Introduction
Tooth development is associated with the continuous
interaction between mesenchymal cells and epithelial cells
(1,2). As the hardest component of teeth,
mammalian enamel serves an essential role in mastication and
occlusion. In mammals, enamel development is categorized into
various stages: Initiation, bud, cap and bell. Amelogenesis begins
with the proliferation of restricted epithelia, which forms dental
lamina, and subsequently invaginates into adjacent mesenchyme
cells; these cells develop into a bud structure (referred to as the
bud stage) (3,4). Following bud generation, tooth germ
cells differentiate into inner enamel epithelium (IEE) cells and
outer enamel epithelium (OEE) cells. Preameloblasts are provided by
IEE cells and further develop into mature ameloblasts, which
secrete enamel matrix proteins, including amelogenin, X-linked
(Amelx), ameloblastin (Ambn) and enamelin
(Enam); these secreted proteins form enamel (5). In rodent mature molars, ameloblasts
are not present and are replaced by mineralized enamel, whereas in
mature incisors, the dental epithelial stem cells in the labial
cervical loop (laCL) maintain the capacity of self-renewal and
differentiation, thus presenting immense potential for tooth
regeneration (6,7).
Mammalian tooth development involves a complex
signaling network, which includes various growth factors and
transcription factors during the developmental process. Various
transcription factors, including paired box 9, paired like
homeodomain 2, Msh homeobox 1 (Msx1), Msh homeobox 2
(Msx2) and distal-less homeobox 2, are expressed in dental
epithelium and mesenchyme, and participate in odontogenesis
(8). B-cell CLL/lymphoma 11B
(Bcl11b) (also known as COUP-TF-interacting protein 2) is a
C2H2 zinc finger transcription factor, which has been demonstrated
to serve crucial roles in the thymus (9), central nervous system (10), and during epidermal development
(11). Mice with Bcl11b
knockdown die perinatally, thus indicating an essential role for
Bcl11b in vertebrate development (12). It has previously been reported that
Bcl11b serves functions during tooth development, including
ameloblast formation (13), and
asymmetric development of the laCL and lingual cervical loop (liCL)
(14). Early genetic studies
demonstrated that Bcl11b knockout mice exhibited delayed
odontogenesis at the bud stage and subsequent cap and bell stages,
as well as thinner enamel (13).
Furthermore, abnormal development occurred at the laCL with
improper differentiation of ameloblasts, and at the lingual side
with ectopic formation of ameloblast-like cells in Bcl11b
knockout mice (14).
The present study aimed to generate an overview
regarding the regulatory effects of Bcl11b in enamel
development. Initally, the protein expression levels of Bcl11b were
detected, thus revealing that Bcl11b was expressed throughout rat
embryonic odontogenesis. Notably, Bcl11b expression was not
detected in the epithelia or epithelial-originated tissues of
postnatal rat teeth; however, it was continuously expressed in
murine postnatal tooth germ cells. Combined with a previous report
regarding the Bcl11b expression pattern in mice (5), the present study potentially
indicated that Bcl11b may not be essential in postnatal
amelogenesis, based on the parallel morphology of rat and mouse
teeth but different Bcl11b expression profile during late
development. Furthermore, the expression levels of various
enamel-associated genes were detected, including Amelx,
Ambn, Enam, kallikrein-related peptidase 4
(Klk4), matrix metallopeptidase 20 (Mmp20) and
Msx2, which were regulated by Bcl11b. These results
suggest that Bcl11b has principal functions in embryonic
odontogenesis, particularly in dental epithelial development but
less noticeably in postnatal tooth development.
Materials and methods
Cell culture and transient
transfection
HAT-7 cells are dental epithelial cells originating
from rat cervical loop epithelium, which were provided by Professor
Hidemitsu Harada's Laboratory (Department of Oral Anatomy and
Developmental Biology, Osaka University Graduate School of
Dentistry, Osaka, Japan). The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare, Logan, UT, USA) and 1%
penicillin/streptomycin. Three small interfering (si)RNAs (si1, si2
and si3) that were designed to specifically target various sites of
Bcl11b (NM_001277288.1), as well as a negative control
siRNA, were purchased from RiboBio Co., Ltd. (Guangzhou, China).
Msx2 cDNA open reading frame (ORF) clone (cat. no. RR202436)
and pCMV6-Entry plasmid (cat. no. PS100001) were purchased from
OriGene Technologies, Inc. (Beijing, China). Cells were cultured in
6-well plates in culture medium for 24 h at 37°C prior to
transfection. siRNA was transfected at a concentration of 50 nM
into HAT-7 cells (70–80% confluence) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc). HAT-7 cells were
harvested 72 h post-transfection. In addition, 2 µg Msx2 ORF
plasmid was transfected using MegaTran 1.0 (cat. no. TT200002;
OriGene Technologies, Inc.) into cells (70–80% confluence) in a
60-mm culture dish cultured in DMEM/F12 supplemented with 10% FBS.
