Introduction
Parkinson's disease (PD), which is a degenerative
disorder of the central nervous system, is the second most
prevalent neurodegenerative disorder. PD affects 1% of individuals
>65 years old and 4% of individuals >80 years old worldwide
(1). The predominant pathological
features associated with PD are death of dopaminergic neurons
within the substantia nigra pars compacta, cytoplasmic accumulation
of α-synuclein and other proteins in Lewy bodies and Lewy neuritis
(2,3). At the molecular level, mutations in
~9 genes, including SNCA, PINK1 and UCHL1, have been reported to be
responsible for familial forms of PD (4). However, these mutations have only
been detected in <5% patients with PD, thus suggesting that
additional genes may contribute to the susceptibility of patients
to the disease (5–7). Therefore, the exact mechanisms
underlying the pathogenesis of PD require further
investigation.
Recently, the authors of the present study have
extended their research in microRNA (miRNA) expression profiles to
determine the underlying mechanisms and functions of miRNA in PD.
miRNAs are RNA molecules, 18–24 nucleotides long, which are
associated with numerous biological processes, including
development, apoptosis and proliferation. miRNAs serve a crucial
role in the regulation of gene expression at the
post-transcriptional level by binding to the 3′-untranslated region
of the target mRNA (8,9). It has been hypothesized that >50%
of human genes are regulated by miRNAs and the regulatory
mechanisms are highly conserved among invertebrates and
vertebrates. Furthermore, associations between miRNAs and
neurodegenerative diseases, particularly PD, have been reported in
brain and blood samples (10–12).
The present study conducted a comprehensive analysis of global
miRNA and mRNA expression profiles of normal individuals and
patients with PD. A total of 88 miRNAs and 391 mRNAs were
dysregulated in patients with PD. In addition, a network of
miRNA-gene associations, which is linked to dysfunctional genes and
signaling pathways associated with PD pathology, was
established.
Materials and methods
mRNA and miRNA expression
datasets
In the present study, the mRNA expression dataset
GSE7621 (13) and the miRNA
expression dataset GSE16658 (14)
were downloaded from the National Center for Biotechnology
Information Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). In the GSE7621
dataset, 16 substantia nigra samples from patients with PD and nine
control samples were included, which were analyzed using the
Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa
Clara, CA, USA). In the GSE16658 dataset, expression profiles were
obtained from peripheral blood mononuclear cells from 19 patients
with PD and 13 normal controls, and were quantified using the
miRCURY LNA microRNA Array, v.10.0-hsa, mmu & rno (Exiqon A/S,
Vedbæk, Denmark).
Microarray data mining
Expression data preprocessing, including background
correction and normalization, was performed for both mRNA and miRNA
microarrays using the robust multi-array algorithm based on affy
package (www.bioconductor.org/packages/release/bioc/html/affy.html)
of R. Furthermore, using the limma package (www.bioconductor.org/packages/release/bioc/html/limma.html),
differentially expressed genes (DEGs) and miRNAs (DEMs) were
identified using an adjusted P-value <0.05 and a fold-change
≥1.414 as criteria.
Functional enrichment analysis
Based on the Database for Visualization, Annotation
and Integrated analysis (DAVID; david.abcc.ncifcrf.gov/) (15), the enriched Gene Ontology (GO)
terms for DEGs were identified. In the present study, only the GO
terms that were enriched in ≥5 genes were included.
Construction of miRNA-gene regulatory
network
starBase (starbase.sysu.edu.cn/) is a web-based tool, which was
developed by Li et al (16), and can be used to determine
hundreds of thousands of RNA-RNA [such as miRNA-mRNA, long
non-coding RNA (lncRNA)-miRNA and lncRNA-mRNA] and protein-RNA
(such as transcription factor-mRNA) regulatory relationships from
108 CLIP-Seq experiments. In the present study, target genes of the
DEMs were screened for using starBase. Furthermore, Cytoscape
(17) was used to visualize the
miRNA-gene regulatory network.
