Introduction
As a type cancer occurring in the rectum or colon,
colorectal cancer (CRC) is also termed rectal cancer or colon
cancer (1). Patients with CRC
often have symptoms of persistent tiredness, altered bowel
movements, blood in stools and weight loss (2). With a mortality rate of ≥200,000 per
year in Europe (3), CRC is the
most common type of cancer and ranks as at second leading cause of
cancer-associated mortality (4).
Several studies have been performed to investigate
the effects of genes on CRC. For example, cyclooxygenase-2, which
is mediated by vascular endothelial growth factor, can function in
the tumor angiogenesis of CRC (5).
In mutations of the catalytic subunit α of
phosphatidylinositol-3-kinase (PIK3CA), all mutations are
functionally active in colon cancer, therefore, it may be
associated with carcinogenesis (6). Mutations in Kirsten-ras, adenomatous
polyposis coli and p53 induce the transition from healthy
colonic epithelia to CRC (7). The
epigenetic loss of function of secreted frizzled-related protein
may provide constitutive WNT signaling, which is essential for
downstream mutations of complement in the progression of CRC
(8). As an epigenetic alteration,
loss of imprinting can affect the insulin-like growth factor II
gene and may be a useful predictor for the risk of CRC (9).
In 2012, Khamas et al (10) performed oligonucleotide microarray
analysis to identify the differentially expressed genes (DEGs)
between CRC samples and normal samples. Using the same data as that
used by Khamas et al (10),
the present study aimed to further identify the DEGs and CRC genes.
A sub-network of the DEGs and seed genes were obtained from the
downloaded protein-protein interaction (PPI) network, termed the
CRC.PPI. In addition, random walk with restart (RWR) analysis was
performed to identify the 50 nodes with the top affinity scores in
the CRC.PPI. The potential functions of the DEGs included in the 50
key nodes were then analyzed using Gene Ontology (GO) and pathway
enrichment analyses. In addition, drug-gene interaction analysis
was performed to identify interactions between antineoplastic drugs
and genes.
Materials and methods
Microarray data
The expression profile of GSE32323, deposited by
Khamas et al (10) was
downloaded from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), which was based on the
platform of the GPL570 (HG-U133_Plus_2) Affymetrix Human Genome
U133 Plus 2.0 array (Affymetrix, Inc., Santa Clara, CA, USA). The
GSE32323 profile included a collection of 17 pairs of matched
cancer-normal colorectal tissue samples. The CRC samples were
obtained from the American Type Culture Collection (Manassas, VA,
USA) and the Cell Resource Center for Biomedical Research of Tohoku
University (Sendai, Japan). The cells were cultured in media
obtained from Sigma-Aldrich; Merck Millipore (Darmstadt, Germany)
or Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA),
supplemented with 100 µg/ml streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 10% heat-inactivated
fetal bovine serum (Nichirei Biosciences, Tokyo, Japan).
Identification of DEGs and CRC
genes
Following downloading of the GSE32323 profile, the
microarray data were preprocessed using the Affy package (11) in Bioconductor (www.ncbi.nlm.nih.gov/geo/) and the Affy probe
annotation file from Brain Array Lab (brainarray.mhri.med.umich.edu/brainarray/default.asp).
In brief, the process included background correction, quantile
normalization, probe summarization, and log2 conversion. The mean
values of probes mapped with the same gene were obtained as
ultimate gene expression values. Subsequently, the Linear Models
for Microarray Data (Limma) package in R (www.bioconductor.org/packages/release/bioc/html/limma.html)
(12) was used to screen the DEGs
of the CRC samples, compared with the normal samples. An adjusted
P-value of P<0.05 and |logfold-change|>1 were used as the
cut-off criteria. The CRC genes were identified from a search using
the Online Mendelian Inheritance in Man (OMIM) database (www.ncbi.nlm.nih.gov/omim) (13) and were then annotated as seed
genes.
PPI network construction and RWR
analysis
The PPI network was downloaded from the Search Tool
for the Retrieval of Interacting Genes database (string-db.org/) (14). The sub-network of the DEGs and seed
genes was then obtained and termed the CRC.PPI. The RWR algorithm
was originally used for image segmentation (15). With a restarting probability (r) of
0.9, the R package dnet (16) was
used to perform RWR analysis to identify the top 50 nodes with the
highest affinity scores in the CRC.PPI. The affinity score referred
to the proximity between two nodes and the parameter ‘r’ is the
probability of moving to seed nodes.
