Introduction
Lung cancer is one of the most common types of
malignant tumor worldwide, and has become the leading contributor
to rates of mortality in China during the past decade (1–3).
Non-small cell lung cancer (NSCLC) accounts for ~80% of all cases
of lung cancer, and includes squamous cell carcinoma,
adenocarcinoma and large cell carcinoma. Compared with small cell
carcinoma, the growth and division of NSCLC cells are more rapid,
and their diffusion and metastasis occur at a relatively early
stage (4). In cases of NSCLC,
>50% of patients are found to be in the middle and advanced
stages, and the 5 year survival rate is relatively low (4–6). The
pathogenesis of NSCLC remains to be fully elucidated. Therefore, it
is important to identify novel genes or factors, which regulate the
progression of NSCLC.
Chronic hypoxia is an often slow, insidious
reduction in tissue oxygenation. Hypoxia-inducible factors (HIFs)
are expressed at high levels in response to hypoxia, and
transcriptionally upregulates a series of genes causing metabolic
alterations (7,8). The decrease of oxygen supply usually
results in significant functional changes of normal and cancerous
cells. In normal cells, including neurons and myocytes, chronic
hypoxia has been shown to cause a series of disorders in the cells
and can even induce their apoptosis (9,10).
However, the roles of hypoxia in the functions of cancer cells
differ substantially. It has been reported that chronic hypoxia has
a promoting effect on cancer progression through regulating
cancerous cell proliferation, metastasis and angiogenesis (11–13).
MicroRNAs (miRs) have emerged as being important in
physiological and pathological biological processes. miR-191 is a
conserved miRNA, which has been investigated in detail and has been
identified as being transcribed from an intronic region of the
gene, DALR anti-codon binding domain containing 3 (14). Induced by several master
transcription factors, miR-191 has been reported to be abnormally
expressed in >20 types of cancer and numerous other diseases,
including obesity, type II diabetes, pulmonary hypertension and
Alzheimer's disease (15,16). miR-191 regulates cell
proliferation, differentiation, apoptosis and migration by
targeting important transcription factors, chromatin remodelers and
cell cycle-associated genes (15).
Several studies have demonstrated that miR-191 is a valuable
biomarker for cancer diagnosis and prognosis (17,18).
A number of reports have shown that miR-191 can be induced by
HIF-1α and has a contributory action in the progression of breast,
hepatic and pancreatic cancers (19–21).
In patients with lung cancer, miR-191 has been found to be
upregulated, suggesting that it may be involved in progression of
lung cancer (22). However, a
previous in vitro study indicated that the overexpression of
miR-191 did not lead to changes in cell cycle, proliferation,
xenograft formation or in the chemosensitivity of A549 lung
adenocarcinoma and BEAS-2B normal lung cell lines (23). Whether miR-191 has any effects on
the progression of NSCLC remains to be elucidated
In the present study, the expression of miR-191 was
detected in the NSCLC tissues and adjacent matched normal tissues,
as well as in human NSCLC cell lines and human normal lung cells,
under both normal and hypoxic condition. Following this, miR-191
was overexpressed or silenced in A549 NSCLC cells under normal or
hypoxic condition, and the exact role of miR-191 in cell
proliferation and migration of NSCLC cells, and the potential
mechanism were investigated. It was found that miR-191 had a
significant impact on hypoxic NSCLC cells, however, not under
normal conditions. The present study provided a novel insight into
the regulation of NSCLC progression.
Materials and methods
Ethical statement and sampling
In the present study, 96 patients with middle- or
late-stage NSCLC were recruited, comprising 32 patients with
squamous cell cancer (aged 50±11 years, 16 men and 16 women), 32
patients with adenocarcinoma (aged 49±10 years, 16 men and 16
women) and 32 patients with large cell carcinoma (aged 52±12 years,
16 men and 16 women). Cancerous tissue and matched adjacent normal
tissues were sampled from each subject during minimal invasive
surgery. Tumor diameter ranged between 3 and 7 cm. The present
study was approved by the Ethical Committee of Zhengzhou University
(Zhengzhou, China), and all the subjects provided written informed
consent prior to participation in the investigation.