Results
DEGs and DEMs
The preprocessed mRNA and miRNA expression values
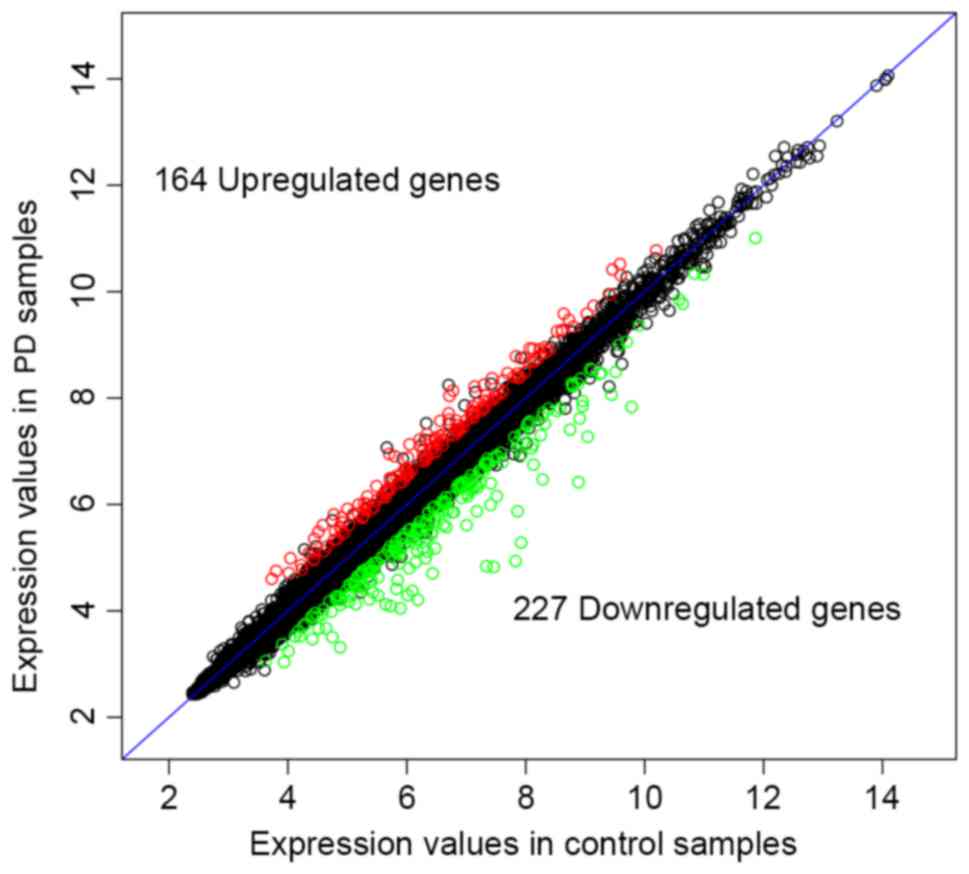
were used to screen DEGs and DEMs. Briefly, 164 up- and 227
downregulated DEGs (Fig. 1), and 2
up- and 86 downregulated DEMs were identified in patients with PD
compared with the normal controls. The top 10 up- and downregulated
DEGs and DEMs are presented in Tables
I and II, respectively.
 | Table I.Top 10 most differentially expressed
genes. |
Table I.
Top 10 most differentially expressed
genes.
| Gene | logFC | P-value |
|---|
| BANP | 0.735 |
4.576×10−7 |
| LCOR | 0.611 |
3.126×10−6 |
| RCC2 | 0.547 |
6.552×10−6 |
| ABCA11P | −0.762 |
6.795×10−6 |
| GATS | −0.825 |
7.366×10−6 |
| RBM3 | −1.521 |
1.022×10−6 |
| SLC18A2 | −2.638 |
1.087×10−6 |
| BANF1 | 0.542 |
1.199×10−6 |
| DNAJB6 | 1.241 |
1.435×10−6 |
| SSSCA1 | −0.825 |
1.583×10−6 |
 | Table II.Top 10 most differentially expressed
miRNAs. |
Table II.
