Introduction
Bacterial infectious pneumonia is one of the major
causes of mortality in neonates, particularly when they suffer from
ventilator-associated pneumonia (VAP). However, it is difficult to
detect the pathogens involved in pneumonia (1–3).
Thus, it is important to understand the diversity and associations
among microflora to improve clinical guidelines for detecting the
pathogens of bacterial infectious pneumonia and VAP (4).
16S rDNA polymerase chain reaction (PCR)-denaturing
gradient gel electrophoresis (DGGE) is one of the important methods
used to investigate the diversity of microflora, and the gold
standard for bacterial detection and classification is sequence
analysis based on 16S rDNA (5–7).
Thus, the present study focused on examining the diversity of the
microflora in the lower respiratory tract in order to elucidate the
reasons neonates suffer from bacterial infectious pneumonia and
VAP.
Materials and methods
Ethics statement
Prior to commencement of the present study,
agreement was obtained from the Medical Ethics Committee of
Chongqing Medical University (Chongqing, China), and the parents of
the neonates provided signed informed consent prior to the
experiment.
Patient selection
Newborns from the neonatal intensive care unit of
the Children's Hospital of Chongqing Medical University between
January 2012 and December 2012 were included in the present study.
Newborn patients suffering from bacterial infectious pneumonia
without VAP (IP group) and those suffering from bacterial
infectious pneumonia combined with VAP (IVAP group) were included
as the experimental groups, and those suffering from RDS without
VAP (RDS group), were included as a positive control group. The
negative control group comprised filter-sterilized double-distilled
water for the extraction of DNA and subsequent PCR (7). There were 19 patients in the IP
group, including 15 male and 4 female, and the gestational age was
37.1±3.3 weeks. There were 8 patients in the IVAP group, including
7 male and 1 female, the gestational age was 30.3±3.9 weeks.
The following exclusion criteria were used for RDS:
Intrauterine infection, infectious diseases prior to intubation,
and mothers with a history of infections or who had used
antibiotics during the last month of pregnancy.
The following diagnostic criteria were used for VAP:
Presence of rales or knock turbidity, with emerging purulent
sputum, positive blood culture or epidemic strains isolated by
endotracheal suction, and the presence of emerging pulmonary
infiltrates, consolidation, cavities or pleural effusions, as
indicated by X-ray examination (8–10).
Sample collection
Following 1, 3 and 5 days of ventilation, sputum
samples were collected from the patients. The suction tube of the
sputum culture collector was placed deep into the collection tube
and, using negative pressure, 1–2 ml of sputum was aspirated. If
the sputum thickness prohibited sputum collection, 1–3 ml of
sterile saline was injected into the endotracheal tube, followed by
five breathing cycles of the patients. Once the patient's oxygen
saturation recovered, aspiration of sputum recommenced. All
specimens were stored at −20°C (11).
DNA extraction
The sputum samples were centrifuged at 4°C and 1,000
× g for 1 min, following which the supernatant was removed
and the pellet was resuspended in 2 ml of sterile saline. The
sample was mixed and centrifuged again, in accordance with the
above methods. Following two washes with sterile saline, the sample
was analyzed using the Mini BEST Bacterial Genomic DNA Extraction
kit (Ver2.0; Takara Biotechnology Co., Ltd., Dalian, China), in
accordance with the manufacturer's protocol The negative control
groups contained filter-sterilized double-distilled water for the
extraction of DNA and PCR.
PCR amplification
The bacterial universal primers, designed according
to the conserved V3 region of the bacterial 16S rDNA gene were as
follows: 357, forward
5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCAGGGGCCTACGGGAGGCAGCAG-3′
(including the 37 bp ‘GC’ cap) and 518, reverse
5′-ATTACCGCGGCTGCTGG-3′. The amplifications were performed using an
Eppendorf PCR machine (Eppendorf, Hamburg, Germany). The reaction
volume was 50 µl and included 6 µl template DNA, 25 µl Premix Taq
Version 2.0 (Takara Bio, Inc., Otsu, Japan), 0.5 µl each primer and
18 µl sterile ddH2O. The following reaction conditions
were used: Initial denaturation at 94°C for 5 min; 10 cycles of
denaturation at 94°C for 30 sec, annealing at 61–56°C
(−0.5°C/cycle) and extension at 72°C for 1 min; 25 cycles of
denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec and
extension at 72°C for 1 min; and a final extension at 72°C for 7
min. A 2% agarose gel, with 1X TAE (Tris base, acetic acid and
EDTA) and 4S Green (Shanghai Biological Engineering Co., Ltd.,
Shanghai, China), was used to resolve the PCR products from the 5
µl. The target bands were ~195 bp in size. The remaining PCR
products were stored at −20°C.
DGGE
A DCode system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used to analyze the DGGE images. Each PCR
product (25 µl) was separated on an 8% polyacrylamide gel with a
35–65% linear gradient of urea and formamide by electrophoresis at
85 V and 60°C for 16 h. SYBR Green I (Tektronix Biotechnology Co.,
Ltd., Beijing, China) was used to stain the gel, and a Herolab
UVT-20 M/W ultraviolet transilluminator was used to the image the
gel. All the bands were excised, washed twice with 500 µl sterile
ddH2O, mashed, placed in 30 µl nuclease-free water and
stored at 4°C overnight to elute the DNA. The supernatants were
amplified with primers lacking the ‘GC’ cap under the same reaction
conditions as described above. A 0.8% agarose gel was prepared with
1X TAE and 4S Green (Shanghai Biological Engineering Company). All
the amplification products were electrophoresed at 110 V for 20
min. An Agarose Gel DNA Purification k (version 2.0; Takara Bio,
Inc.) was used to recover the DNA in the target bands and the DNA
was stored at −20°C (11).