Transfected cells were harvested after a 72 h culture at 37°C in an
incubator containing 5% CO2. RNA and protein samples
were extracted from all transfected samples.
Animals, histology and
immunohistochemistry
C57BL/6 mice and Sprague Dawley rats were
commercially purchased from the Animal Centre (Sichuan University,
Sichuan, China). Animal care and handling was conducted in
compliance with the guidelines of the Ethics Committee of West
China College of Stomatology (Sichuan University) for animal
research. All experimental protocols were approved by the Sichuan
University Science Animal Care and Use Committee. Embryos and
mandibles from postnatal offspring were collected at various time
points, which were then used for sectioning and subsequent
immunostaining. The first detection of a vaginal plug was
considered embryonic day (E) 0.5 and the date of birth was
considered postnatal day (P) 0.5. All tissues were treated with 4%
paraformaldehyde and the postnatal mandibles were treated with 10%
EDTA for demineralization. Subsequently, the tissues were
dehydrated with a sequential concentration of alcohol and finally
with xylene. The tissues were embedded in paraffin and were cut
into 5–7 µm sections.
To examine Bcl11b protein expression by
immunohistochemistry, sections were dried overnight in a 60°C
incubator, were dehydrated with sequential concentrations of
alcohol, and were treated with 3% hydrogen peroxide for 15 min at
room temperature to block endogenous peroxidase activity.
Subsequently, the sections were incubated in slightly boiling 0.1 M
sodium citrate buffer at 95°C for 15 min. Sections were then
incubated with goat serum working solution (ZSGB-BIO, Beijing,
China) for 30 min, followed by 1 h (37°C) and then overnight (4°C)
incubations with anti-Bcl11b antibody (1:300; cat. no. 12120; Cell
Signaling Technology, Inc., Danvers, MA, USA) and anti-Sonic
hedgehog (Shh; 1:500; cat. no. 06-1106; EMD Millipore, Billerica,
MA, USA), which was used as a positive control. Following primary
antibody incubation, the slides were treated with horseradish
peroxidase-labeled anti-rabbit secondary antibody (ChemMate
EnVision Detection kit; Dako, Carpinteria, CA, USA) for 30 min at
37°C. Avidin biotin complex and 3,3′-diaminobenzidine substrate
were used as a reporting system, and the nuclei were stained with
hematoxylin. Finally images were captured under a light microscope
(BX43F; Olympus Corporation, Tokyo, Japan).
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assays were performed according to the
manufacturer's protocol of the ChIP assay kit (cat. no. 17-10086;
Upstate; Merck Millipore, Darmstadt, Germany), and all reagents
were provided in the kit. Briefly, HAT-7 cells were plated in 100
mm dishes and harvested the next day at ~90% confluence. Cells were
treated with a final concentration of 1% formaldehyde, 10X glycine
was added to quench unreacted formaldehyde. After two washes with
PBS, cells were collected for lysis and subsequent nuclear lysis.
The DNA/chromatin complex obtained from nuclear lysis were
sonicated into fragments and the protein/DNA complex was
immunoprecipitated with Bcl11b antibody. Finally purified DNA was
obtained and underwent polymerase chain reaction (PCR).
All PCR analyses were conducted as follows: Initial
denaturation at 94°C for 3 min followed by 32 cycles at 94°C for 20
sec, 59°C for 30 sec and 72°C for 30 sec, and final extension at
72°C for 2 min. The 2X PCR Master Mix (Thermo Fisher Scientific,
Inc.) was used to conduct the PCR reaction. Primers (forward
CTGAGGAAACACAAGACCAA and reverse GCGATGGAGAGGTACTGT) were used to
amplify the Msx2 promoter. GAPDH primer (forward
TATGACTCTACCCACGGCAAG and reverse TACTCAGCACCAGCATCACC) was used as
a control primer. Size of sonicated DNA fragments were confirmed by
2% agarose gel electrophoresis.
To ensure the ChIP assay had been successfully
performed, anti-RNA Polymerase II was used as a positive control
and normal mouse immunoglobulin G was used to replace the Bcl11b
antibody as a negative control, thus revealing nonspecific
immunoprecipitation. GAPDH primer was used as a primer
control.
RNA preparation and reverse
transcription-quantitative (RT-q)PCR
RNA extraction was performed using RNAiso (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocol. RT was performed using the First Strand
cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). qPCR was
conducted using SYBR Green I kit (Thermo Fisher Scientific, Inc.),
and all quantification cycle (Cq) values were normalized to
GAPDH levels (15). For
qPCR, a reaction volume of 10 µl was prepared as follows: 5 µl SYBR
Green I master mix, 100 ng cDNA, 0.2 µl forward and reverse primers
(0.4 µM final concentration), and water. Cycling conditions were as
follows: Denaturation at 95°C for 30 sec, followed by 40 cycles at
95°C for 5 sec and 60°C for 30 sec, and finally 1 cycle at 95°C for
15 sec, 60°C for 1 min and 95°C for 15 sec. All qPCR products were
examined by melt curve analysis and 2% agarose gel electrophoresis.