Functional and pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; david.abcc.ncifcrf.gov/) is a tool for agglomerating a
large quantity of gene/protein symbols into gene clusters (17). Gene Ontology (GO) analysis is used
to predict the potential functions of large-scale transcriptomic
data or genomic data (18). The
Kyoto Encyclopedia of Genes and Genomes database is a base for
functional analysis combing with networks of genes and other
molecules (19). Using the DAVID
online tool, GO functional and pathway enrichment analyses were
performed for the DEGs in the PPI network of the top 50 nodes.
P<0.05 was used as the cut-off criterion.
Drug-gene interaction analysis
The Drug-Gene Interaction database (dgidb.genome.wustl.edu/) (20) was used to identify interactions
between antineoplastic drugs and genes. The actions of the drugs
inhibit genes expressed at high levels and promote genes expressed
at low levels.
Results and Discussion
DEG analysis
Compared with the normal samples, there were 1,640
DEGs screened in the CRC samples, including 850 upregulated genes
and 790 downregulated genes. The CRC genes or seed genes from the
OMIM database are listed in Table
I, including cyclin D1 (CCND1) and aurora kinase A
(AURKA).
 | Table I.Seed genes from an Online Mendelian
Inheritance in Man database search. |
Table I.
Seed genes from an Online Mendelian
Inheritance in Man database search.
| Gene/locus | ID | Name |
|---|
| PLA2G2A | 5320 | Phospholipase A2,
group IIA (platelets, synovial fluid) |
| NRAS | 4893 | Neuroblastoma RAS
viral (v-ras) oncogene homolog |
| ODC1 | 4953 | Ornithine
decarboxylase 1 |
| CTNNB1 | 1499 | Catenin
(cadherin-associated protein), β1, 88 kDa |
| PIK3CA | 5290 |
Phosphatidylinositol-4,5-bisphosphate
3-kinase, catalytic subunit α |
| FGFR3 | 2261 | Fibroblast growth
factor receptor 3 |
| TLR2 | 7097 | Roll-like receptor
2 |
| APC |
324 | Adenomatous
polyposis coli |
| MCC | 4163 | Mutated in
colorectal cancers |
| PTPN12 | 5782 | Protein tyrosine
phosphatase, non-receptor type 12 |
