Introduction
Chondrocyte differentiation, proliferation,
secretion and apoptosis are considered to be critical in bone
development and for the maintenance of joint function (1). In the bone microenvironment, a number
of growth factors are involved in regulating chondrocyte metabolism
(2).
G-protein-coupled receptor kinase interacting
protein-1 (GIT1) is considered to serve a vital function in bone
development and growth (3–6). Menon et al (3) demonstrated that GIT1 may function as
a key regulator of bone mass in vivo by regulating
osteoclast function, and suggested that it may be a potential
target for osteoporosis therapy. GIT1, a protein-binding partner of
G protein-coupled receptor kinase 2, was discovered by a yeast
two-hybrid technique (7) and is
constitutively expressed in mammals and birds. GIT1 is primarily
located at focal adhesion points and at cytoplasmic structures
within cells, such as inclusion bodies (8). The function of GIT1 was initially
determined to involve regulating the function of cell surface
G-protein coupled receptors in cells (9). However, a recent study has
demonstrated that GIT1 may serve a role in regulating cytoskeletal
dynamics during cell growth and migration processes (10). In particular, GIT1 binds to a
number of cytoskeletal proteins, such as paxillin and focal
adhesion kinase, and is regulated by Src to promote cell migration
(10).
MicroRNAs (miRNAs) have been a focus of research in
the field of osteoarticular disease (11). An increasing number of studies have
indicated that miRNAs serve an important role in regulating cell
differentiation and extracellular matrix secretion in bone and
chondroid tissue generation and metabolism processes, and are
involved in the regulation of multiple signaling pathways in the
bone and joint tissues (12,13).
For example, miR-140 is specifically expressed in the cartilage of
mouse embryos during long and flat bone development, and can
suppress histone deacetylase 4 expression to maintain the
chondrocyte phenotype (14). Kim
et al (15) indicated that
miR-221 may regulate cell proliferation by negatively regulating
mouse double minute 2 homolog, thereby inhibiting Slug degradation
during the chondrogenesis of limb mesenchymal cells.
To date, a limited number of studies have
investigated miRNAs that target GIT1, and no research conducted
thus far has investigated GIT1 and its associated miRNAs in
chondrocytes. Therefore, the aim of the present study was to
investigate the interaction between GIT1 and miRNAs in
chondrocytes.
Materials and methods
Cell culture
The CHON-002 human chondrocyte cell line was
purchased from the American Type Culture Collection (Manassas, VA,
USA), and cultured in Dulbecco's Modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 0.1 mg/ml G-418 solution (Sigma-Aldrich;
Merck-Millipore, Darmstadt, Germany).
Expression vector construction and
transfection
TRIzol reagent (Thermo Fisher Scientific, Inc.) was
used to extract RNA from chondrocytes (1×107 cells),
according to the manufacturer's instructions. This was followed by
reverse transcription-polymerase chain reaction (RT-PCR) to amplify
the coding region of GIT1. The primers used for GIT1 amplification
are listed in Table I. The product
was digested with KpnI and EcoRI restriction
endonucleases (Takara Biotechnology Co., Ltd., Dalian, China), and
then cloned into pcDNA3.1 vectors (Thermo Fisher Scientific, Inc.),
sequenced and verified in a 3730xl DNA Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Cells
(1×105) were seeded in 6-well culture plates and
cultured until they reached ~70% confluence, before expression
vectors were transfected using Lipofectamine 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. The concentration of the GIT1
transfection vector used was 4 µg/well. The GIT1 small-interfering
RNA (siRNA; cat. no. sc-35477; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), miR-195 mimics and inhibitors (Shanghai
GenePharma Co., Ltd., Shanghai, China) were all transfected into
cells at a concentration of 50 nM/well.
 | Table I.Primer sequences used for the purposes
of the present study. |
Table I.
Primer sequences used for the purposes
of the present study.
| Analysis | Name | Sense primer sequence
(5′3′) | Antisense primer
sequence (5′-3′) | Product size
(bp) |
|---|
| GIT1 CDS
amplification | GIT1 CDS |
GGGGTACCGCCACCATGTCCCGAAAGGGGCCG |
CGGAATTCTCACTGCTTCTTCTCTCGGGTG |
|
| RT-qPCR | GAPDH |
GGTATCGTGGAAGGACTC |
GTAGAGGCAGGGATGATG | 128 |
|
| GIT1 |
ATGTATGAACCTGGCTCTG |
TGAATAGATGGCGTCGTC | 114 |
|
| miR-195 RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCCAATAT |
|
|
|
| miR-195 |
ACACTCCAGCTGGGTAGCAGCACAGAAATATT |
CTCAACTGGTGTCGTGGA | 71 |
|
| U6
RT |
AACGCTTCACGAATTTGCGT |
|
|
|
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT | 94 |
| GIT1 3′-UTR
amplification | GIT1 3′-UTR |
CCGCTCGAGCCTCTCTCCCCACACCCTCA |
ATAAGAATGCGGCCGCTAACAGCTCATGGTCACTTCTTTAT |
RT-quantitative PCR (RT-qPCR)
Total RNA was isolated from cultured cell samples
using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions, and the mirVana miRNA Isolation
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used
to purify miRNAs. Target gene expression levels of GIT1 and miR-195
were measured using RT-qPCR. cDNA was synthesized by reverse
transcription using random and oligo-dT primers (Promega
Corporation, Madison, WI, USA) or specific primers for miRNA-195
(Takara Biotechnology Co., Ltd.), together with the GoScript
Reverse Transcription System (Promega Corporation). qPCR was
performed using the GoTaq qPCR Master Mix (Promega Corporation) and
an ABI PRISM® 7500 Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The thermocycling conditions were as
follows: 95°C for 2 min; and 40 cycles of 95°C for 15 sec and 60°C
for 32 sec. The primer sequences are shown in Table I. GAPDH served as a control for
GIT1 expression and U6 as a control for miR-195. To measure miRNA
expression, specific primers for miRNA-195 and U6 were used. Three
independent experiments were conducted for each sample. Data were
analyzed by comparing the 2−ΔΔCq values (16).