Cloning and sequencing
A PMD18-T Vector system (Takara Bio, Inc.) was used
to clone the PCR amplicons into a plasmid, and the Escherichia
coli DH5α-competent cells (Tiangen, China) were transformed by
the resulting clones. Luria Broth (LB) media containing ampicillin
was used to culture the cells overnight at 37°C, following which 1
ml of this liquid was sent to Shanghai Biological Engineering Co.,
Ltd. for sequencing. The Basic Local Alignment Search Tool
(blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare
the results with the nucleotide databases in the National Center
for Biotechnology Information GenBank (www.ncbi.nlm.nih.gov/).
Bacterial culture
The collected sputum oscillating fluid samples were
inoculated onto Columbia blood agar plates and separate
Haemophilus influenzae plates, and were cultured for 18–48 h
at 37°C. Gram staining was used to stain the resulting colonies,
which were identified using a MicroScan WalkAway-40 (Siemens AG,
Berlin, Germany) automated bacterial identification and
susceptibility instrument.
Analysis of diversity and
similarity
The numbers and the similarity of the bands from the
DGGE images were measured using Quantity One version 4.62 (Bio Rad
Laboratories, Inc.) software. The unweighted pair group method with
arithmetic averages was used to analyze the cluster maps, and
BIO-DAP version 2.0 software was used to calculate the
Shannon-Wiener diversity index (Shannon-Wiener index) (11–13).
Statistical analysis
The data were analyzed using SPSS 17.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Data with a normal
distribution are expressed as the mean± standard deviation.
Comparisons between two groups were performed using an independent
sample t-test, and comparisons between several groups were
performed using one-way analysis of variance or a pairwise least
significant difference t-test when the variance was homogeneous.
When the variance was heterogeneous, a non-parametric test was
used. Data without a normal distribution are expressed as P50
(P25-P75), and were analyzed using the non-parametric test method.
Count data were analyzed using a χ2 test when n≥40 and
T≥1, and the extract method was used when n<40 or T<1.
P<0.05 was considered to indicate a statistically significant
difference. The pairwise comparisons of multiple count data were
performed when the test level was fixed.
Results
Clinical characteristics
A total of 42 newborn patients were suitable for
participation in the present study due to meeting the inclusion
criteria, which included 15 patients in the RDS group, 19 in the IP
group and eight in the IVAP group. The clinical data for the
experimental groups (IP group and IVAP group) are listed in
Table I. Cefoxitin was used
empirically in the patients in the RDS group. The patients in of
the IP and IVAP groups with positive culture results were treated
according to the susceptibility data, and those with negative
culture results were treated according to empirical treatments. The
patients in the IVAP group were considered to be VAP within 1.9–6.5
days (P50=3.2) following intubation. The gestational age was lower
and the intubation duration was longer for the patients in the IVAP
group, compared with those in the IP group.
 | Table I.Clinical characteristics of patients
in the IP and IVAP groups. |
Table I.
Clinical characteristics of patients
in the IP and IVAP groups.
| Characteristic | IP (n=19) | IVAP (n=8) | Statistical
analysis | P-value |
|---|
| Gender (male) | 15 | 7 | – | 1.000a |
| Gestational age
(weeks)c | 37.1±3.3 | 30.3±3.9 | t=8.550 | 0.007 |
| Birth weight
(g)c | 2,773±450 | 1,876±667 | t=3.871 | 0.060 |
| Total intubation
duration [P50 (P25-P75), days] | 4.25
(2.90–5.10) | 14.70
(8.10–17.70) | – | 0.000b |
| Sputum culture
results |
|
| – | 0.000a |
| Negative (n) | 18 | 6 | – | – |
| Normal flora
(n) | 7 | 3 | – | – |
| Klebsiella
pneumoniae subspecies (n) | 4 | 9 | – | – |
| Acinetobacter
baumannii (n) | 0 | 3 | – | – |
| Pseudomonas
aeruginosa (n) | 0 | 2 | – | – |
| Antibiotic |
Cefamandole/Cefoperazone,
sulbactam/Panipenem, betamipron/Cefpiramide/Piperacillin,
tazobactam/Latamoxef |
Cefamandole/Cefoperazone,
sulbactam/Panipenem, betamipron/Cefpiramide/Piperacillin,
tazobactam/Tienam/Metronidazole/Amphotericin/Fluconazole/Ciprofloxacin/Vancomycin | – | – |
| Prognosis |
|
| – | 1.000a |
| Improved and
recovered (n) | 15 | 6 | – | – |
| Succumbed to
mortality (n) | 4 | 2 | – | – |
A total of 73 sputum samples were sent for culture
in the Children's Hospital of Chongqing Medical University, and the
detection ratio was 54.4%. The detection ratio for the IP group was
lower, compared with for the IVAP group.
Sample collection, DNA extraction and
PCR
A total of 73 samples were collected. The target
bands (~195 base pairs) were obtained from 73 sputum samples
(82.0%), which are presented in Table
II. No target band was generated from the negative control
group samples.
 | Table II.Groupings based on the DNA
amplification of 73 sputum samples. |
Table II.