| PDGFRL | 5157 | Platelet-derived
growth factor receptor-like |
| RAD54B | 25788 | RAD54 homolog B (S.
cerevisiae) |
| TLR4 | 7099 | Toll-like receptor
4 |
| PTPRJ | 5795 | Protein tyrosine
phosphatase, receptor type, J |
| CCND1 |
595 | Cyclin D1 |
| MLH3 | 27030 | MutL homolog 3 |
| AKT1 |
207 | V-akt murine
thymoma viral oncogene homolog 1 |
| BUB1B |
701 | BUB1 mitotic
checkpoint serine/threonine kinase B |
| TP53 | 7157 | Tumor protein
p53 |
| FLCN | 201163 | Folliculin |
| AXIN2 | 8313 | Axin 2 |
| DCC | 1630 | Deleted in
colorectal carcinoma |
| BAX |
581 | BCL2-associated X
protein |
| AURKA | 6790 | Aurora kinase
A |
| EP300 | 2033 | E1A binding protein
p300 |
PPI network analysis
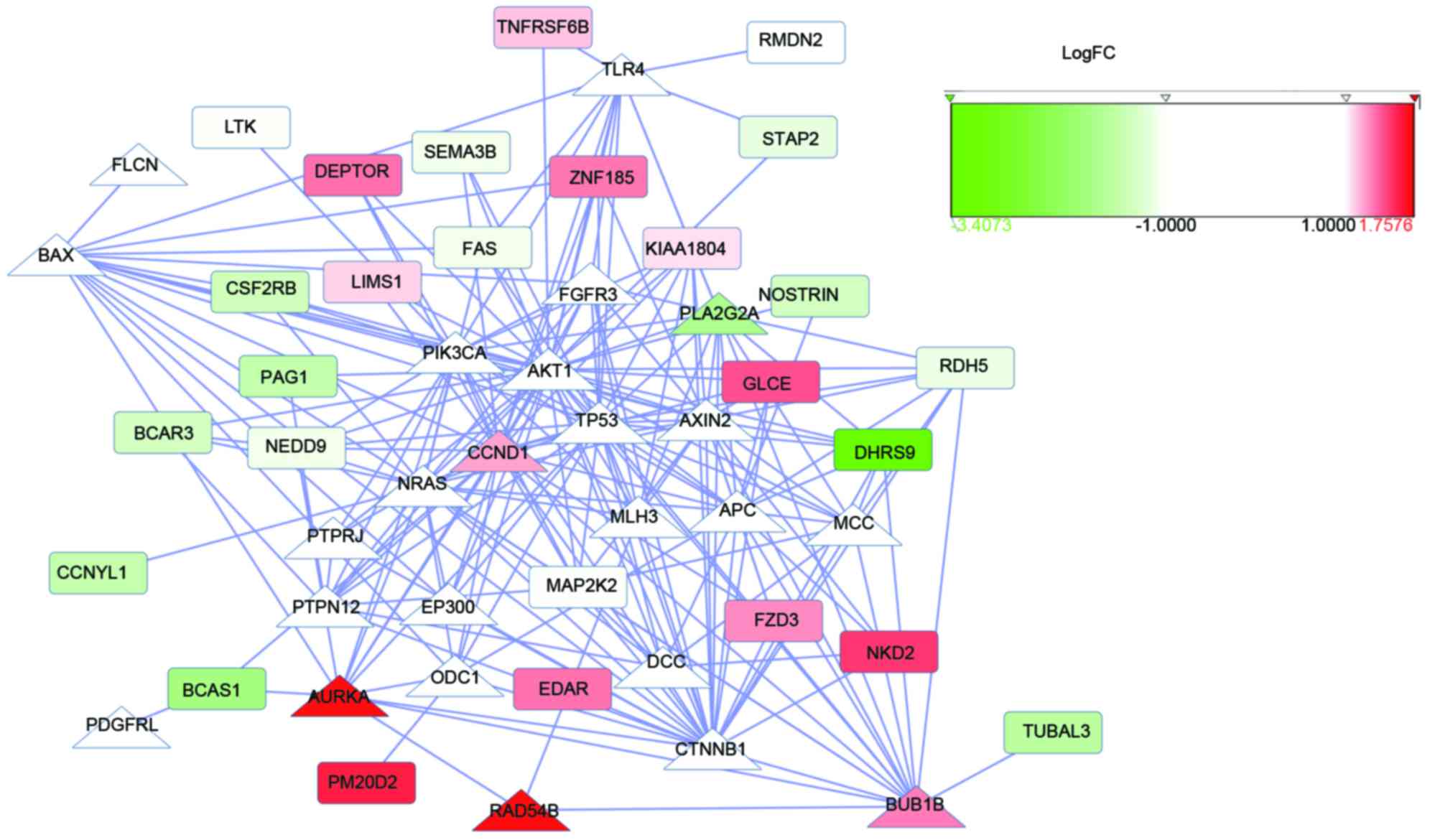
Following downloading of the PPI network and
obtaining the CRC.PPI of the DEGs and seed genes, RWR analysis was
performed and nodes with the top 50 affinity scores, which included
DEP domain-containing MTOR-interacting protein (DEPTOR), breast
carcinoma amplified sequence-1 (BCAS1), neural precursor cell
expressed developmentally downregulated 9 (NEDD9),
mitogen-activated protein kinase kinase 2, (MAP2K2) in the CRC.PPI
were identified. The PPI network of the top 50 nodes with the
highest affinity scores, which comprised 14 upregulated genes, 17
downregulated genes and 19 non-DEGs, had 224 interactions. These
interactions included DEPTOR-CCND1, AURKA-BCAS1, CCND1-BCAS1,
CCND1-NEDD9 and CCND1-MAP2K2 (Fig.
1). The DEGs included in the top 50 nodes are listed in
Table II. The enriched functions
of the 31 DEGs in the PPI network of the top 50 nodes were
primarily associated with cell cycle, including cell cycle phase
(P=0.005651), M phase (P=0.019763) and intracellular signaling
cascade (P=0.019784), as shown in Table III. The enriched pathway for the
31 DEGs in the PPI network of the top 50 nodes included
cytokine-cytokine receptor interaction (P=0.04602; Table III).
 | Table II.31 differentially expressed genes in
the protein-protein interaction network of the top 50 nodes. |
Table II.