Western blot analysis
Total protein was extracted by incubating cells
(1×106) in radioimmunoprecipitation assay (RIPA) buffer
(cat. no. sc-24948; Santa Cruz Biotechnology, Inc.) for 10 min in
an ice bath, prior to loading of 50 µg samples in 10% SDS-PAGE gels
for separation and transferred to nitrocellulose membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Membranes were blocked with
5% non-fat milk for 1 h at room temperature and then incubated with
antibodies against GIT1 (cat. no. 2919; dilution, 1:1,000; Cell
Signaling Technology, Inc., Danvers, MA, USA) or GAPDH (cat. no.
2118; 1:1,000; Cell Signaling Technology, Inc.) in 5% non-fat milk
overnight at 4°C. Immunoreactive proteins were visualized using
incubation with horseradish peroxidase-conjugated IgG secondary
antibodies (cat. no. 7074; dilution, 1:7,000–8,000; Cell Signaling
Technology, Inc.) at room temperature for 1 h and enhanced
chemiluminescence reagents (Pierce; Thermo Fisher Scientific,
Inc.). Images were analyzed using Image-Pro Plus 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA). Each band was scanned with
background correction, and values were expressed as the mean ±
standard deviation.
Dual luciferase assay
The software applications miRanda (http://www.microrna.org/microrna/getGeneForm.do),
TargetScanHuman (http://www.targetscan.org/vert_71/), miRBase
(http://www.mirbase.org/) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
predicted that GIT1 has miR-195a-3p binding sites. The GIT1 3′-UTR
was cloned into the psiCHECK-2 vector (Promega Corporation), and
the seed region of the miR-195 binding site in the 3′-UTR was
mutated using the QuikChange II Site-Directed Mutagenesis kit
(Agilent Technologies, Inc., Santa Clara, CA, USA). For the
luciferase assay, 2×104 HEK293A cells (American Type
Culture Collection) were seeded in 24-well dishes and were cultured
until they reached 80% confluence. Cells were transfected with
psiCHECK-2 (containing the wild-type GIT1 3′-UTR or the mutated
form) together with miR-195 mimics using the Lipofectamine 2000
transfection agent (Invitrogen; Thermo Fisher Scientific, Inc.).
Cells were analyzed at 24 h post-transfection. Firefly and
Renilla luciferase activities were quantified in cell
lysates using the Dual-Luciferase Reporter assay kit (Promega
Corporation) on a Glomax 20/20 luminometer (Promega Corporation)
according to the manufacturer's instructions. Luciferase readings
were corrected to background readings and firefly luciferase values
were normalized to Renilla values in order to determine the
transfection efficiency. Samples were analyzed in triplicate and
three independent experiments were conducted.
Immunoprecipitation
A total of 1×107 HEK 293A cells were
lysed in RIPA buffer containing 1 mM protease inhibitors (Roche
Diagnostics GmbH, Mannheim, Germany) and centrifuged at 10,000 ×
g for 10 min at 4°C. For immunoprecipitation, 5 ml of
supernatant from monoclonal anti-Ago2 antibody (cat. no. 2897;
dilution, 1:50; Cell Signaling Technology, Inc.) was coupled to 80
µl Protein-G-Sepharose beads (GE Healthcare Life Sciences,
Chalfont, UK). Beads were subsequently incubated with 10 ml HEK 293
lysate for 5 h under constant rotation at 4°C. Following
incubation, the beads were washed three times with Tris-buffered
saline. The beads were then washed once with phosphate-buffered
saline (PBS). Co-immunoprecipitated RNA was extracted using phenol:
chloroform: isoamyl alcohol (25:24:1; cat. no, 15593-031;
Invitrogen; Thermo Fisher Scientific, Inc.). The RNA pellet was
used for RT-qPCR analysis of GIT1 expression, using the
aforementioned methods.