The primers used are listed in Table
I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Accession
number | Primer sequence
(5′-->3′) |
|---|
| Bcl11b | NM_001277288.1 | F:
GATCGGCAAGGAGGTGTA |
|
|
| R:
CATCATTAGTCAGCAAGTGTTC |
| Msx2 | NM_012982 | F:
CTGAGGAAACACAAGACCAA |
|
|
| R:
GCGATGGAGAGGTACTGT |
| Amelx | NM_001271078.1 | F:
AGCTTTTGCTATGCCCCTACC |
|
|
| R:
GATGAGGCTGAAGGGTGTGACT |
| Ambn | NM_012900.1 | F:
CTGCTCCTGTTCCTGTCCCTA |
|
|
| R:
GCTTCCCAACTGTCTCATTGTC |
| Enam | NM_001106001.1 | F:
GGTGTCTTCCCTCTCCCTAAA |
|
|
| R:
AGTGGTTTGCCATTGTCTTTCT |
| Mmp20 | NM_001106800.1 | F:
GCCTTGCTGTCCTTGTCAC |
|
|
| R:
GAGGTGGTAGTTGCTCCTGAAG |
| Klk4 | NM_001004101.1 | F:
CCGAACTACAATGACCCTTCTT |
|
|
| R:
TCAGATGCTACCGAGAGATTCA |
| GAPDH | NM_017008.4 | F:
TATGACTCTACCCACGGCAAG |
|
|
| R:
TACTCAGCACCAGCATCACC |
Protein extraction and western
blotting
Protein extraction was conducted using a total
protein extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China). Protein samples were treated with 4X loading buffer and
incubated at 100°C for 5–10 min. Briefly, protein concentration was
determined using the Bicinchoninc Acid Protein Assay kit ((Nanjing
KeyGen Biotech Co., Ltd.). Equal amounts of protein (30 ng) for
each sample were separated by 8% SDS-PAGE and transferred to
polyvinylidene difluoride membranes (Bio-Rad, Laboratories, Inc.,
Hercules, CA, USA). The membranes were then incubated with 5%
fat-free milk at room temperature for 2 h, followed by primary
antibody incubation at 4°C overnight. After three washes, the
membranes were incubated with appropriate secondary antibodies at
room temperature for 2 h. Finally, Immobilon Western
Chemiluminescent Horseradish Peroxidase Substrate (WBKLS0500; EMD
Millipore) was used for visualization combined with the LAS-3000
imaging system (Bio-Rad Laboratories, Inc.). The primary antibodies
used for western blotting were as follows: Anti-Bcl11b (1:1,000;
cat. no. 12120; Cell Signaling Technology, Inc.), anti-Msx2 (1:500;
cat. no. sc-17731; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA), anti-amelogenin (1:500; cat. no. sc-32892; Santa Cruz
Biotechnology, Inc.), anti-ameloblastin (1:500; cat. no. sc-50534;
Santa Cruz Biotechnology, Inc.), anti-Klk4 (1:500; cat. no.
sc-20622; Santa Cruz Biotechnology, Inc.), anti-Mmp20 (1:1,000;
cat. no. ab76109; Abcam, Cambridge, UK) and anti-β-actin (1:2,000;
cat. no. ab3280; Abcam). In addition, anti-mouse, anti-rabbit and
anti-goat antibodies (1:10,000; cat. nos. ZB-2305, ZB-2301 and
ZB-2306; ZSGB-BIO) were used as secondary antibodies.
Cell proliferation assay
HAT-7 cells were seeded in a 48-well plate,
transfected with Bcl11b-specific siRNA for 48 h, and were
incubated with 5-ethynyl-2′-deoxyuridine (EdU; final concentration,
20 µM; RiboBio Co., Ltd.) for 2 h at 37°C. Images were acquired
under a Leica DMI6000 B inverted fluorescence microscope (Leica
Microsystems, Wetzlar, Germany). The number of fluorescent cells
was counted and used to calculate proliferation rate (fluorescent
cells/total cells).
Statistical analysis
Student's t-test was used to perform all statistical
analyses, which were analyzed using Excel 2010 (Microscoft
Corporation, Redmond, WA, USA). Data are presented as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference (16).