31 differentially expressed genes in
the protein-protein interaction network of the top 50 nodes.
| Gene | Seed | Affinity score | logFC | P-value |
|---|
| PLA2G2A | Y | 0.033061 | −1.790530 | 2.46E-02 |
| CCND1 | Y | 0.032774 | 1.195653 | 5.43E-06 |
| BUB1B | Y | 0.032755 | 1.290377 | 3.10E-03 |
| AURKA | Y | 0.032712 | 1.748420 | 3.98E-05 |
| RAD54B | Y | 0.032643 | 1.757562 | 1.64E-08 |
| LTK | N | 0.001651 | −1.023600 | 1.24E-05 |
| RDH5 | N | 0.001267 | −1.146630 | 1.34E-08 |
| NOSTRIN | N | 0.001110 | −1.379460 | 1.42E-04 |
| DHRS9 | N | 0.001109 | −3.407330 | 9.77E-05 |
| RMDN2 | N | 0.001097 | −1.010210 | 7.66E-04 |
| PAG1 | N | 0.000960 | −1.535700 | 2.55E-05 |
| SEMA3B | N | 0.000917 | −1.108180 | 7.87E-07 |
| BCAS1 | N | 0.000903 | −1.881610 | 1.78E-06 |
| MAP2K2 | N | 0.000881 | −1.020110 | 3.50E-07 |
| CSF2RB | N | 0.000838 | −1.414110 | 8.49E-05 |
| TUBAL3 | N | 0.000837 | −1.631620 | 2.32E-03 |
| STAP2 | N | 0.000829 | −1.175130 | 5.35E-03 |
| CCNYL1 | N | 0.000828 | −1.516030 | 3.86E-06 |
| NEDD9 | N | 0.000808 | −1.110480 | 6.60E-05 |
| BCAR3 | N | 0.000777 | −1.380050 | 8.69E-09 |
| FAS | N | 0.000768 | −1.110550 | 1.40E-04 |
|
KIAA1804 | N | 0.001365 | 1.058162 | 2.43E-03 |
| EDAR | N | 0.001124 | 1.319916 | 2.78E-04 |
| DEPTOR | N | 0.001112 | 1.325986 | 4.94E-05 |
| LIMS1 | N | 0.000946 | 1.088253 | 7.41E-09 |
| ZNF185 | N | 0.000912 | 1.31012 | 2.84E-06 |
| NKD2 | N | 0.000910 | 1.495638 | 4.17E-08 |
| FZD3 | N | 0.000898 | 1.25656 | 2.96E-05 |
| GLCE | N | 0.000843 | 1.419823 | 7.12E-06 |
|
TNFRSF6B | N | 0.000829 | 1.121187 | 3.31E-03 |
| PM20D2 | N | 0.000822 | 1.602442 | 1.00E-06 |
 | Table III.Enriched functions and pathways for
the 31 differentially expressed genes in the protein-protein
interaction network of the top 50 nodes. |
Table III.
Enriched functions and pathways for
the 31 differentially expressed genes in the protein-protein
interaction network of the top 50 nodes.