BrdU cell proliferation assay
A BrdU assay was used to investigate the roles of
miR-195 and GIT1 on cell proliferation. Briefly, the cultured cells
(1×105 chondrocytes) were seeded into 6-well plates and
incubated for 24 h before miR-195 mimics and inhibitors, plasmids
or siRNAs were transfected into cells using the Lipofectamine 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
A BrdU Cell Proliferation Assay kit (Cell Signaling Technology,
Inc.) was used to determine cell proliferation according to the
manufacturer's instructions. Following transfection and incubation
for 48 h, the medium was removed and cells were labeled with 10 mM
BrdU for 3 h at 37°C. Cells were fixed and incubated with
peroxidase-conjugated anti-BrdU antibody for 90 min at room
temperature. Subsequently, the peroxidase substrate
(3,3′,5,5′-tetramethylbenzidine) was added, and BrdU incorporation
was quantitated by differences in absorbance at wavelength of 370
subtract wavelength of 492 nm. Cell proliferation was expressed as
the mean percentage relative to the control values (set at
100%).
Cell migration examination
Cell migration rates were measured using a transwell
chamber (BD Biosciences, Franklin Lakes, NJ, USA) containing
Matrigel. The trypsinized chondrocytes were diluted to a final
concentration of 2×106 cells/ml in serum-free media, and
100 µl cell suspension was added into the upper chamber and 0.6 ml
DMEM with 10% FBS was added into the lower chamber. Chambers were
incubated at 5% CO2 and 37°C for 6 h. Following removal
of the medium, cells were fixed on the lower side of the insert
filter by incubating with 4% paraformaldehyde (Sigma-Aldrich;
Merck-Millipore) for 15 min, and cells that did not migrate on the
upper side of the filter membrane were removed with a cotton swab.
The cells on the lower side of the insert filter were stained with
0.1% Crystal Violet (Sigma-Aldrich; Merck-Millipore) for 10 min.
The number of the cells on the lower side of the filter were then
visualized and counted under a microscope (IX-70; Olympus
Corporation, Tokyo, Japan) after washing with PBS (Gibco; Thermo
Fisher Scientific, Inc.). The cells that had migrated through the
membrane were stained and counted, and chondrocyte migration was
expressed as the percentage of the total number of cells that had
migrated.
Statistical analysis
Experiments were performed in triplicate and results
are expressed as mean ± standard deviation. Statistical analyses
were performed using the SPSS statistical software package
(version, 17.0; SPSS, Inc., Chicago, IL, USA). Differences between
control and treated groups were analyzed using non-parametric
Mann-Whitney U tests. P<0.01 was considered to indicate a
statistically significant difference.
Results
miR-195 targets GIT1
Multiple miRNAs may target the same gene, and an
miRNA can also target multiple genes, for example, miR-425,
miR-376a and miR-138 may target GIT1, and miR-195 can target