Top 10 most differentially expressed
miRNAs.
| miRNA | logFC | P-value |
|---|
| hsa-miR-374a | −0.978 |
9.140×10−6 |
| hsa-miR-335 | −0.831 |
8.720×10−6 |
| hsa-miR-126* | −1.593 |
2.234×10−6 |
| hsa-miR-199a-3p | −1.126 |
1.650×10−6 |
| hsa-miR-199a-5p | −0.951 |
2.769×10−6 |
| hsa-miR-151-3p | −0.614 |
2.320×10−5 |
| hsa-miR-29b | −1.236 |
4.915×10−5 |
| hsa-miR-126 | −0.985 |
4.713×10−5 |
| hsa-miR-151-5p | −0.719 |
4.702×10−5 |
| hsa-miR-30b | −0.827 |
6.737×10−6 |
Functional enrichment analysis
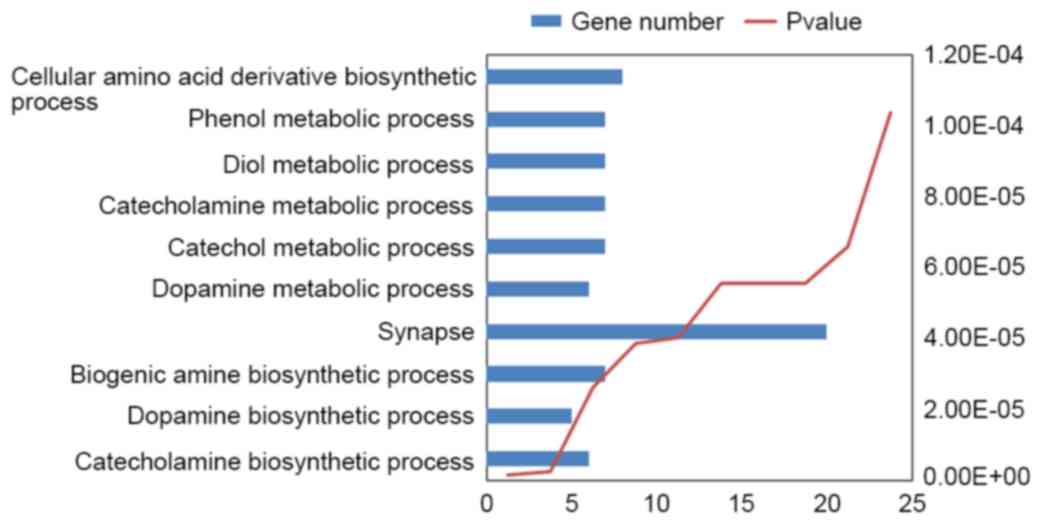
Using DAVID, 96 enriched GO terms were obtained for
DEGs. The 10 most significantly enriched GO terms, including
cellular amino acid derivative biosynthetic, phenol metabolic, diol
metabolic, catecholamine metabolic, catechol metabolic, dopamine
metabolic, synapse, biogenic amine biosynthetic, dopamine
biosynthetic and catecholamine biosynthetic processes, according to
their P-value, are presented in Fig.
2.