| Category | ID | Description | Genes (n) | Gene symbol | P-value |
|---|
| GO | 0022403 | Cell cycle
phase | 5 | CCND1, BUB1B,
NEDD9, AURKA, RAD54B | 0.005651 |
| GO | 0022402 | Cell cycle
process | 5 | CCND1, BUB1B,
NEDD9, AURKA, RAD54B | 0.016449 |
| GO | 0000279 | M phase | 4 | BUB1B, NEDD9,
AURKA, RAD54B | 0.019763 |
| GO | 0007242 | Intracellular
signaling cascade | 7 | CCND1, MAP2K2,
KIAA1804, BUB1B, AURKA, PAG1, BCAR3 | 0.019784 |
| GO | 0016055 | Wnt receptor
signaling pathway | 3 | CCND1, NKD2,
FZD3 | 0.022988 |
| GO | 0000278 | Mitotic cell
cycle | 4 | CCND1, BUB1B,
NEDD9, AURKA | 0.026838 |
| GO | 0006468 | Protein amino acid
phosphorylation | 5 | CCND1, LTK, MAP2K2,
KIAA1804, AURKA | 0.028349 |
| GO | 0048545 | Response to steroid
hormone stimulus | 3 | CCND1, AURKA,
FAS | 0.045081 |
| GO | 0007049 | Cell cycle | 5 | CCND1, BUB1B,
NEDD9, AURKA, RAD54B | 0.045692 |
| KEGG | 04060 | Cytokine-cytokine
receptor interaction | 4 | TNFRSF6B, CSF2RB,
EDAR, FAS | 0.046018 |
Drug-gene interaction analysis
The interactions between antineoplastic drugs and
the 31 DEGs in the CRC.PPI network were screened. Only three DEGs
(CCND1, AURKA and DEPTOR), had interactions
with their corresponding antineoplastic drugs.
For the analysis of drug-gene interactions in the
present study, a total of 1,640 DEGs were screened in the CRC
samples, compared with normal samples, which included 850
upregulated genes and 790 downregulated genes. The obtained seed
genes included CCND1 and AURKA. The enriched
functions for the 31 DEGs in the PPI network of the top 50 nodes
were primarily associated with cell cycle. According to the
drug-gene interaction analysis, only three DEGs (CCND1,
AURKA and DEPTOR) had interactions with their
corresponding antineoplastic drugs.
DEPTOR, which is negatively correlated with
tumor progression and the activity of mammalian target of rapamycin
complex 1 (mTORC1) in CRC, may function as a marker for
prognosis and treatment of CRC (21). The PI3K/Akt/mTOR signaling pathway
is one of the primary mechanisms involved in maintaining tumor
progression and metastasis, and mTOR activation/inhibition
functions in CRC (22). It has
been reported that DEPTOR may be closely correlated with
CRC. The variants in CCND1 may have potential associations
with familial CRC (23), and the
CCND1 A870G polymorphism may increase the risk of CRC
(24). The epidermal growth factor
A61G and CCND1 A870G polymorphisms may be valuable molecular
biomarkers for the prognosis of patients with CRC treated with the
single-agent, cetuximab (25).
Thus, the expression levels of CCND1 may be associated with
CRC. The PPI network of the top 50 nodes in the present study also
showed CCND1 had an interaction with DEPTOR, indicating that
CCND1 may be involved in CRC by mediating DEPTOR.
Independent of established clinicopathological
variables, AURKA can act as a molecular marker for
predicting clinical outcome in patients with liver metastasis of
CRC (26). AURKA, which is
located on chromosome 20q, is involved in adenoma-to-carcinoma
progression and may function as an indicator of poor prognosis
(27–33). NABC1, also known as
BCAS1, and its variant, NABC15B, are downregulated in
CRC (34). These findings suggest
that AURKA and BCAS1 may be associated with CRC. In
the PPI network of the top 50 nodes in the present study, it was
also found that BCAS1 interacted with AURKA, suggesting that
BCAS1 may also be involved in CRC through regulating
AURKA.
As a scaffolding protein, NEDD9, also termed
human enhancer of filamentation 1 (HEF1) is involved in
mediating several cellular processes, including cell cycle
progression, cellular attachment, apoptosis, motility and
inflammation (35,36). Prostaglandin E2 (PGE2)
promotes the expression of HEF1 and then induces cell cycle
progression, and HEF1 is an important downstream mediator of
the activity of PGE2 in the progression of CRC (37). The activation and overexpression of
extracellular signal-regulated kinase/MAPK is important in
the progression of CRC and can be a molecular target for the
treatment of the cancer (38). It
has been reported that MAP2K inhibitors can be of use in CRC
therapy (39). Therefore, the
dysregulation of NEDD9 and MAP2K2 may affect CRC. In
the PPI network of the top 50 nodes in the present study, CCND1 was
also found to have interactions with BCAS1, NEDD9 and MAP2K2,
indicating that CCND1 may also function in CRC through
BCAS1, NEDD9 and MAP2K2.