ZNF367, HDGF and CHEK1,. The present study used common
bioinformatic algorithms to predict miRNAs that target GIT1.
Previous studies have demonstrated that miR-195 serves a pivotal
role in osteogenesis and bone development (17–19),
however, there has been less research into the effect of other
miRNAs on bone development, thus, the present study selected
miR-195 for further investigation. As shown in Fig. 1A and B, the GIT1 3′-UTR was
observed to contain miR-195 binding sites. In the present study,
dual-luciferase reporter assay was used to verify binding of
miR-195 to GIT1. As shown in Fig.
1B, luciferase expression levels decreased significantly
following transfection of cells with vectors containing GIT1 3′-UTR
clones plus miR-195 mimics (P<0.01). Conversely, luciferase
expression levels were not significantly altered when the binding
site was mutated (Fig. 1B). This
demonstrated that miR-195 may bind to a site in the GIT1 3′-UTR,
which suggests that GIT1 may be a target of miR-195.
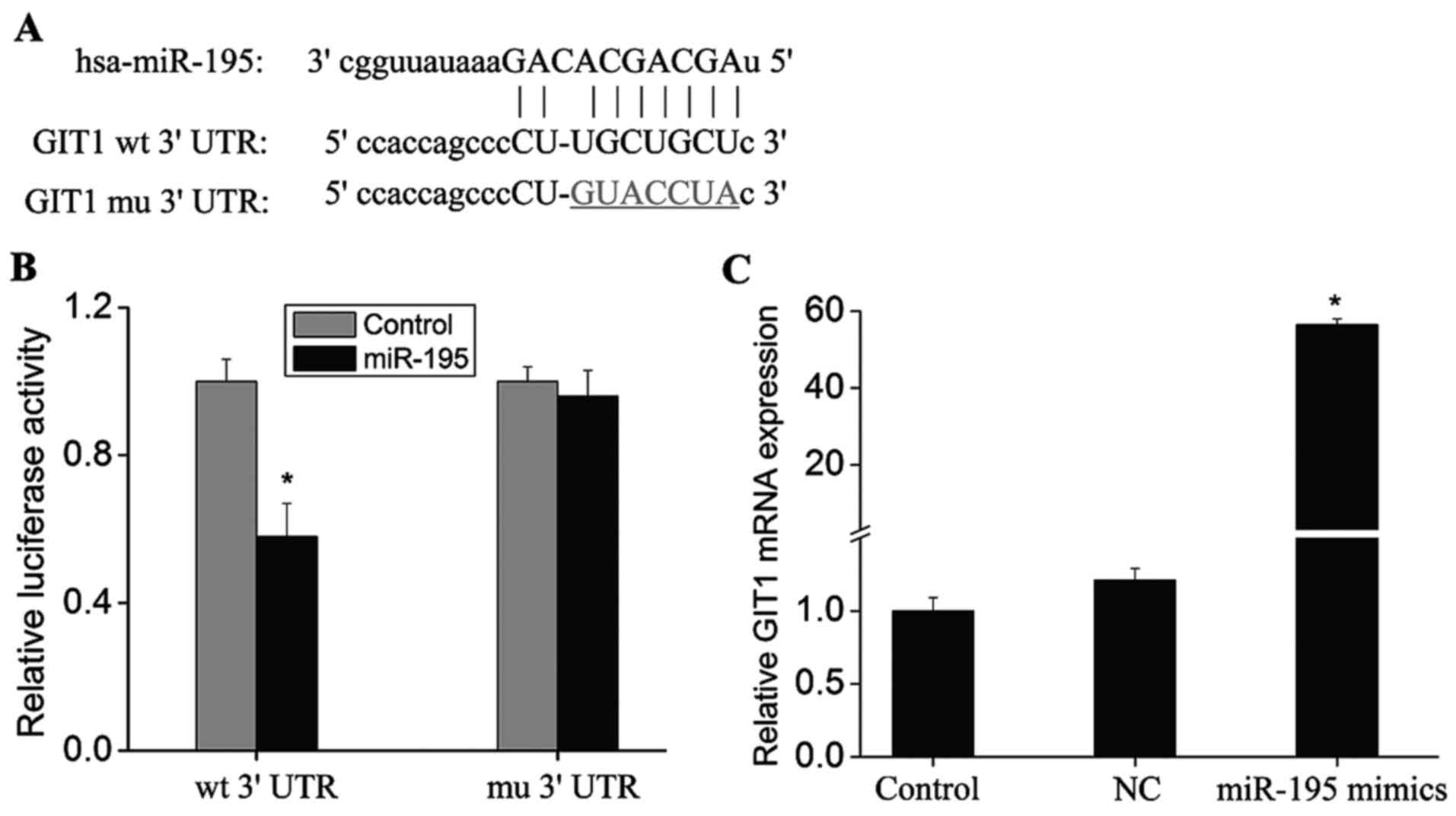 | Figure 1.Prediction of miR-195 target site in
GIT1 3′-UTR. (A) miR-195 target sites in the conservative sequence
of the GIT1 3′-UTR were identified using common bioinformatic
algorithms (miRanda, TargetScanHuman, miRBase and miRWalk), as
indicated by capital letters. The putative miR-195 binding site was
mutated (underlined nucleotides). (B) Luciferase expression in
HEK293A cells transfected with vectors containing the wt GIT1
3′-UTR or mu GIT1 3′-UTR together with miR-195 mimics or controls.
(C) The Ago2 monoclonal antibody was immobilized on
Protein-G-Sepharose beads and incubated with HEK 293 cell lysates
obtained following transfection with miR-195 mimics. After
stringent washing, co-immunoprecipitated Ago-bound RNAs were
subject to RT-qPCR to detect GIT1 mRNA expression. Data are
presented as the mean ± standard deviation (n=3). *P<0.01 vs.
the controls. miR-195, microRNA-195; GIT1, G-protein-coupled
receptor kinase interacting protein-1; UTR, untranslated region;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; NC, negative control miRNA transfection group; wt,
wild-type; mu, mutant. |
Ago2 is a core component of the RNA-induced
silencing complex that associates with miRNAs and their mRNA
targets (20). Therefore,
immunopurification of Ago2 under the appropriate conditions may
retain associated miRNAs and mRNAs, thereby allowing miRNA targets
to be identified. A monoclonal antibody against Ago2 was
immobilized on Protein-G-Sepharose beads and incubated with HEK 293
cell lysates. Following stringent washing, the
co-immunoprecipitated Ago-bound RNAs were extracted and subject to
RT-qPCR analysis in order to detect GIT1 mRNA expression. Following
transfection of miR-195 mimics, GIT1 mRNA levels were significantly
higher when compared with controls (Fig. 1C; P<0.01). This further
demonstrated that miR-195 may target and regulate GIT1
expression.