Groupings based on the DNA
amplification of 73 sputum samples.
| Day | RDS | IP | IVAP |
|---|
| 1 | 10 | 15 | 7 |
| 3 | 7 | 12 | 6 |
| 5 | 5 | 6 | 5 |
| Total | 22 | 33 | 18 |
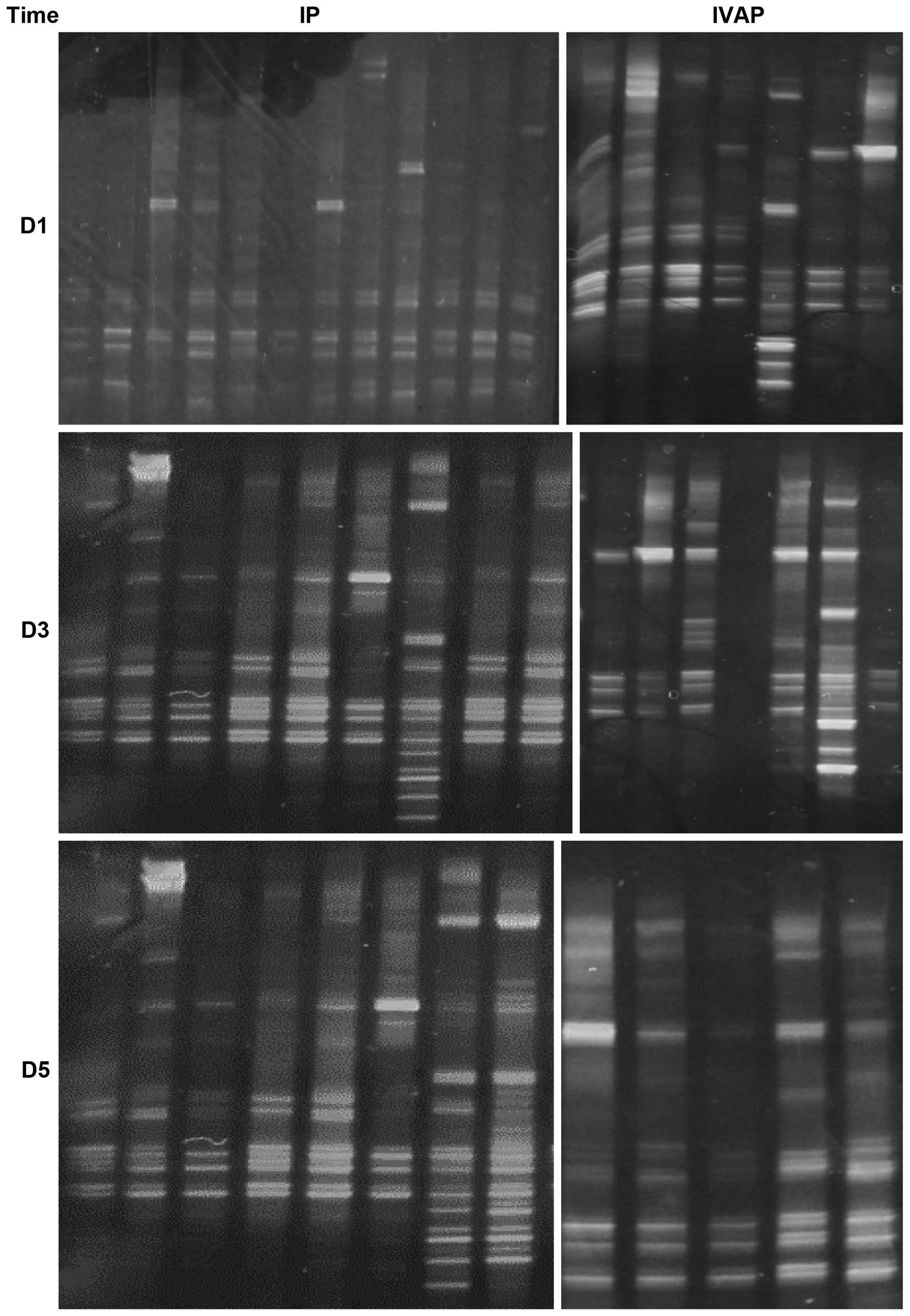
DGGE images
Visible bands were found in all 73 sputum samples,
as shown in Fig. 1.
Sequencing results
A total of 11 species were detected following the
isolation, cloning and sequencing of the DGGE bands (Table III).
 | Table III.Sequencing results of the bands in
the denaturing gradient gel electrophoresis images. |
Table III.
Sequencing results of the bands in
the denaturing gradient gel electrophoresis images.
| NCBI BLAST
result | Accession
number | Identity (%) |
|---|
| Serratia
sp. | KC182731.1 | 100 |
|
Achromobacter sp. | HE613447.1 | 100 |
| Klebsiella
sp. | KC354804.1 | 100 |
|
Staphylococcus sp. | JX849039.1 | 100 |
|
Acinetobacter sp. | KC245151.1 | 99 |
|
Streptococcus sp. | JX861486.1 | 100 |
| Pseudomonas
sp. | KC415769.1 | 100 |
| Macrococcus
sp. | HQ238716.1 | 100 |
|
Brevundimonas sp. | JX950099.1 | 99 |
| Actinomyces
sp. | HM854563.1 | 99 |
| Schlegel
sp. | AY538706.1 | 99 |
Analysis of diversity
The bacterial diversity of a sample is determined by
the number of bands in a lane; if the former is high, the latter is
high (14). In the present study,
the number of bands in each group were determined, as shown in
Fig. 2. As indicated by the
results, with prolonged intubation, alterations in the diversity
was observed in each group, as follows: The diversity of the RDS
group initially increased and then leveled off, whereas the IP
group showed a gradual increase in diversity. The IVAP group
initially decreased, followed by an increase. Comparison between
the groups within the same time period indicated no differences in
diversity within the first day of intubation, and the levels of
diversity were in the order of RDS group > IP group > IVAP
group on days 1–5 post-intubation. The data of the Shannon-Wiener
index, shown in Fig. 3, showed
that the overall trends and associations were consistent with the
above levels of diversity.