In conclusion, the present study performed a
comprehensive bioinformatics analysis of genes, which may be
involved in CRC. A total of 1,640 DEGs were screened in the CRC
samples, compared with normal samples. It was found that
DEPTOR, AURKA, CCND1, BCAS1,
NEDD9 and MAP2K2 may be correlated with CRC. However,
further confirmation is required to elucidate their mechanisms of
action in CRC.
References
|
1
|
Kune GA, Kune S and Watson LF: Colorectal
cancer risk, chronic illnesses, operations and medications: Case
control results from the melbourne colorectal cancer study. Cancer
Res. 48:4399–4404. 1988.PubMed/NCBI
|
|
2
|
Majumdar SR, Fletcher RH and Evans AT: How
does colorectal cancer present?; symptoms, duration, and clues to
location. Am J Gastroenterol. 94:3039–3045. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Bouvier AM, Foschi R, Hackl M,
Larsen IK, Lemmens V, Mangone L and Francisci S: Progress in
colorectal cancer survival in Europe from the late 1980s to the
early 21st century: The EUROCARE study. Int J Cancer.
131:1649–1658. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Parkin DM and Steliarova-Foucher
E: Estimates of cancer incidence and mortality in Europe in 2008.
Eur J Cancer. 46:765–781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cianchi F, Cortesini C, Bechi P, Fantappiè
O, Messerini L, Vannacci A, Sardi I, Baroni G, Boddi V, Mazzanti R
and Masini E: Up-regulation of cyclooxygenase 2 gene expression
correlates with tumor angiogenesis in human colorectal cancer.
Gastroenterology. 121:1339–1347. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ikenoue T, Kanai F, Hikiba Y, Obata T,
Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, et al:
Functional analysis of PIK3CA gene mutations in human colorectal
cancer. Cancer Res. 65:4562–4567. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith G, Carey FA, Beattie J, Wilkie MJ,
Lightfoot TJ, Coxhead J, Garner RC, Steele RJ and Wolf CR:
Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways
to colorectal cancer. Proc Natl Acad Sci USA. 99:9433–9438. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, van
Engeland M, et al: Epigenetic inactivation of SFRP genes allows
constitutive WNT signaling in colorectal cancer. Nat Genet.
36:417–422. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui H, Cruz-Correa M, Giardiello FM,
Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR and
Feinberg AP: Loss of IGF2 imprinting: A potential marker of
colorectal cancer risk. Science. 299:1753–1755. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khamas A, Ishikawa T, Shimokawa K, Mogushi
K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H and Sugihara
K: Screening for epigenetically masked genes in colorectal cancer
Using 5-Aza-2′-deoxycytidine, microarray and gene expression
profile. Cancer Genomics-Proteomics. 9:67–75. 2012.PubMed/NCBI
|
|
11
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
13
|
Hamosh A, Scott AF, Amberger JS, Bocchini
CA and McKusick VA: Online Mendelian Inheritance in Man (OMIM), a
knowledgebase of human genes and genetic disorders. Nucleic acids
Res. 33:D514–D517. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang H and Gough J: A
disease-drug-phenotype matrix inferred by walking on a functional
domain network. Mol Biosyst. 9:1686–1696. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang H and Gough J: The ‘dnet’ approach
promotes emerging research on cancer patient survival. Genome Med.
6:642014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sherman BT, da W Huang, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for Gene Ontology
analysis. BMC Proc. 3 Suppl 4:S102009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Griffith M, Griffith OL, Coffman AC,
Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB,
Eldred JM, et al: DGIdb: Mining the druggable genome. Nat Methods.