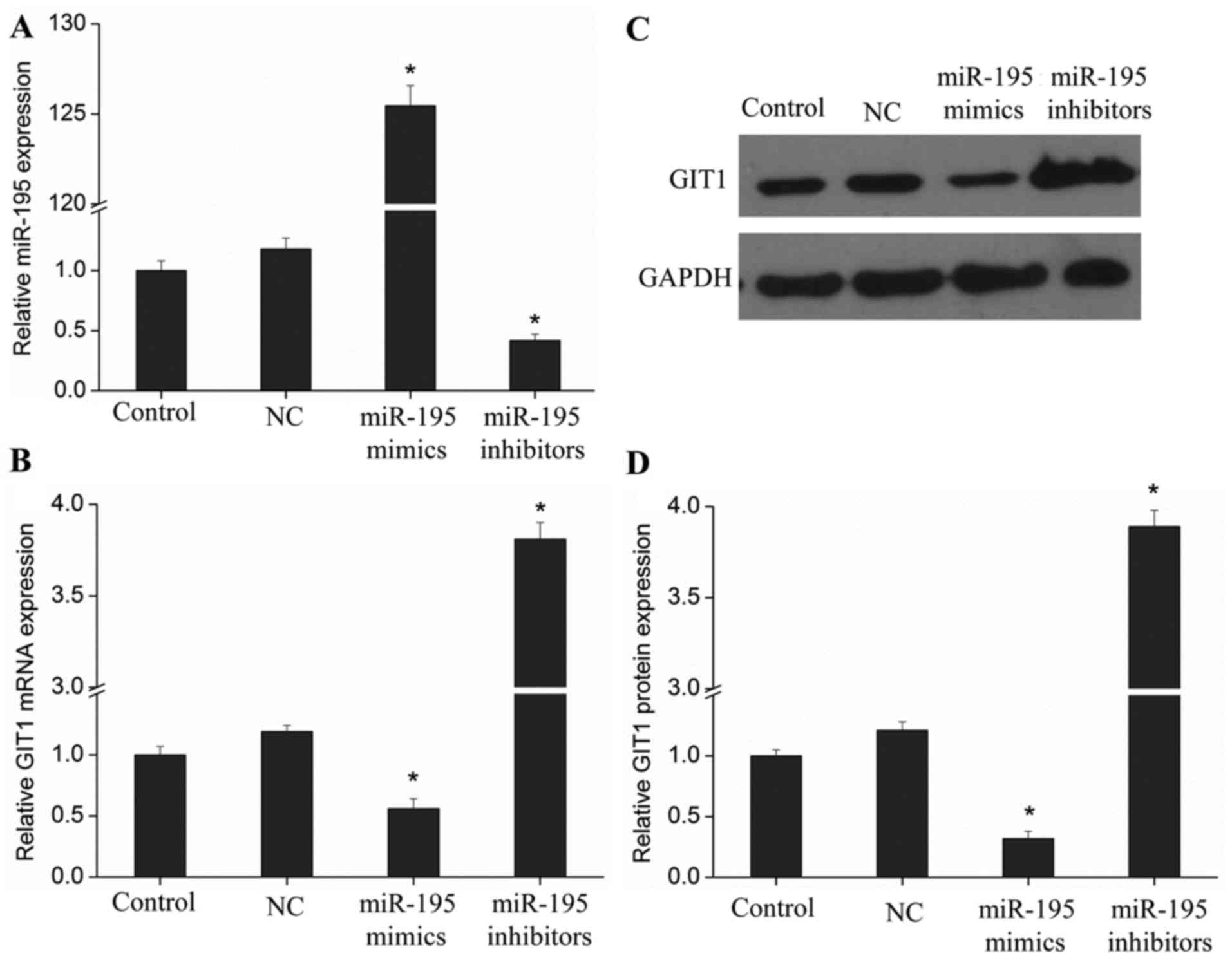
miR-195 inhibits the expression of
GIT1 in chondrocytes
As GIT1 may be a target gene of miR-195, it is
formally possible that miR-195 may regulate the expression of GIT1
in chondrocytes. In the current study, miR-195 mimics and
inhibitors were transfected into human chondrocytes, and miR-195
and GIT1 expression was measured. The results indicated that a
significant increase in miR-195 expression was associated with a
significant downregulation in GIT1 mRNA and protein expression
levels when compared with controls (Fig. 2; P<0.01). By contrast, when
miR-195 expression was suppressed, GIT1 mRNA and protein expression
increased significantly when compared with controls (Fig. 2B-D; P<0.01). These results
suggest that miR-195 may regulate GIT1 expression in
chondrocytes.
miR-195 inhibits chondrocyte
proliferation through targeted regulation of GIT1 expression
The results presented so far suggest that miR-195
targets and regulates the expression of GIT1 in chondrocytes.
However, the role and association of this interaction with the
biological behavior of chondrocytes requires further investigation.
In the present study, a BrdU assay was performed in order to
investigate the effect of miR-195 on chondrocyte proliferation. As
shown in Fig. 3, chondrocyte
proliferation increased significantly when miR-195 expression was
suppressed with miR-195 inhibitors, as well as following
overexpression of GIT1 compared with the control group (P<0.01).
By contrast, transfection with miR-195 mimics or GIT1 siRNA
demonstrated the opposite effect on chondrocyte proliferation
compared with the control group (Fig.
3). These results demonstrate that chondrocyte proliferation
may be inhibited by miR-195, but promoted by GIT1 expression. When
miR-195 and GIT1 overexpression vectors were co-transfected, the
inhibitory effect of miR-195 on chondrocyte proliferation was
significantly attenuated (Fig. 3;
P<0.01). However, upon co-transfection with miR-195 and GIT1
overexpression vectors containing the wild-type 3′-UTR sequence,
miR-195-mediated inhibition of chondrocyte proliferation was
unaffected (Fig. 3). These results
demonstrate that miR-195 may be involved in mediating chondrocyte
proliferation by regulating GIT1 expression.