miRNA-gene regulation network
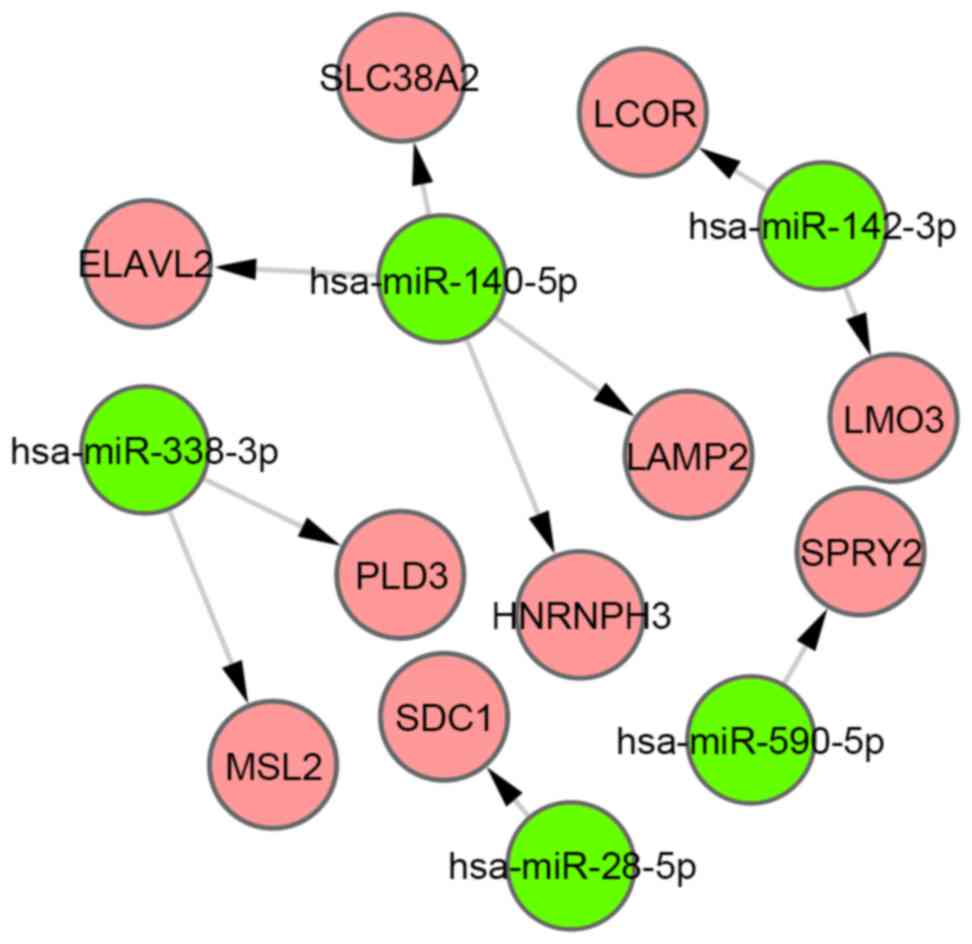
Using starBase, 620 target genes of DEMs were
identified, and 10 overlaps were identified between these target
genes and DEGs. Furthermore, 10 miRNA-gene pairs (Fig. 3) were obtained among the 10
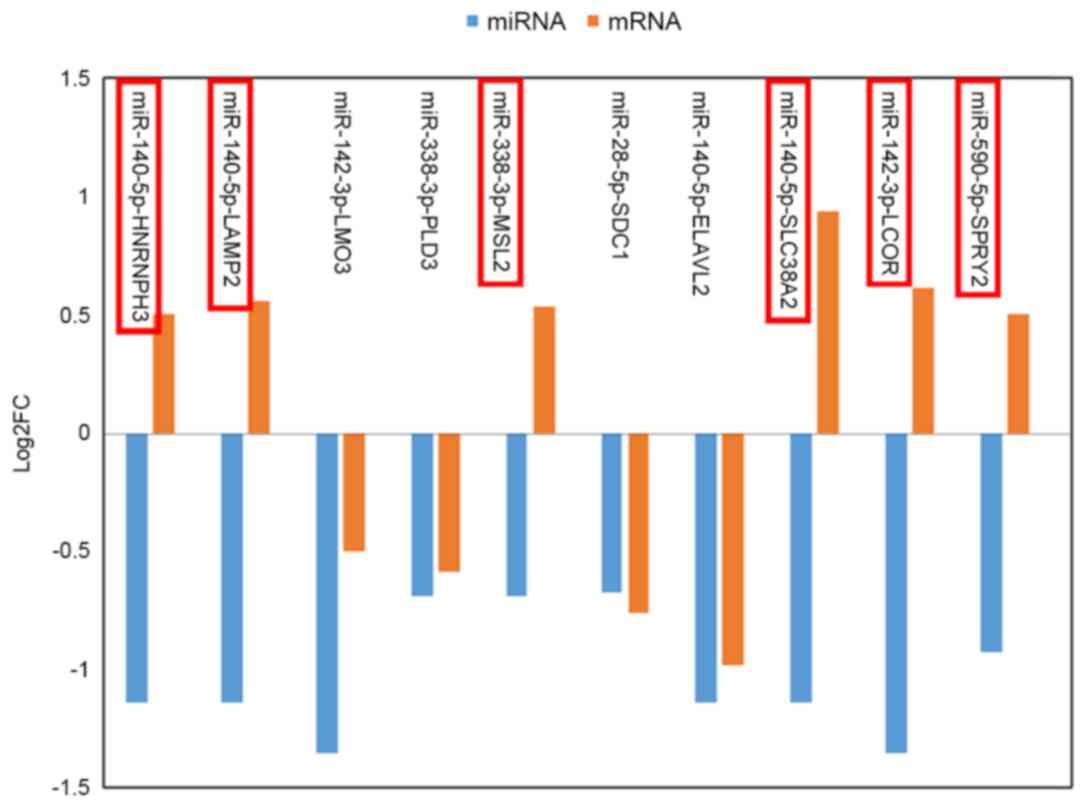
overlapped genes and 5 DEMs. Six of these miRNA-gene pairs (which
are indicated by a red border in Fig.
4) exhibited an inverse tendency with regards to changes in
miRNA and mRNA expression.
Discussion
miRNAs are regulatory molecules, which were
initially described in 1993 (18),
and have been hypothesized to serve a critical role in gene
expression associated with the pathogenesis of PD (19). PD is a multifactorial disorder that
is characterized by the loss of dopaminergic neurons within the
substantia nigra, and is associated with genetic and environmental
factors. Numerous studies have addressed whether the alterations in
miRNA expression in the brain are involved in the development and
progression of PD (11,14,20).
Furthermore, it has previously been suggested that miRNAs are able
to specifically regulate the expression of known PD-associated
genes and gene products, including leucine-rich repeat kinase and
alpha-synuclein (21,22). However, their detailed mechanistic
functions in vivo remain to be elucidated.
In the present study, 32 miRNA expression profiles
and 25 mRNA expression profiles, including patients with PD and
normal controls, were selected from the original datasets. A total
of 164 up- and 227 downregulated DEGs, and 2 up- and 86
downregulated DEMs were identified in patients with PD compared
with normal controls. A GO analysis revealed that several
significantly enriched terms were identified in the biological
process category, including catecholamine biosynthetic process,
dopamine biosynthetic process and biogenic amine biosynthetic
process, which were markedly over-represented in patients with PD.
Previous studies have reported that dopamine-dependent
neurotoxicity of α-synuclein induced selective neurodegeneration in
PD (23). Furthermore, it has
previously been suggested that regulatory enzymes of catecholamine
synthesis are important in PD (24). According to this functional
analysis and previous reports, the present study suggests that it
may be of therapeutic interest for particular elements of these
biological processes to be further researched.
