterization to immunotherapy. Clin Rev
Allergy Immunol. 47:344–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Sun B, Huang Y, Lin X, Zhao D, Tan
G, Wu J, Zhao H, Cao L, Zhong N, et al: China Alliance of research
on respiratory allergic disease: A multicentre study assessing the
prevalence of sensitizations in patients with asthma and/or
rhinitis in China. Allergy. 64:1083–1092. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thomas WR, Heinrich TK, Smith WA and Hales
BJ: Pyroglyphid house dust mite allergen. Prot Pept Lett.
14:943–953. 2007. View Article : Google Scholar
|
|
10
|
Vrtala S, Huber H and Thomas WR:
Recombinant house dust mite allergens. Methods. 66:67–74. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lynch NR, Thomas WR, Garcia NM, Di Prisco
MC, Puccio FA, L'opez RI, Hazell LA, Shen HD, Lin KL and Chua KY:
Biological activity of recombinant Der p 2, Der p 5 and Der p 7
allergens of the house-dust mite Dermatophaogides pteronyssinus.
Int Arch Allergy Immunol. 114:59–67. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomas WR, Smith WA, Hales BJ, Mills KL
and O'Brien RM: Characterization and immunobiology of house dust
mite allergens. Int Arch Allergy Immunol. 129:1–18. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weghofer M, Grote M, Dall'Antonia Y,
Fernández-Caldas E, Krauth MT, van Hage M, Horak F, Thomas WR,
Valent P, Keller W, et al: Characterization of folded recombinant
Der p 5, a potential diagnostic marker allergen for house dust mite
allergy. Int Arch Allergy Immunol. 147:101–109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin KL, Hsieh KH, Thomas WR, Chiang BL and
Chua KY: Characterization of Der p 5 allergen, cDNA analysis, and
IgE-mediated reactivity to the recombinant protein. J Allergy Clin
Immunol. 94:989–996. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui Y, Zhou Y, Ma G, Yang L, Wang Y and
Shi W: Cloning, bioinformatics analysis, and expression of the dust
mite allergen Der f 5 of Dermatophagoides farinae. Braz J Med Biol
Res. 45:746–752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu-bao C, Zhou Y, Weihong S, Guifang M,
Yang L and Yungang W: Cloning, expression, and analysis of the
group 2 allergen from Dermatophagoides farinae from China. An Acad
Bras Cienc. 82:941–951. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui YB, Jiang Y, Ji Y, Zhou Y, Yu L, Wang
N, Yang L and Zhang C: Cloning, expression, and analysis of a cDNA
coding for the Dermatophagoides farinae group 21 (Der f 21)
allergen. Am J Transl Res. 6:786–792. 2014.PubMed/NCBI
|
|
18
|
Pande J, Szewczyk MM and Grover AK: Phage
display: Concept, innovations, applications and future. Biotechnol
Adv. 28:849–858. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ponomarenko J, Bui HH, Li W, Fusseder N,
Bourne PE, Sette A and Peters B: ElliPro: A new structure-based
tool for the prediction of antibody epitopes. BMC Bioinformatics.
9:5142008. View Article : Google Scholar : PubMed/NCBI
|