Results
Bcl11b expression pattern in early
tooth germ cells
To determine the expression pattern of Bcl11b in rat
teeth, the present study detected Bcl11b protein expression during
the initiation stage and in postnatal tooth germ cells. The results
demonstrated that Bcl11b was highly expressed in the thickened
dental lamina at E12.5 (Fig. 1A),
as detected by anti-Bcl11b immunohistochemistry; however,
expression levels were much lower in the mesenchyme (Fig. 1A'). Bcl11b was also expressed in
the tooth bud at E15.5 (Fig. 1B)
as well as in the adjacent mesenchyme. During the later cap and
bell stages, Bcl11b exhibited high expression in OEE cells,
cervical loops, and stratum intermedium (SI) cells; however, levels
were reduced in IEE cells and ameloblasts, and expression was
negative in the mesenchyme and stellate reticulum cells (Fig. 1C and C'). In the neonatal molars,
Bcl11b was detected in the IEE, OEE and SI cells (Fig. 1D and D'), similar to the late cap
stage. However, no obvious expression was detected in the postnatal
mineralizing tooth (Fig. 1E and
F), which was confirmed by positive control Shh expression
(Fig. 1F and F') and
nuclear-specific location of Bcl11b. In addition, Bcl11b was
detected in mineralized mouse teeth and was restricted to the root
area (Fig. 1G and G'). Msx2 was
negatively expressed in embryonic tooth germs at E18.5 (Fig. 1H); however, it was strongly
expressed in mature ameloblasts at P1.5 (Fig. 1H').
 | Figure 1.Bcl11b expression pattern in rat
teeth. Bcl11b immunostaining appeared brown in all panels colored
by 3,3′-diaminobenzidine. Bcl11b expression in rat developing
molars at (A) E12.5, (B) E15.5, (C) E18.5 and (D) P0.5 (D). (A',
B', C', D' and G') are partial magnifications of (A, B, C, D and G)
respectively, as indicated by red rectangles. No detectable Bcl11b
expression was detected in (E) P2.5 molars, and (F) P8.5 molars and
incisors. Sonic hedgehog expression was used as a positive control
in (E') P2.5 molars, and (F') P8.5 molars and incisors. Bcl11b was
expressed in mineralized mouse teeth at (G and G') P14. Msx2 was
more strongly expressed in (H') mature ameloblasts (red arrow)
compared with in (H) embryonic tooth germs. [Scale bars: (A, B, E,
E', F, F') 200 µm; (C, D, G', H, H') 100 µm; (A', B', C', D') 50
µm; (G) 500 µm]. Bcl11b, B-cell CLL/lymphoma 11B; oe, oral
epithelium; de, dental epithelium; dm, dental mesenchyme; oee,
outer enamel epithelium; iee, inner enamel epithelium; a,
ameloblast; sr, stellate reticulum; p, papilla; cl, cervical loop;
si, stratum intermedium; E, embryonic day; P, postnatal day. |
Knockdown of Bcl11b expression
inhibits HAT-7 cell proliferation
The present study analyzed the effects of
Bcl11b expression on dental epithelial cell proliferation
using the EdU assay. The results indicated that transfection with
Bcl11b-specific siRNA statistically attenuated HAT-7
proliferation (Fig. 2). The
proliferative rate was represented by EdU-positive cells. These
results suggest that Bcl11b may promote dental epithelial cell
proliferation during tooth development.
Bcl11b regulates the expression of
enamel-associated genes and proteins
The present study aimed to determine whether
Bcl11b exerts an effect on enamel-associated gene and
protein expression. Two efficient siRNAs: Si1 and Si2, were
screened from the three siRNA molecules designed to knockdown
Bcl11b expression (Fig.
3A). Following transfection of HAT-7 cells with
Bcl11b-specific siRNA the expression levels of enamel matrix
genes and proteins, including Amelx, Ambn and
Enam, were significantly downregulated. In addition, two
enamel matrix enzymes, Klk4 and Mmp20, exhibited
slight decreases at the gene and protein level (Fig. 3B-D); however, these findings were
not significant (P=0.074 and 0.087).
 | Figure 3.Enamel-associated genes and proteins
were downregulated following knockdown of Bcl11b. (A) Si1
and Si2 were efficient siRNA molecules, as determined by western
blotting. Total protein was extracted from HAT-7 cells 72 h
post-transfection. (B) Quantitative polymerase chain reaction and
(C and D) western blotting demonstrated that enamel-associated
genes and proteins were downregulated by Bcl11b knockdown.
Statistical significance was determined using Student's t-test.
*P<0.05, **P<0.005, ***P<0.001, n=3. ns, no significance;
Bcl11b, B-cell CLL/lymphoma 11B; Amelx/AMGN,
amelogenin, X-linked; Ambn/AMBN, ameloblastin; Enam,
enamelin; Mmp20, matrix metallopeptidase 20; Klk4,
kallikrein related peptidase 4; SiN, negative control
siRNA-transfected cells; Si1/2/3, Bcl11b siRNA-transfected
cells. siRNA, small interfering RNA. |
Msx2 is associated with enamel-related
gene regulation
As shown in Fig.