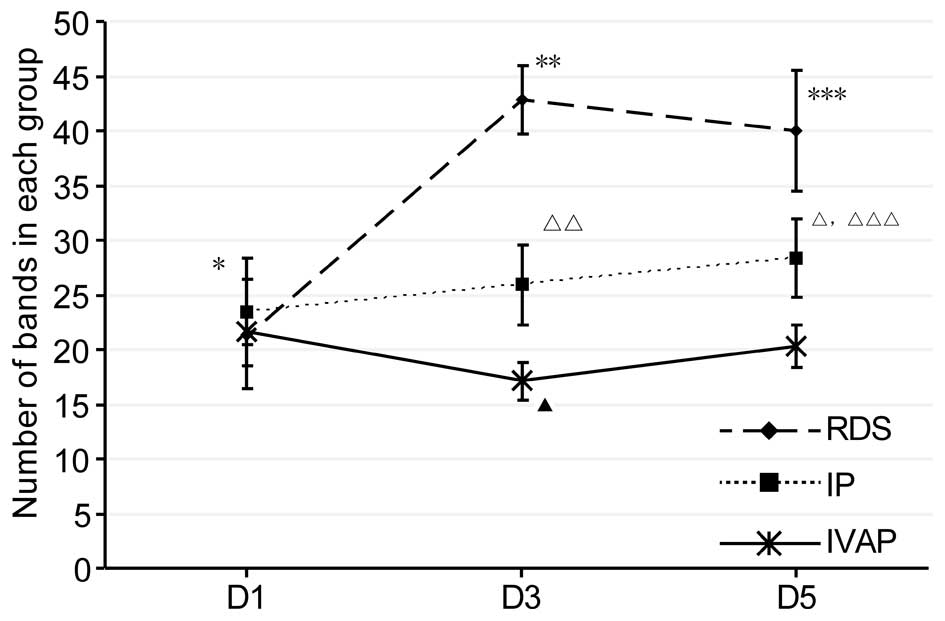 | Figure 2.Number of bands in each group. Data
are expressed as the mean ± standard deviation. RDS, respiratory
distress syndrome; IP, bacterial infectious pneumonia; IVAP,
bacterial infectious pneumonia with ventilator-associated
pneumonia; D, day. *P<0.05, RDS D1 vs. RDS D3 and RDS D5 groups;
**P<0.05, RDS D3 vs. IP D3 and IVAP D3 groups; ***P<0.05, RDS
D5 vs. IP D5 and IVAP D5 groups; △P<0.05, IP D5 vs.
IP D1 and IP D5 groups; △△P<0.05, IP D3 vs. IVAP D3
group; △△△P<0.05, IP D5 vs. IVAP D5 group,
▲P<0.05, IVAP D3 vs. IVAP D1 and IVAP D5 groups. |
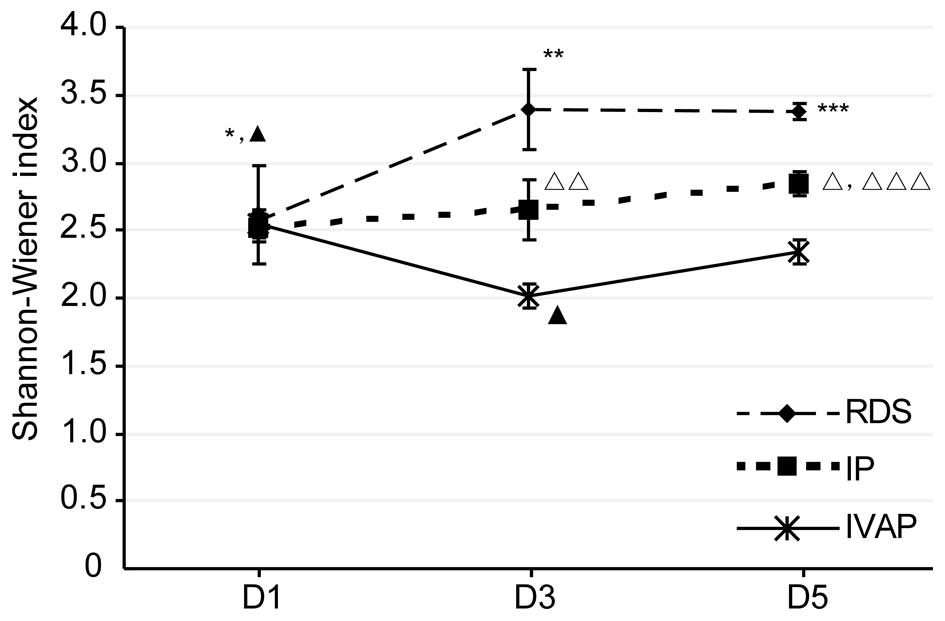 | Figure 3.Shannon-Wiener Index for each group.
Data are expressed as the mean ± standard deviation. RDS,
respiratory distress syndrome; IP, bacterial infectious pneumonia;
IVAP, bacterial infectious pneumonia with ventilator-associated
pneumonia; D, day. *P<0.05, RDS D1 vs. RDS D3 and RDS D5 groups;
**P<0.05, RDS D3 vs. IP D3 and IVAP D3 groups; ***P<0.05, RDS
D5 vs. IP D5 and IVAP D5 groups; △P<0.05, IP D5 vs.
IP D1 and IP D5 groups; △△P<0.05, IP D3 vs. IVAP D3
group; △△△P<0.05, IP D5 vs. IVAP D5 group,
▲P<0.05, IVAP D1 vs. IVAP D3 and IVAP D5 groups;
▲▲P<0.05, IVAP D3 vs. IVAP D5 group. |
Similarity analysis of microflora
As shown in Fig. 4,
the similarity results of each group were examined. With prolonged
intubation, there was no significant change in the similarity of
the RDS group in the lower respiratory tract, whereas the
similarity levels of the other two groups were maintained at an
initial increased level. Comparison between the groups within the
same time period revealed no differences in similarity within the
first day of intubation, whereas the similarity levels on days 1–5
post-intubation were in the order, IVAP group > IP group >
RDS group.