The present study also identified numerous
regulatory pairs between DEMs and their target genes, including
miR-140-5P-heterogeneous nuclear ribonucleoprotein H3,
miR-140-5p-lysosomal associated membrane protein 2,
miR-338-3p-male-specific lethal 2 homolog (Drosophila),
miR-140-5p-solute carrier family 38 member 2, miR-142-3p-ligand
dependent nuclear receptor corepressor and miR-590-5p-sprouty
homolog 2 (Drosophila) (SPRY2). SPRY2 is a protein-coding
gene that is involved in thanatophoric dysplasia and Legius
syndrome (25). The encoded
protein contains a carboxyl-terminal cysteine-rich domain, which is
essential for the inhibitory activity on receptor tyrosine kinase
signaling proteins and controls various biological processes of the
central nervous system. In immature neurons, overexpression of wild
type Spry2 suppresses neurite formation and neurofilament light
chain expression; however, inhibition of Spry2 induces multiple
neuritis (26). A genome-wide
association study analyses confirmed that the most associated locus
was located in chromosome 13q31 and contained LOC729479 and SPRY2
(27). An association study of
heterogeneous nuclear ribonucleoprotein (hnRNP)-A1 and its
regulatory miRNA miR-590 was conducted in 274 patients with
frontotemporal lobar degeneration and 287 patients with Alzheimer's
disease (AD) compared with 344 age- and gender-matched controls.
The relative expression levels of miR-590 were decreased in
patients with AD compared with the controls (0.685±0.080 vs.
0.931±0.111; P=0.079), and the expression values of miR-590 and
hnRNP-A1 mRNA were negatively correlated (r=−0.615; P=0.0237).
Notably, the present study demonstrated that miR-590 levels were
decreased in patients with PD. These results indicated that the
diminished expression of miR-590 and the overexpression of its
target gene SPRY2 may have an essential role in the progression of
PD. In addition, miR-142-3p has been hypothesized to target genes
including methylenetetrahydrofolate dehydrogenase (NADP+
dependent) 2 (miRanda), SOX6 (TargetScan and PicTar) and SOX11
(TargetScan), among others, which have key roles in neural
development (28). Furthermore, in
a previous study, transfection of precursor miR-338 into the axons
of primary sympathetic neurons decreased the mRNA and protein
expression levels of cytochrome c oxidase IV, resulting in
decreased mitochondrial activity (29). This mitochondrial dysfunction and
subsequent increased oxidative stress leads to neuronal damage that
may result in the development of PD. The present study may
therefore hypothesize that miR-338 may be important in the
pathological process of PD.
In conclusion, the results of the present study are
consistent with those of the aforementioned studies, and may be a
valuable resource regarding the understanding of miRNA functions in
the pathophysiology of PD. However, the observed associations
remain controversial; therefore, more studies with larger sample
sizes across diverse populations and further molecular biology
experiments are required.
References
|
1
|
Mayeux R: Epidemiology of
neurodegeneration. Annu Rev Neurosci. 26:81–104. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spillantini MG, Schmidt ML, Lee VM,
Trojanowski JQ, Jakes R and Goedert M: Alpha-synuclein in Lewy
bodies. Nature. 388:839–840. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spillantini MG, Crowther RA, Jakes R,
Hasegawa M and Goedert M: alpha-Synuclein in filamentous inclusions
of Lewy bodies from Parkinson's disease and dementia with lewy
bodies. Proc Natl Acad Sci USA. 95:6469–6473. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wood-Kaczmar A, Gandhi S and Wood NW:
Understanding the molecular causes of Parkinson's disease. Trends
Mol Med. 12:521–528. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tenreiro S, Reimão-Pinto MM, Antas P, Rino
J, Wawrzycka D, Macedo D, Rosado-Ramos R, Amen T, Waiss M,
Magalhães F, et al: Phosphorylation modulates clearance of
alpha-synuclein inclusions in a yeast model of Parkinson's disease.
PLoS Genet. 10:e10043022014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clarimón J and Kulisevsky J: Parkinson's
disease: From genetics to clinical practice. Curr Genomics.