4A, knockdown of Bcl11b significantly downregulated the
expression of Msx2. To determine whether Bcl11b
regulated enamel expression through Msx2, the binding of
Bcl11b to the promoter region of Msx2 was determined
in HAT-7 cells using a ChIP assay. Msx2 promoter enrichment
in Bcl11b immunoprecipitation was determined using PCR
(Fig. 4B and C). As shown in
Fig. 4C Msx2 was
significantly enriched in Bcl11b immunoprecipitation,
compared with the anti-polymerase II positive control. These
results indicate that Msx2 is a target gene of the
transcription factor Bcl11b (Fig. 4B and C).
 | Figure 4.Msx2 regulates
enamel-associated genes. Msx2 is a target gene of the
transcription factor Bcl11b, as verified by ChIP assay. (A)
Knockdown of Bcl11b led to a significant decrease in
Msx2 expression. *P<0.05, **P<0.005, n=3. (B) PCR
products of ChIP assay were verified by 2% agrose gel
electrophoresis. Bcl11b, anti-Bcl11b antibody; input, cell lysate;
Positive control, anti-RNA polymerase II antibody; GAPDH,
GAPDH primer; pBcl11b, Bcl11b promoter primer;
Bcl11b, Bcl11b primer. (C) Analysis of relative fold
enrichment of ChIP. Quantitative PCR result between the
Bcl11b experimental group and the positive control group.
Student's t-test was used for statistical analysis. *P<0.05,
n=3. (D) Relative expression of enamel-associated genes following
Msx2 overexpression. *P<0.05, ***P<0.001, n=3. ns, not
significant; Bcl11b, B-cell CLL/lymphoma 11B; Msx2,
Msh homeobox 2; Amelx, amelogenin, X-linked; Ambn,
ameloblastin; Enam, enamelin; Mmp20, matrix
metallopeptidase 20; Klk4, kallikrein related peptidase 4;
PCR, polymerase chain reaction. |
The present study also transfected an Msx2
ORF cDNA clone into HAT-7 cells; pCMV6-Entry plasmid was used as a
control. The results demonstrated that all enamel-associated genes,
including Ambn, Amelx, Enam and Klk4,
were downregulated by overexpression of Msx2 (Fig. 4D). Notably, Bcl11b was also
significantly decreased. These results suggest that a feedback loop
may exist between Bcl11b and Msx2. However, further
experiments are required.
Discussion
The present study demonstrated that Bcl11b
regulates enamel-associated gene and protein expression, via its
transcriptional target Msx2, thus suggesting that
Bcl11b serves a role in sustaining differentiation of
epithelial cells.
Bcl11b-null mice exhibited a reduction in
early tooth germ size, as well as aberrant ameloblast proliferation
and differentiation, and reduced stellate reticulum (13). Furthermore, asymmetric development
of incisor laCL and liCL was disrupted by ectopic proliferation of
epithelial cells, thus suggesting that Bcl11b was crucial
for epithelial cell proliferation and differentiation (14). At the molecular level,
Bcl11b is expressed at all stages throughout embryonic
odontogenesis in mice and rats, particularly in the epithelium;
however, its expression is reduced during late postnatal rat tooth
development (Fig. 1). Notably,
Bcl11b protein was continuously expressed in mouse teeth until
adulthood (Fig. 1G and G'). In
mineralized mouse molars, Bcl11b appeared in preameloblasts,
ameloblasts, dental follicle cells (DFCs) and periodontal ligament
cells (PLCs). In mineralized mouse incisors, Bcl11b was similarly
located in preameloblasts, ameloblasts, DFCs and PLCs (data not
shown); and in the adult cervical loop area it was strongly
expressed in OEE of the laCL and liCL, whereas it was lowly
expressed in IEE and mature ameloblasts (5). Mouse and rat teeth share similarities
in shape and regulation pattern during odontogenesis, with a
different expression pattern of Bcl11b. However, teeth in newborn
Bcl11b mutant mice (P0) exhibited indistinguishable
differences in morphogenesis and mineralization compared with wild
type mice (13), which may suggest
a nonessential role for Bcl11b in late embryonic and
postnatal tooth development. Furthermore, the present study
revealed that in Bcl11b siRNA-transfected HAT-7 cells,
proliferation was reduced compared with the control, which was
consistent with the reduced proliferation of cells in the laCL of
P21 Bcl11b mutant mouse incisors (5). These findings may suggest that
Bcl11b maintains the proliferation of non-differentiated
epithelial cells.
Msx2, together with Msx1 and
Msx3, belongs to the Msx homeobox gene family, lack
of which may cause defects in tooth cusp morphogenesis and
amelogenesis (17,18). Msx2 protein was markedly expressed
in secretory ameloblasts (Fig.