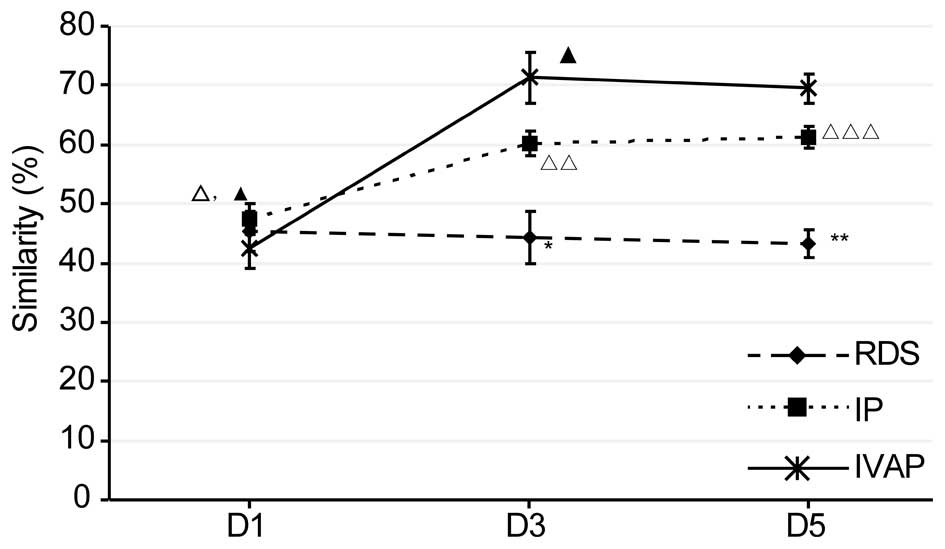 | Figure 4.Similarity of the sputum in each
group. Data are expressed as the mean ± standard deviation. RDS,
respiratory distress syndrome; IP, bacterial infectious pneumonia;
IVAP, bacterial infectious pneumonia with ventilator-associated
pneumonia; D, day. *P<0.05, RDS D3 vs. IP D3 and IVAP D3 groups;
**P<0.05, RDS D5 vs. IP D5 and IVAP D5 groups;
△P<0.05, IP D1 vs. IVAP D3 group;
△△P<0.05, IP D3 vs. IVAP D3 group;
△△△P<0.05, IP D5 vs. IVAP D5 group,
▲P<0.05, IVAP D1 vs. IVAP D3 and IVAP D5 groups. |
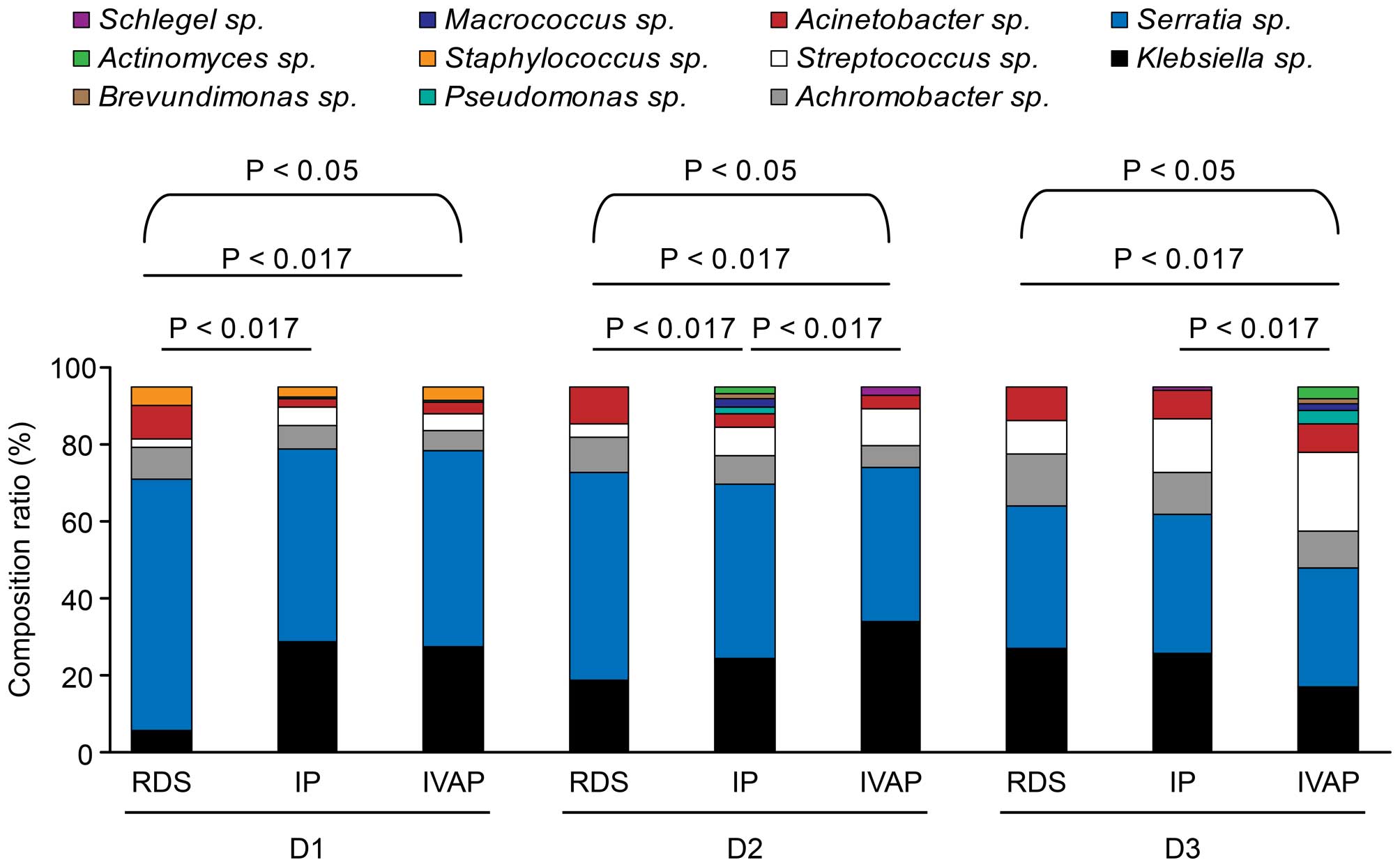
Composition of the microflora
The variety of the genera in each group and their
composition ratios are shown in Table
IV and Fig. 5.
 | Table IV.Composition ratios of the genera in
each group. |
Table IV.