 | Figure 3.Role of miR-195 and GIT1 in
chondrocyte proliferation. Chondrocytes were transfected with
GIT1-expression vectors, miR-195 mimics, miR-195 inhibitors or GIT1
siRNA and cultured for 72 h. Cell proliferation was measured using
a BrdU assay. The results are presented as the mean ± standard
deviation, (n=3). *P<0.01 vs. the control. miR-195,
microRNA-195; GIT1, G-protein-coupled receptor kinase interacting
protein-1; NC, negative control; UTR, untranslated region; wt,
wild-type; mut, mutant; siRNA, small-interfering RNA. |
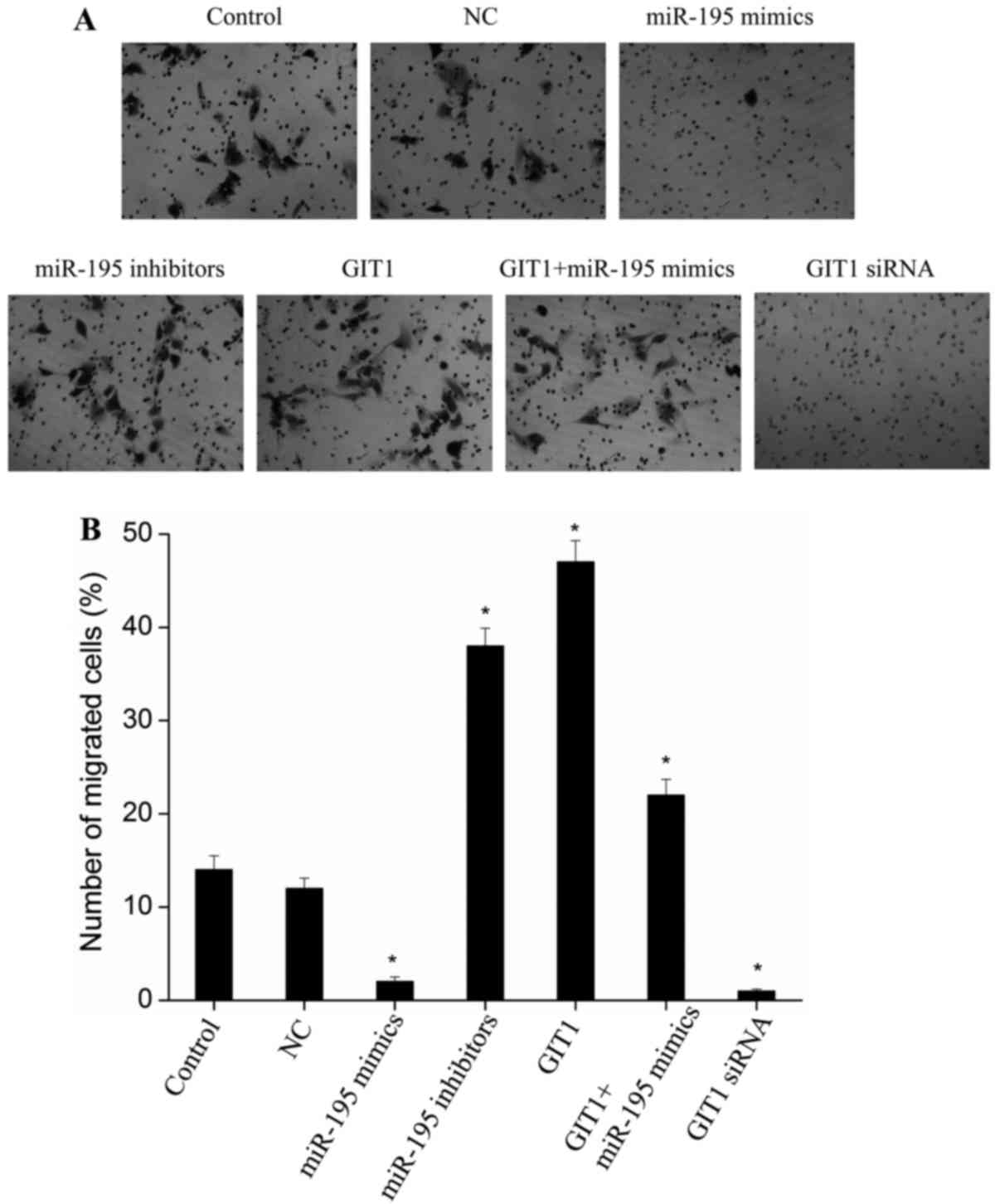
miR-195 inhibits chondrocyte migration
through targeted regulation of GIT1 expression
Previous studies have demonstrated that a key
function of GIT1 is to promote cell migration (21,22).
Therefore, due to the observed putative role of miR-195 in
regulating GIT1, it is possible that miR-195 may suppress cell
migration. As shown in Fig. 4,
upon transfection of miR-195 mimics in chondrocytes, the cell
migration capacity decreased significantly when compared with
controls (P<0.01). A similar effect was observed following
transfection of cells with GIT1 siRNA (Fig. 4; P<0.01). Following transfection
of miR-195 inhibitors, the migration capacity of chondrocytes
increased significantly compared with the control group
(P<0.01). This was similar to the migration capacity observed
following transfection of cells with GIT1 expression vectors
(Fig. 4; P<0.01).
Co-transfection of miR-195 mimics with GIT1 expression vectors
attenuated the inhibitory effect of miR-195 on cell migration
(Fig. 4). These results suggest
that miR-195 may inhibit chondrocyte migration by regulating GIT1
expression.
Discussion
Chondrocytes are located in cartilage lacunae and
possess supportive and protective roles in joint movement and
weight-bearing. In addition, chondrocytes respond to wounds, stress
and external stimuli to initiate cell repair and proliferation
processes (1,2). The proliferation of chondrocytes is
affected by various growth factors, cytokines and additional
external conditions including, mechanical pressure and alterations
in cell density (1,2).