1H'), and has been reported to control expression of the
extracellular matrix gene laminin 5α3 to mediate terminal
ameloblast differentiation (17).
These findings suggested that Msx2 serves a significant role
in enamel development. The present study demonstrated that the
expression of enamel-associated genes, such as Amelx,
Ambn, Enam and Mmp20, were decreased
post-transfection with Msx2 overexpression DNA (Fig. 4D), and Bcl11b expression was
also downregulated. Previous studies have demonstrated that
Ambn prevented Msx2 expression (19) and Msx2 transcriptionally
suppressed Ambn (20), thus
presenting a reciprocal regulatory mechanism between Msx2
and Ambn. A dose-dependent relationship has also been
identified between Msx2 and enamel-related genes. In
Msx2−/− mice, Amelx and Enam were
significantly decreased, whereas in Msx2+/−
heterozygous mice Amelx exhibited increased expression
(18). Furthermore, knockdown of
Bcl11b also led to diminished Msx2 expression
(Fig. 4A), and results of a ChIP
assay suggested that Msx2 was a target gene of the
transcription factor Bcl11b (Fig. 4B) (13). Therefore it may be hypothesized
that a feedback regulatory network exists between the two molecules
for proper expression and function during amelogenesis (Fig. 5). In addition, Msx2 is
highly expressed in osteoclasts and regulates bone resorption in
the alveolar compartment (21),
which may provide evidence regarding the regulatory mechanism
between Msx2 and Bcl11b, since Bcl11b was
continuously observed in the alveolar areas in the postnatal
secretory and mature stages (data not shown).
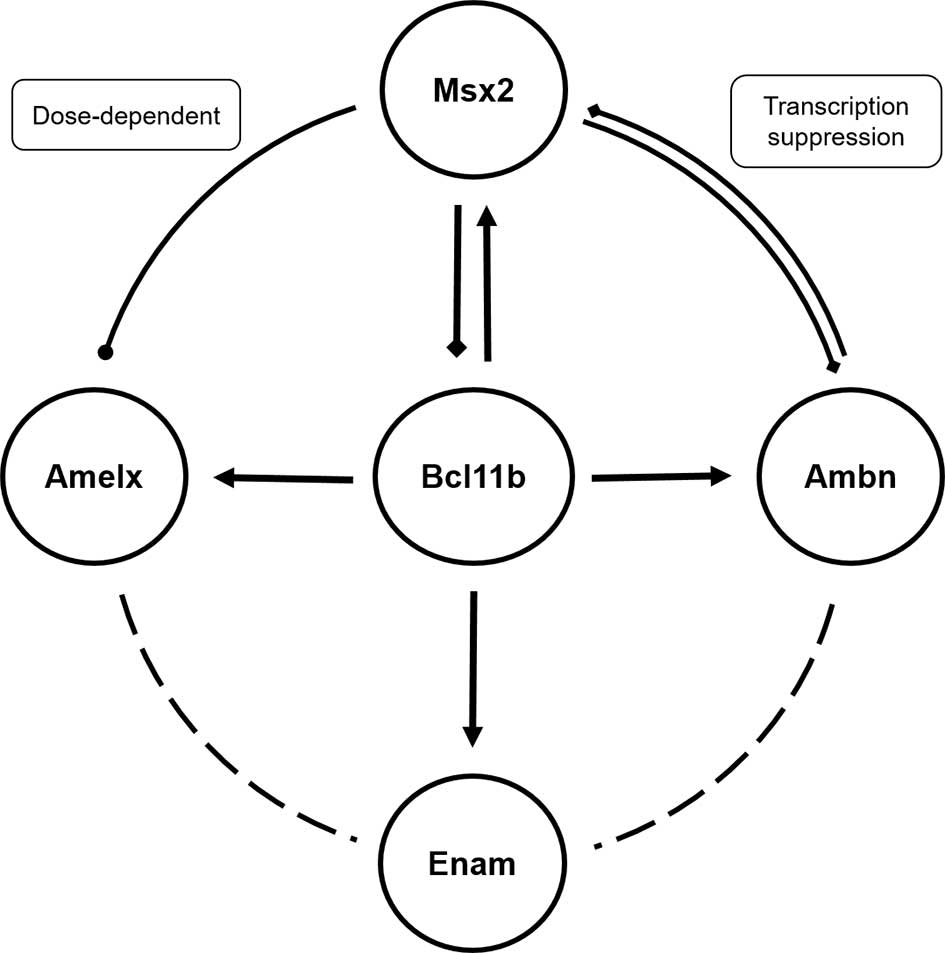 | Figure 5.Regulatory network among
Bcl11b, Msx2 and enamel matrix proteins.