Composition ratios of the genera in
each group.
|
| RDS | IP | IVAP |
|---|
|
|
|
|
|
|---|
| Genus | D1 | D3 | D5 | D1 | D3 | D5 | D1 | D3 | D5 |
|---|
| Serratia
(%) | 5.5 | 19.5 | 27.9 | 14.8 | 25.4 | 27.0 | 16.5 | 35.3 | 15.4 |
|
Achromobacter (%) | 69.1 | 57.2 | 39.5 | 52.9 | 47.8 | 37.9 | 54.2 | 42.4 | 28.5 |
| Klebsiella
(%) | 9.1 | 9.7 | 14.0 | 6.6 | 8.0 | 11.8 | 5.2 | 6.1 | 8.7 |
|
Staphylococcus (%) | 2.0 | 3.4 | 9.3 | 5.3 | 7.9 | 14.4 | 4.7 | 10.0 | 19.1 |
|
Acinetobacter (%) | 9.1 | 10.3 | 9.3 | 2.1 | 3.4 | 7.8 | 3.2 | 4.1 | 6.6 |
|
Streptococcus (%) | 0 | 0 | 0 | 0.3 | 1.9 | 0 | 0.5 | 0 | 3.1 |
| Pseudomonas
(%) | 5.3 | 0 | 0.0 | 17.9 | 0 | 0 | 15.7 | 0 | 0 |
| Macrococcus
(%) | 0 | 0 | 0 | 0 | 2.5 | 0 | 0 | 0 | 5.8 |
|
Brevundimonas (%) | 0 | 0 | 0 | 0 | 1.5 | 0 | 0 | 0 | 7.3 |
| Actinomyces
(%) | 0 | 0 | 0 | 0 | 1.7 | 0 | 0 | 0 | 5.7 |
| Schlegel
(%) | 0 | 0 | 0 | 0 | 0 | 1.1 | 0 | 2.1 | 0 |
| χ2 |
| 38.230 |
|
| 32.661 |
|
| 43.623 |
|
| P-value |
| 0.000 |
|
| 0.000 |
|
| 0.000 |
|
In total, 11 bacterial genera were detected, and
dynamic alterations were observed in the composition and
constituent ratios of the bacterial genera in the three groups with
prolonged intubation. In the three groups, six genera were
detected: Klebsiella sp., Serratia sp.,
Streptococcus sp., Achromobacter sp.,
Acinetobacter sp. and Staphylococcus sp.
Staphylococcus sp. on the first day of intubation. With
prolonged intubation, the composition ratio of Streptococcus
sp. gradually increased, whereas Serratia sp. exhibited a
trend of gradual reduction. Pseudomonas sp. was not present
in the RDS group.
The composition ratios of the bacterial genera in
the three groups were compared following the same duration of
intubation No differences were observed between the IP group and
IVAP group on the first day of intubation. The composition ratios
of Serratia sp. and Acinetobacter sp. in the RDS
group were higher, compared with those in the IP group and IVAP
group, whereas the composition ratios of Klebsiella sp.,
Achromobacter sp. and Streptococcus sp. in the RDS
group were lower, compared with those in the other two groups. The
IVAP group showed the highest composition ratios of
Klebsiella sp. and Streptococcus sp. on days 1–3
post-intubation, followed by the IP group and the RDS group, which
exhibited the lowest ratio. The composition ratios of
Serratia sp. and Achromobacter sp. were in the order
IVAP group < IP group < RDS group, whereas the constituent
ratio of Acinetobacter sp. was in the order IP group <
IVAP group < RDS group. On days 3–5 post-intubation, no
differences were observed between the RDS group and IP group,
however, the IVAP group showed lower constituent ratios of
Serratia sp., Achromobacter sp., Acinetobacter
sp. and Klebsiella sp., and a higher composition ratio of
Streptococcus sp., compared with the RDS group and IP
group.
Comparison of the detection rate
between the two methods
In the present study, the detection rate determined
using the 16S rDNA PCR-DGGE cloning-sequencing method was 82.0%
(73/89), whereas the rate determined using the culture method was
54.4% (χ2=17.092; P<0.001), which showed that the
former method had higher efficiency, compared with the latter
method.
Discussion
Associations between the microfloral
diversity of the respiratory tract and neonatal infectious
pneumonia with concurrent VAP. No differences were observed in the
microfloral diversity among the RDS, IP and IVAP groups in the
first day of intubation
This was likely due to the low number of bacteria
entering the lower respiratory tract through the throat and
endotracheal tubing in the early stages of intubation in the
patients with RDS, resulting in a low level of diversity. The
reduced diversity of the respiratory microflora may also have been
due to infection in the patients with IP and IVAP. In addition, no
VAP complications were present in the patients with IVAP within the
first day of intubation; thus, the microfloral diversity in the
patients with IVAP was equivalent to that in the patients with IP.
Therefore, microfloral diversity was found to decrease in the RDS,
IP and IVAP groups in the first day of intubation, with no
difference in diversity observed among the three groups.