An increasing number of studies have demonstrated
that GIT1 demonstrates an important role in bone growth and
development (3–6). For instance, Xiao et al
(4) suggested that
platelet-derived growth factor regulates chondrocyte proliferation
through activation of the ERK1/2 signaling pathway via upregulation
of GIT1 expression and Rac1 phosphorylation. In addition, an
investigation into miRNA function was demonstrated to be involved
in the differentiation and formation of human bone and joint
tissues, including osteoblasts, osteoclasts and chondrocytes
(12,13). Therefore, miRNAs may be an
important focus of research concerning joint disease prevention and
treatment. A previous study demonstrated that miR-1 regulates
aggrecan expression in human chondrocytes, and is involved in
regulating chondrocyte phenotypic stability (23). In addition, miR-1 serves an
important regulatory role in the late differentiation of
chondrocytes, and in maintaining the integrity of cartilage tissues
(23).
The present study investigated the miRNAs that
target GIT1 and demonstrated that miR-195 may target and regulate
GIT1 due to the identification of a putative binding site in the
GIT1 3′-UTR. To date, studies concerning miR-195 function in tumors
have made significant progress (24–26).
Zhou et al (24) indicated
that miR-195 inhibited non-small cell lung cancer cell
proliferation, migration and invasion by targeting the MYB
proto-oncogene. However, studies investigating the role of miR-195
in bone growth and development are rare. According to the results
of the current study, miR-195 demonstrated an inhibitory effect on
the expression of GIT1 in chondrocytes, and may affect chondrocyte
proliferation and migration by regulating GIT1.
According to the results of the current study,
miR-195 inhibits chondrocyte proliferation and migration, and one
pathway by which miR-195 may mediate this effect is through
regulating GIT1. Consistent with these observations, the inhibitory
effect of miR-195 on cell proliferation in additional cell types
has been reported previously. Sekiya et al (27) demonstrated that downregulation of
cyclin E1 expression by miR-195 accounted for the
interferon-β-induced inhibition of hepatic stellate cell
proliferation. In addition, Wang et a (28) demonstrated that miR-195 inhibited
the proliferation and growth, and induced apoptosis of endometrial
stromal cells by targeting the fractalkine gene. Grünhagen et
al (17) identified the
miR-497~195 cluster, a member of the miR-15 family, as being
strongly upregulated during postnatal bone development in
vivo, and late differentiation stages of primary osteoblasts
cultured in vitro. Early expression of miR-195-5p was
observed to inhibit osteoblast differentiation and mineralization.
Using microarray and RT-qPCR analyses, miR-195-5p was observed to
alter the gene regulatory network of osteoblast differentiation,
and impair the induction of bone morphogenetic protein responsive
genes. In addition, Bai et al (18) demonstrated that miR-195
significantly increased apoptosis and downregulated
hypoxia-inducible factor 1-α mRNA expression simultaneously in
hypoxic chondrocytes. According to the results of the present
study, miR-195 was observed to inhibit chondrocyte cell
proliferation and migration, potentially through regulating GIT1
expression.
In conclusion, miR-195 may target and regulate the
expression of GIT1 in chondrocytes. In addition, miR-195 inhibited
the proliferation and migration of chondrocytes, likely through the
targeted regulation of GIT1 expression. The results of the current
study may provide a rationale for investigating the regulatory
effects and underlying mechanisms of miRNAs in bone and chondrocyte
tissues, and may provide a novel approach for understanding
osteoarticular diseases.
Acknowledgements
The present study was supported by the Health Bureau
of Wuxi City Foundation for Youths (grant no. Q201407).
References
|
1
|
Adams CS and Shapiro IM: The fate of the
terminally differentiated chondrocyte: Evidence for
microenvironmental regulation of chondrocyte apoptosis. Crit Rev
Oral Biol Med. 13:465–473. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ashraf S, Cha BH, Kim JS, Ahn J, Han I,
Park H and Lee SH: Regulation of senescence associated signaling
mechanisms in chondrocytes for cartilage tissue regeneration.
Osteoarthritis Cartilage. 24:196–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Menon P, Yin G, Smolock EM, Zuscik MJ, Yan
C and Berk BC: GPCR kinase 2 interacting protein 1 (GIT1) regulates
osteoclast function and bone mass. J Cell Physiol. 225:777–785.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiao J, Chen X, Xu L, Zhang Y, Yin Q and
Wang F: Regulation of chondrocyte proliferation through
GIT1-Rac1-mediated ERK1/2 pathway by PDGF. Cell Biol Int.