14:560–567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chartier-Harlin MC, Kachergus J, Roumier
C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J,
Hulihan M, et al: Alpha-synuclein locus duplication as a cause of
familial Parkinson's disease. Lancet. 364:1167–1169. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dorval V, Mandemakers W, Jolivette F,
Coudert L, Mazroui R, De Strooper B and Hébert SS: Gene and
MicroRNA transcriptome analysis of Parkinson's related LRRK2 mouse
models. PLoS One. 9:e855102014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cardo LF, Coto E, De Mena L, Ribacoba R,
Moris G, Menéndez M and Alvarez V: Profile of microRNAs in the
plasma of Parkinson's disease patients and healthy controls. J
Neurol. 260:1420–1422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Margis R, Margis R and Rieder CR:
Identification of blood microRNAs associated to Parkinsonĭs
disease. J Biotechnol. 152:96–101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lesnick TG, Papapetropoulos S, Mash DC,
Ffrench-Mullen J, Shehadeh L, De Andrade M, Henley JR, Rocca WA,
Ahlskog JE and Maraganore DM: A genomic pathway approach to a
complex disease: Axon guidance and Parkinson disease. PLoS Genet.
3:e982007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martins M, Rosa A, Guedes LC, Fonseca BV,
Gotovac K, Violante S, Mestre T, Coelho M, Rosa MM, Martin ER, et
al: Convergence of miRNA expression profiling, α-synuclein
interacton and GWAS in Parkinson's disease. PLoS One. 6:e254432011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
16
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mouradian MM: MicroRNAs in Parkinson's
disease. Neurobiol Dis. 46:279–284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miñones-Moyano E, Porta S, Escaramís G,
Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I,
Estivill X and Martí E: MicroRNA profiling of Parkinson's disease
brains identifies early downregulation of miR-34b/c which modulate
mitochondrial function. Hum Mol Genet. 20:3067–3078. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cho HJ, Liu G, Jin SM, Parisiadou L, Xie
C, Yu J, Sun L, Ma B, Ding J, Vancraenenbroeck R, et al:
MicroRNA-205 regulates the expression of Parkinson's
disease-related leucine-rich repeat kinase 2 protein. Hum Mol
Genet. 22:608–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Doxakis E: Post-transcriptional regulation
of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem.
285:12726–12734. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu J, Kao SY, Lee FJ, Song W, Jin LW and
Yankner BA: Dopamine-dependent neurotoxicity of alpha-synuclein: A
mechanism for selective neurodegeneration in Parkinson disease. Nat
Med. 8:600–606. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goldstein M and Lieberman A: The role of
the regulatory enzymes of catecholamine synthesis in Parkinson's
disease. Neurology. 424(4 Suppl 4): 8–12, 41–48. 1992.
|
|
25
|
Guo C, Degnin CR, Laederich MB, Lunstrum
GP, Holden P, Bihlmaier J, Krakow D, Cho YJ and Horton WA: Sprouty
2 disturbs FGFR3 degradation in thanatophoric dysplasia type II: A
severe form of human achondroplasia. Cell Signal. 20:1471–1477.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gross I, Armant O, Benosman S, De Aguilar
JL, Freund JN, Kedinger M, Licht JD, Gaiddon C and Loeffler JP:
Sprouty2 inhibits BDNF-induced signaling and modulates neuronal
differentiation and survival. Cell Death Differ. 14:1802–1812.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Simón-Sánchez J, Van Hilten JJ, van De
Warrenburg B, Post B, Berendse HW, Arepalli S, Hernandez DG, De Bie
RM, Velseboer D, Scheffer H, et al: Genome-wide association study
confirms extant PD risk loci among the Dutch. Eur J Hum Genet.
19:655–661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miska EA, Alvarez-Saavedra E, Townsend M,
Yoshii A, Sestan N, Rakic P, Constantine-Paton M and Horvitz HR:
Microarray analysis of microRNA expression in the developing
mammalian brain. Genome Biol. 5:R682004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aschrafi A, Schwechter AD, Mameza MG,
Natera-Naranjo O, Gioio AE and Kaplan BB: MicroRNA-338 regulates
local cytochrome c oxidase IV mRNA levels and oxidative
phosphorylation in the axons of sympathetic neurons. J Neurosci.
28:12581–12590. 2008. View Article : Google Scholar : PubMed/NCBI
|