On days 1–3 post-intubation, the RDS group showed
the highest level of microfloral diversity, followed by the IP
group and the IVAP group, which had the lowest level of diversity.
The patients with IVAP had the lowest microfloral diversity as they
were afflicted with more severe infections, namely VAP, in addition
to their preceding pneumonia. Only one type of pneumonia was
present in the patients with IP, who showed an intermediate level
of respiratory microfloral diversity (7). These findings indicated that more
severe pneumonia was associated with reduced microfloral
diversity.
The microfloral diversity among the three groups at
3–5 days post-intubation was comparable with the diversity at 1–3
days post-intubation. The microfloral diversity of the IP group
increased, but remained lower than that of the RDS group,
indicating more severe infection and poorer overall prognoses in
the patients of the IP group. Thus, patients with IP may require
additional time to recover completely. The diversity of the IVAP
group remained the lowest, which was likely to be associated with
the highest severity of infection and the inhibitory effects of
antibiotics.
In the present study, the microfloral similarity was
highest in the IVAP group, followed by the IP group and RDS group.
This finding indicated that a higher severity of infection caused
higher levels of microfloral similarity and more marked inhibitory
effects on microfloral diversity (14).
Association between the composition of
respiratory microflora and pneumonia
A total of 11 bacterial genera were detected in the
lower respiratory tract, and the microfloral constituent ratio
exhibited common features among the three groups. One common
feature was the dynamic changes observed in the number, type and
constituent ratios of the bacterial genera in the three groups
following prolonged intubation, and another common feature was that
six genera were shared by the three groups.
Compared with the RDS group, microfloral imbalance
was observed in the IP and IVAP groups, manifested as follows: i)
Changes in the composition of common bacterial genera. Within 1 day
post-intubation, increased ratios of Klebsiella sp.,
Streptococcus sp. and Achromobacter sp., and reduced
ratios of Serratia sp. and Acinetobacter sp.
suggested possible infectious pneumonia. On days 1–3
post-intubation, the composition of Klebsiella sp. and
Streptococcus sp. were the highest in the IVAP group,
followed by the IP group and RDS group; however, the composition of
Serratia sp., Achromobacter sp. and
Acinetobacter sp. were lowest in the IVAP group, higher in
the IP group and highest in the RDS group. Therefore, the increased
composition of Klebsiella sp. and Streptococcus sp.,
together with the reduced ratios of Serratia sp. and
Acinetobacter sp. suggested possible VAP complications in
the patients with IP. At 3–5 days post-intubation, no differences
in the constituent ratios were observed between the RDS group and
IP group, which was likely to be due to certain patients with IP
showing improvement in their condition and certain RDS patients
showing concurrent infection. In addition, the IVAP group exhibited
lower constituent ratios of Serratia sp.,
Achromobacter sp., Acinetobacter sp.,
Klebsiella sp. and Streptococcus sp., compared with
those of the RDS and IP groups. This observation was likely
associated with the suppression from antibiotics, disease outcome
and other types of bacteria present in the IVAP group. 2)
Pseudomonas sp., Brevundimonas sp.,
Actinomyces sp. and other genera were detected in the IP and
IVAP groups, indicating changes in bacterial composition with
infectious pneumonia, the suppression of dominant bacteria and an
increase of opportunistic pathogenic bacteria (15,16).
In the present study, Klebsiella subspecies
and Pseudomonas aeruginosa were detected in the bacterial
culture. In addition, the compositions of Klebsiella sp., to
which Klebsiella subspecies belong, were higher in the IP
and IVAP groups, compared with the RDS group. Pseudomonas
sp., to which Pseudomonas aeruginosa belongs, was not
detected in the RDS group. This suggested that, when a bacterial
strain is present in the culture and the constituent ratio of its
bacterial genus is higher, compared with that of the control group,
or the bacterial genus is newly detected, this bacterial strain is
likely a pathogenic bacterial strain. However, although
Acinetobacter baumannii was also detected in the culture in
the present study, the constituent ratio of its bacterial genus,
Acinetobacter sp., was lower in the IP and IVAP groups,
compared with the RDS group. This finding was inconsistent with the
above results. Acinetobacter baumannii is a multi-drug
resistant opportunistic pathogen, and it is frequently found in the
lower respiratory tract of patients with severe pneumonia,
Guillain-Barre syndrome or traumatic brain injury who are supported
with mechanical ventilation (17,18).
The majority of the patients exhibited improvements in their
condition with continued treatment of the originally prescribed
antibiotics, or even without the administration of antibiotics,
which indicated that the Acinetobacter baumannii present in
the culture only colonized the respiratory tract and was not
pathogenic. This suggested that, when a bacterial strain is present
in culture and the constituent ratio of its bacterial genus is
higher, compared with that of the control group, the bacterial
strain is most likely not pathogenic. This conclusion requires
comprehensive analysis based on clinical manifestations and other
laboratory examination, however the analysis of changes in
constituent ratios of bacterial genera in the lower respiratory
tract can assist in determining the condition of pneumonia and
whether a bacteria strain is pathogenic.
Comparison between the 16S
rDNA-PCR-DGGE cloning-sequencing method and culture method
In the present study, 11 bacterial genera were
detected in sputum samples using the 16S rDNA-DGGE
cloning-sequencing method. The three bacterial strains detected in
the sputum culture, Klebsiella pneumoniae subspecies,
Acinetobacter baumannii and Pseudomonas aeruginosa,
belong to Klebsiella sp., Acinetobacter sp. and
Pseudomonas sp., respectively, indicating consistent results
using the two methods.