Bcl11b promoted expression of enamel matrix proteins
Amelx, Ambn and Enam, and Msx2. Msx2
regulated Amelx and Ambn in a dose-dependent manner. In turn,
Msx2 inhibited enamel matrix protein expression, thus
indicating the existence of a negative feedback loop between
Msx2 and Bcl11b. Bcl11b, B-cell CLL/lymphoma
11B; Msx2, Msh homeobox 2; Amelx, amelogenin,
X-linked; Ambn, ameloblastin; Enam, enamelin. |
Amelogenin is the most abundant component of the
enamel matrix, and together with other non-amelogenin matrix
proteins, ameloblastin and enamelin, is directly responsible for
enamel formation (22). Aberrant
expression of these proteins results in grievous amelogenesis
imperfecta, which is characterized by disorganized hypoplastic
enamel, thinner enamel and chalky-white discoloration (23–25).
Adhesion of ameloblastin to ameloblasts maintains the
differentiation fate of secretory ameloblasts but inhibits
proliferation (23). Bcl11b
is a transcriptional suppressor in the central nervous, tumor and
cutaneous systems (11,26); however, it has been reported that
Bcl11b-null mice exhibit defects in enamel (13) and knockdown of Bcl11b in the
present study induced decreased enamel matrix expression. A
previous study reported that Bcl11b was detected on promoter
regions of amelogenin, enamelin and laminin 5α3, which control
ameloblast terminal differentiation (13,17).
Furthermore, it has been reported that Msx2 acts as a
dose-dependent transcriptional suppressor that regulates amelogenin
expression through antagonistic protein-protein interaction with
CCAAT/enhancer-binding protein α, which binds to its homologous
region on the mouse amelogenin promoter (27). Taken together, Bcl11b
positively regulates enamel development, and its role in ameloblast
differentiation may be mediated via Msx2 and enamel matrix
proteins. In addition, the present data revealed that Bcl11b
appeared to positively regulate Mmp20 and Klk4, and
their expression is consistent with enamel proteins; thus
suggesting that enamel proteinase expression relies on their enamel
matrix substrates.
In conclusion, there is a complex regulatory network
among Bcl11b, Msx2 and enamel matrix proteins
(Fig. 5). Bcl11b serves a
critical role in early epithelial development and amelogenesis
(13,14), and promotes the expression of
enamel matrix proteins, which are deemed to be markers of
ameloblasts. These findings suggested that Bcl11b has a role
in mediating ameloblast differentiation. However, in the present
study, Bcl11b was revealed to possess a spatiotemporal
differential regulatory role during rat enamel development. These
results indicated that postnatal Bcl11b may not be as
essential as it is during embryonic tooth development.
Acknowledgements
The present study was supported by the Nature
Science Foundation of China (grant no. 81271119), the Program for
New Century Excellent Talents by the State Education Commission of
China (grant no. NCET-13-0385), the Key Technology R&D Program
of Sichuan Province of China (grant nos. 13ZC0971, 2013GZX0158 and
13ZC0979) and the Basic Research Program of Sichuan Province of
China (grant no. 2013JY0019).
References
|
1
|
Lumsden AG: Spatial organization of the
epithelium and the role of neural crest cells in the initiation of
the mammalian tooth germ. Development. 103:(Suppl). S155–S169.
1988.
|
|
2
|
Maupin K, Droscha C and Williams B: A
Comprehensive overview of skeletal phenotypes associated with
alterations in Wnt/β-catenin signaling in humans and mice. Bone
Res. 1:27–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bei M: Molecular genetics of tooth
development. Curr Opin Genet Dev. 19:504–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang YD, Chen Z, Song YQ, Liu C and Chen
YP: Making a tooth: Growth factors, transcription factors and stem
cells. Cell Res. 15:301–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katsuragi Y, Anraku J, Nakatomi M,
Ida-Yonemochi H, Obata M, Mishima Y, Sakuraba Y, Gondo Y, Kodama Y,
Nishikawa A, et al: Bcl11b transcription factor plays a role in the
maintenance of the ameloblast-progenitors in mouse adult maxillary
incisors. Mech Dev. 130:482–492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitsiadis TA, Lardelli M, Lendahl U and
Thesleff I: Expression of Notch 1, 2 and 3 is regulated by
epithelial-mesenchymal interactions and retinoic acid in the
developing mouse tooth and associated with determination of
ameloblast cell fate. J Cell Biol. 130:407–418. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harada H, Kettunen P, Jung HS, Mustonen T,
Wang YA and Thesleff I: Localization of putative stem cells in
dental epithelium and their association with Notch and FGF
signaling. J Cell Biol. 147:105–120. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Z, Yu M and Tian W: An inductive
signalling network regulates mammalian tooth morphogenesis with
implications for tooth regeneration. Cell Prolif. 46:501–508.