10:1209–1210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lai1 EY, Chen ZG, Zhou X, Fan XR, Wang H,
Lai PL, Su YC, Zhang BY, Bai XC and Li YF: DEPTOR Expression
negatively correlates with mTORC1 activity and tumor progression in
colorectal cancer. Asian Pac J Cancer Prev. 15:4589–4594. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Francipane MG and Lagasse E: mTOR pathway
in colorectal cancer: An update. Oncotarget. 5:492014.PubMed/NCBI
|
|
23
|
Grünhage F, Jungck M, Lamberti C, Berg C,
Becker U, Schulte-Witte H, Plassmann D, Rahner N, Aretz S,
Friedrichs N, et al: Association of familial colorectal cancer with
variants in the E-cadherin (CDH1) and cyclin D1 (CCND1) genes. Int
J Colorectal Dis. 23:147–154. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lewis RC, Bostick RM, Xie D, Deng Z,
Wargovich MJ, Fina MF, Roufail WM and Geisinger KR: Polymorphism of
the cyclin D1 gene, CCND1, and risk for incident sporadic
colorectal adenomas. Cancer Res. 63:8549–8553. 2003.PubMed/NCBI
|
|
25
|
Zhang W, Gordon M, Press OA, Rhodes K,
Vallböhmer D, Yang DY, Park D, Fazzone W, Schultheis A, Sherrod AE,
et al: Cyclin D1 and epidermal growth factor polymorphisms
associated with survival in patients with advanced colorectal
cancer treated with Cetuximab. Pharmacogenet Genomics. 16:475–483.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goos JA, Coupe VM, Diosdado B, Delis-Van
Diemen PM, Karga C, Beliën JA, Carvalho B, van den Tol MP, Verheul
HM, Geldof AA, et al: Aurora kinase A (AURKA) expression in
colorectal cancer liver metastasis is associated with poor
prognosis. Br J Cancer. 109:2445–2452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bischoff JR, Anderson L, Zhu Y, Mossie K,
Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et
al: A homologue of Drosophila aurora kinase is oncogenic and
amplified in human colorectal cancers. EMBO J. 17:3052–3065. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hermsen M, Postma C, Baak J, Weiss M,
Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W,
et al: Colorectal adenoma to carcinoma progression follows multiple
pathways of chromosomal instability. Gastroenterology.
123:1109–1119. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aust DE, Muders M, Köhler A, Schmidt M,
Diebold J, Müller C, Löhrs U, Waldman FM and Baretton GB:
Prognostic relevance of 20q13 gains in sporadic colorectal cancers:
A FISH analysis. Scand J Gastroenterol. 39:766–772. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakao K, Mehta KR, Fridlyand J, Moore DH,
Jain AN, Lafuente A, Wiencke JW, Terdiman JP and Waldman FM:
High-resolution analysis of DNA copy number alterations in
colorectal cancer by array-based comparative genomic hybridization.
Carcinogenesis. 25:1345–1357. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Postma C, Terwischa S, Hermsen MA, Van der
Sijp JR and Meijer GA: Gain of chromosome 20q is an indicator of
poor prognosis in colorectal cancer. Cell Oncol. 29:73–75.
2007.PubMed/NCBI
|
|
32
|
Carvalho B, Postma C, Mongera S, Hopmans
E, Diskin S, van De Wiel MA, Van Criekinge W, Thas O, Matthäi A,
Cuesta MA, et al: Multiple putative oncogenes at the chromosome 20q
amplicon contribute to colorectal adenoma to carcinoma progression.
Gut. 58:79–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sillars-Hardebol AH, Carvalho B, De Wit M,
Postma C, Delis-van Diemen PM, Mongera S, Ylstra B, van De Wiel MA,
Meijer GA and Fijneman RJ: Identification of key genes for
carcinogenic pathways associated with colorectal
adenoma-to-carcinoma progression. Tumour Biol. 31:89–96. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Correa RG, De Carvalho AF, Pinheiro NA,
Simpson AJ and De Souza SJ: NABC1 (BCAS1): Alternative splicing and
downregulation in colorectal tumors. Genomics. 65:299–302. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim SH, Xia D, Kim SW, Holla V, Menter DG
and DuBois RN: Human enhancer of filamentation 1 is a mediator of
hypoxia-inducible factor-1alpha-mediated migration in colorectal
carcinoma cells. Cancer Res. 70:4054–4063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Singh MK, Cowell L, Seo S, O'Neill G and
Golemis E: Molecular basis for HEF1/NEDD9/Cas-L action as a
multifunctional co-ordinator of invasion, apoptosis and cell cycle.
Cell Biochem Biophys. 48:54–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xia D, Holla VR, Wang D, Menter DG and
DuBois RN: HEF1 is a crucial mediator of the proliferative effects
of prostaglandin E(2) on colon cancer cells. Cancer Res.
70:824–831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yeh JJ, Routh ED, Rubinas T, Peacock J,
Martin TD, Shen XJ, Sandler RS, Kim HJ, Keku TO and Der CJ:
KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for
MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther.
8:834–843. 2009. View Article : Google Scholar : PubMed/NCBI
|