Among the 52 samples collected from the patients
with IP and IVAP and used for detection in sputum culture, 24
samples did not show either bacterial growth or normal microfloral
growth. The bacterial genera detected using the sequencing method,
including Serratia sp., Achromobacter sp.,
Streptococcus sp., Staphylococcus sp.,
Actinomyces sp., Brevundimonas sp.,
Macrococcus sp. and Schlegel sp., were not detected
using the sputum culture method, indicating that the 16S rDNA-DGGE
cloning-sequencing method was more sensitive and detected bacteria,
which were not detected through bacterial culture. These results
showed that the detection efficiency of the 16S rDNA-DGGE
cloning-sequencing method was higher, compared with that of the
culture method. Therefore, the 16S rDNA-DGGE cloning-sequencing
method was considered to be suitable for use as a supplement to the
culture method.
In conclusion, the present study demonstrated that:
i) Microfloral imbalances in the lower respiratory tract of
newborns with bacterial pneumonia caused a reduction in microfloral
diversity, and the increased severity of infection was associated
with the lower diversity. The microfloral diversity was ordered as
follows: IVAP group < IP group < RDS group. ii) Reductions in
microfloral diversity were found in the lower respiratory tract of
newborns with bacterial pneumonia. Increased constituent ratios of
Klebsiella sp. and Streptococcus sp., and reduced
constituent ratios of Serratia sp. and Acinetobacter
sp. provided an early indicator of the occurrence of VAP.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81370744), the
Doctoral Degree Funding from the Chinese Ministry of Education
(grant no. 20135503110009), the State Key Clinic Discipline Project
(grant no. 2011-873) and the subproject of the National Science
& Technology Pillar Program during the Twelfth Five-year Plan
Period in China (grant no. 2012BAI04B05).
References
|
1
|
Peters BM, Jabra-Rizk MA, O'May GA,
Costerton JW and Shirtliff ME: Polymicrobial interactions: Impact
on pathogenesis and human disease. Clin Microbiol Rev. 25:193–213.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Balter M: Taking stock of the human
microbiome and disease. Science. 336:1246–1247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yalaz M, Altun-Köroğlu O, Ulusoy B, Yildiz
B, Akisu M, Vardar F, Ozinel MA and Kültürsay N: Evaluation of
device-associated infections in a neonatal intensive care unit.
Turk J Pediatr. 54:128–135. 2012.PubMed/NCBI
|
|
4
|
Kellenberger E: Exploring the unknown. The
silent revolution of microbiology. EMBO Rep. 2:5–7. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Relman DA, Loutit JS, Schmidt TM, Falkow S
and Tompkins LS: The agent of bacillary angiomatosis. An approach
to the identification of uncultured pathogens. N Engl J Med.
323:1573–1580. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Relman DA, Schmidt TM, MacDermott RP and
Falkow S: Identification of the uncultured bacillus of Whipple's
disease. N Engl J Med. 327:293–301. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cairns S, Thomas JG, Hooper SJ, Wise MP,
Frost PJ, Wilson MJ, Lewis MA and Williams DW: Molecular analysis
of microbial communities in endotracheal tube biofilms. PLoS One.
6:e147592011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grgurich PE, Hudcova J, Lei Y, Sarwar A
and Craven DE: Diagnosis of ventilator-associated pneumonia:
Controversies and working toward a gold standard. Curr Opin Infect
Dis. 26:140–150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Craven DE, Hudcova J and Lei Y: Diagnosis
of ventilator-associated respiratory infections (VARI):
Microbiologic clues for tracheobronchitis (VAT) and pneumonia
(VAP). Clin Chest Med. 32:547–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F and He B: The role of endotracheal
aspirate culture in the diagnosis of ventilator-associated
pneumonia: A meta analysis. Zhonghua Jie He He Hu Xi Za Zhi.
36:27–32. 2013.(In Chinese). PubMed/NCBI
|
|
11
|
Payne MS, Goss KC, Connett GJ,
Kollamparambil T, Legg JP, Thwaites R, Ashton M, Puddy V, Peacock
JL and Bruce KD: Molecular microbiological characterization of
preterm neonates at risk of bronchopulmonary dysplasia. Pediatr
Res. 67:412–418. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Hoenig JD, Malin KJ, Qamar S,
Petrof EO, Sun J, Antonopoulos DA, Chang EB and Claud EC: 16S rRNA
gene-based analysis of fecal microbiota from preterm infants with
and without necrotizing enterocolitis. ISME J. 3:944–954. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hill TC, Walsh KA, Harris JA and Moffett
BF: Using ecological diversity measures with bacterial communities.
FEMS Microbiol Ecol. 43:1–11. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Signoretto C, Bianchi F, Burlacchini G,
Sivieri F, Spratt D and Canepari P: Drinking habits are associated
with changes in the dental plaque microbial community. J Clin
Microbiol. 48:347–356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Codling C, O'Mahony L, Shanahan F, Quigley
EM and Marchesi JR: A molecular analysis of fecal and mucosal
bacterial communities in irritable bowel syndrome. Dig Dis Sci.
55:347–356. 2010. View Article : Google Scholar
|
|
16
|
Noor SO, Ridgway K, Scovell L, Kemsley EK,
Lund EK, Jamieson C, Johnson IT and Narbad A: Ulcerative colitis
and irritable bowel patients exhibit distinct abnormalities of the
gut microbiota. BMC Gastroenterol. 10:1342010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song W, Wu YM, Ji Z, Zhu JJ and Pan SY:
Guillain-Barré syndrome following sepsis after stereotactic
aspiration for spontaneous pontine hemorrhage. Neurol Sci.
3:657–660. 2012. View Article : Google Scholar
|
|
18
|
Reddy D, Morrow BM and Argent AC:
Acinetobacter baumannii infections in a South African paediatric
intensive care unit. J Trop Pediatr. 3:182–187. 2015. View Article : Google Scholar
|