38:695–701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiao J, Chen X, Xu L, Zhang Y, Yin Q and
Wang F: PDGF regulates chondrocyte proliferation through activation
of the GIT1- and PLCγ1-mediated ERK1/2 signaling pathway. Mol Med
Rep. 10:2409–2414. 2014.PubMed/NCBI
|
|
6
|
Zhang LQ, Zhao GZ, Xu XY, Fang J, Chen JM,
Li JW, Gao XJ, Hao LJ and Chen YZ: Integrin-β1 regulates
chondrocyte proliferation and apoptosis through the upregulation of
GIT1 expression. Int J Mol Med. 35:1074–1080. 2015.PubMed/NCBI
|
|
7
|
Premont RT, Claing A, Vitale N, Freeman
JL, Pitcher JA, Patton WA, Moss J, Vaughan M and Lefkowitz RJ:
beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled
receptor kinase-associated ADP ribosylation factor
GTPase-activating protein. Proc Natl Acad Sci USA. 95:14082–14087.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmalzigaug R, Phee H, Davidson CE, Weiss
A and Premont RT: Differential expression of the ARF GAP genes GIT1
and GIT2 in mouse tissues. J Histochem Cytochem. 55:1039–1048.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manabe R, Kovalenko M, Webb DJ and Horwitz
AR: GIT1 functions in a motile, multi-molecular signaling complex
that regulates protrusive activity and cell migration. J Cell Sci.
115:1497–1510. 2002.PubMed/NCBI
|
|
10
|
Zhang H, Webb DJ, Asmussen H and Horwitz
AF: Synapse formation is regulated by the signaling adaptor GIT1. J
Cell Biol. 161:131–42. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pers YM and Jorgensen C: MicroRNA in 2012:
Biotherapeutic potential of microRNAs in rheumatic diseases. Nat
Rev Rheumatol. 9:76–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Qiu M, Dou C, Cao Z and Dong S:
MicroRNAs in Bone Balance and Osteoporosis. Drug Dev Res.
76:235–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jing D, Hao J, Shen Y, Tang G, Li ML,
Huang SH and Zhao ZH: The role of microRNAs in bone remodeling. Int
J Oral Sci. 7:131–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tuddenham L, Wheeler G, Ntounia-Fousara S,
Waters J, Hajihosseini MK, Clark I and Dalmay T: The cartilage
specific microRNA-140 targets histone deacetylase 4 in mouse cells.
FEBS Lett. 580:4214–4217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim D, Song J and Jin EJ: MicroRNA-221
regulates chondrogenic differentiation through promoting
proteosomal degradation of slug by targeting Mdm2. J Biol Chem.
285:26900–26907. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grünhagen J, Bhushan R, Degenkolbe E,
Jäger M, Knaus P, Mundlos S, Robinson PN and Ott CE: MiR-497~195
cluster microRNAs regulate osteoblast differentiation by targeting
BMP signaling. J Bone Miner Res. 30:796–808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bai R, Zhao AQ, Zhao ZQ, Liu WL and Jian
DM: MicroRNA-195 induced apoptosis in hypoxic chondrocytes by
targeting hypoxia-inducible factor 1 alpha. Eur Rev Med Pharmacol
Sci. 19:545–551. 2015.PubMed/NCBI
|
|
19
|
Cai H, Zhao H, Tang J and Wu H: Serum
miR-195 is a diagnostic and prognostic marker for osteosarcoma. J
Surg Res. 194:505–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meister G, Landthaler M, Patkaniowska A,
Dorsett Y, Teng G and Tuschl T: Human Argonaute2 mediates RNA
cleavage targeted by miRNAs and siRNAs. Mol Cell. 15:185–197. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren Y, Yu L, Fan J, Rui Z, Hua Z, Zhang Z,
Zhang N and Yin G: Phosphorylation of GIT1 tyrosine 321 is required
for association with FAK at focal adhesions and for PDGF-activated
migration of osteoblasts. Mol Cell Biochem. 365:109–118. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Penela P, Nogués L and Mayor F Jr: Role of
G protein-coupled receptor kinases in cell migration. Curr Opin
Cell Biol. 27:10–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sumiyoshi K, Kubota S, Ohgawara T, Kawata
K, Nishida T, Shimo T, Yamashiro T and Takigawa M: Identification
of miR-1 as a micro RNA that supports late-stage differentiation of
growth cartilage cells. Biochem Biophys Res Commun. 402:286–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yongchun Z, Linwei T, Xicai W, Lianhua Y,
Guangqiang Z, Ming Y, Guanjian L, Yujie L and Yunchao H:
MicroRNA-195 inhibits non-small cell lung cancer cell
proliferation, migration and invasion by targeting MYB. Cancer
Lett. 347:65–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han K, Chen X, Bian N, Ma B, Yang T, Cai
C, Fan Q, Zhou Y and Zhao TB: MicroRNA profiling identifies MiR-195
suppresses osteosarcoma cell metastasis by targeting CCND1.
Oncotarget. 6:8875–8889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He JF, Luo YM, Wan XH and Jiang D:
Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle,
and apoptosis. J Biochem Mol Toxicol. 25:404–408. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sekiya Y, Ogawa T, Iizuka M, Yoshizato K,
Ikeda K and Kawada N: Down-regulation of cyclin E1 expression by
microRNA-195 accounts for interferon-β-induced inhibition of
hepatic stellate cell proliferation. J Cell Physiol. 226:2535–2542.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Chen H, Fu Y, Ai A, Xue S, Lyu Q
and Kuang Y: MiR-195 inhibits proliferation and growth and induces
apoptosis of endometrial stromal cells by targeting FKN. Int J Clin
Exp Pathol. 6:2824–2834. 2013.PubMed/NCBI
|