2013.PubMed/NCBI
|
|
9
|
Zhang L, Vogel WK, Liu X, Topark-Ngarm A,
Arbogast BL, Maier CS, Filtz TM and Leid M: Coordinated regulation
of transcription factor Bcl11b activity in thymocytes by the
mitogen-activated protein kinase (MAPK) pathways and protein
sumoylation. J Biol Chem. 287:26971–26988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Simon R, Brylka H, Schwegler H,
Venkataramanappa S, Andratschke J, Wiegreffe C, Liu P, Fuchs E,
Jenkins NA, Copeland NG, et al: A dual function of Bcl11b/Ctip2 in
hippocampal neurogenesis. EMBO J. 31:2922–2936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Bhattacharya S, Leid M,
Ganguli-Indra G and Indra AK: Ctip2 is a dynamic regulator of
epidermal proliferation and differentiation by integrating EGFR and
Notch signaling. J Cell Sci. 125:5733–5744. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wakabayashi Y, Watanabe H, Inoue J, Takeda
N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O,
et al: Bcl11b is required for differentiation and survival of
alphabeta T lymphocytes. Nat Immunol. 4:533–539. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Golonzhka O, Metzger D, Bornert JM, Bay
BK, Gross MK, Kioussi C and Leid M: Ctip2/Bcl11b controls
ameloblast formation during mammalian odontogenesis. Proc Natl Acad
Sci USA. 106:4278–4283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kyrylkova K, Kyryachenko S, Biehs B, Klein
O, Kioussi C and Leid M: BCL11B regulates epithelial proliferation
and asymmetric development of the mouse mandibular incisor. PLoS
One. 7:e376702012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuchler U, Schwarze UY, Dobsak T, Heimel
P, Bosshardt DD, Kneissel M and Gruber R: Dental and periodontal
phenotype in sclerostin knockout mice. Int J Oral Sci. 6:70–76.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bei M, Stowell S and Maas R: Msx2 controls
ameloblast terminal differentiation. Dev Dyn. 231:758–765. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Molla M, Descroix V, Aïoub M, Simon S,
Castañeda B, Hotton D, Bolaños A, Simon Y, Lezot F, Goubin G, et
al: Enamel protein regulation and dental and periodontal
physiopathology in MSX2 mutant mice. Am J Pathol. 177:2516–2526.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sonoda A, Iwamoto T, Nakamura T, Fukumoto
E, Yoshizaki K, Yamada A, Arakaki M, Harada H, Nonaka K, Nakamura
S, et al: Critical role of heparin binding domains of ameloblastin
for dental epithelium cell adhesion and ameloblastoma
proliferation. J Biol Chem. 284:27176–27184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolaños A, Hotton D, Ferbus D, Loiodice S,
Berdal A and Babajko S: Regulation of calbindin-D(28k) expression
by Msx2 in the dental epithelium. J Histochem Cytochem. 60:603–610.
2012.PubMed/NCBI
|
|
21
|
Aïoub M, Lézot F, Molla M, Castaneda B,
Robert B, Goubin G, Néfussi JR and Berdal A: Msx2−/−
transgenic mice develop compound amelogenesis imperfecta,
dentinogenesis imperfecta and periodental osteopetrosis. Bone.
41:851–859. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paine ML, Wang HJ and Snead ML: Amelogenin
self-assembly and the role of the proline located within the
carboxyl-teleopeptide. Connect Tissue Res. 44:(Suppl). S52–S57.
2003. View Article : Google Scholar
|
|
23
|
Fukumoto S, Kiba T, Hall B, Iehara N,
Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB and
Yamada Y: Ameloblastin is a cell adhesion molecule required for
maintaining the differentiation state of ameloblasts. J Cell Biol.
167:973–983. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gibson CW, Yuan ZA, Hall B, Longenecker G,
Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington
R, et al: Amelogenin-deficient mice display an amelogenesis
imperfecta phenotype. J Biol Chem. 276:31871–31875. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wright JT, Hart TC, Hart PS, Simmons D,
Suggs C, Daley B, Simmer J, Hu J, Bartlett JD, Li Y, et al: Human
and mouse enamel phenotypes resulting from mutation or altered
expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs.
189:224–229. 2009.PubMed/NCBI
|
|
26
|
Marban C, Redel L, Suzanne S, Van Lint C,
Lecestre D, Chasserot-Golaz S, Leid M, Aunis D, Schaeffer E and
Rohr O: COUP-TF interacting protein 2 represses the initial phase
of HIV-1 gene transcription in human microglial cells. Nucleic
Acids Res. 33:2318–2331. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou YL, Lei Y and Snead ML: Functional
antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in
regulating the mouse amelogenin gene expression is mediated by
protein-protein interaction. J Biol Chem. 275:29066–29075. 2000.
View Article : Google Scholar : PubMed/NCBI
|



