Cell culture and chronic hypoxia
treatment
HBE normal lung cells, SK-MES-1 human lung squamous
cell carcinoma cells, A549 human lung adenocarcinoma cells and
NCI-H460 human lung large cell carcinoma cells were purchased from
Shanghai Institutes for Biological Sciences (Shanghai, China). The
cells were stored in liquid nitrogen and thawed in a 37°C water
bath. The cells were centrifuged at 1,000 g for 7 min, following
which the HBE, SK-MES-1 and A549 cells were suspended in Dulbecco's
modified Eagl's medium containing 4.5 g/l glucose and 4 mmol/l
L-glutamin (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.). The NCI-H460 cells were suspended
in RPMI-1640 containing 4.5 g/l glucose and 4 mmol/l L-glutamin
supplemented with 10% FBS. For treatment, the cells were seeded
into 12-well plates at a density of 105/cm2. The cells were
incubated in a humidified incubator with an atmosphere of 95%
air/5% CO2 at 37°C until adhesion. For hypoxia
treatment, the adherent cells were incubated in a humidified
incubator with an atmosphere of 80% N2/15%
O2/5% CO2 at 37°C.
Transfection
A single-strand miRNA mimic negative control (NC)
and miR-191 mimic were designed, synthesized and confirmed as
effective by RiboBio Co., Ltd. (Guangzhou, China). On reaching 80%
confluence, 6 pmol NC or 6 pmol miR-191 mimic were transfected into
the cell lines using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
medium was replaced every 3 days.
Cell proliferation and migration
analysis
Cell proliferation was evaluated using MTT
(Sigma-Aldrich, St. Louis, MO, USA) and Cell Counting kit-8
(Sigma-Aldrich) assays. Following treatment, the cells
(1×104 cells/well) were incubated at 37°C for 0, 24, 48
and 72 h prior to the addition of MTT reagent to each well at a
final concentration of 0.5 mg/ml and incubation at 37°C for 4 h.
Following removal of the medium, 500 µl dimethyl sulfoxide was
added to each well. The proportion of viable cells were measured by
absorbance at a 550 nm wavelength using a microplate reader (BioRad
Laboratories, Inc., Hercules, CA, USA). The Cell Counting kit-8
assay was performed, according to the manufacturer's protocol.
Migration was detected using a Transwell migration
assay. The cell migration assay was performed using Matrigel (BD
Biosciences, Sparks, MD, USA), which was coated on the upper
surface of the Transwell chamber (Corning, Lowell, MA, USA). The
cells, which migrated through the membrane were fixed with methanol
and stained with crystal violet (Sigma-Aldrich). Images of three
randomly selected fields of the fixed cells were captured, and
cells numbers were counted using a Countess automatic cell counter
(Invitrogen; Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The RNA concentration was quantified using
a spectrophotometer measuring the optical density (OD)260/280 ratio
(1.80–1.95). The integrity of the RNA was confirmed by
electrophoresis on 1.0% agarose gel with ethidium bromide
(Sigma-Aldrich) staining. RT was performed using a SuperScript II
1st Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The reaction was run in a 20 µl system
containing 2 µl SuperScript® RT Enzyme Mix I, 8 µl
SuperScript® Buffer, 2 µl Invitrogen NCode miRNA
universal qPCR primer, 2 µl RNA template, and 6 µl RNase-free
H2O. The system was incubated at 42°C for 30 min and was
subsequently incubated at 85°C for 15 sec to finalize the reaction.
The qPCR reactions were performed using a final sample volume of 25
µl, containing SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan),
0.4 mM of each primer and 200 ng of cDNA template. The mature
miR-191 stem-loop primer and quantitative primers, as well as the
U6 RNA primers, were designed and produced by RiboBio Co., Ltd.).
Primer sequences were as follows: U6, forward:
5′-CGCTTCGGCAGCACATATAC-3′ and reverse: 5′-TTCACGAATTTGCGTGTCAT-3′'
miR-191, forward: 5′-CAACGGAATCCCAAAAGCAGCT-3′ and reverse:
Invitrogen NCode miRNA universal qPCR primer. Each individual
sample was run in triplicate wells. The PCR amplification cycles
were performed using a iQTM5 Multicolor Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc.) and a SYBR Premix Ex Taq II kit
(Invitrogen; Thermo Fisher Scientific, Inc.). The reactions were
initially denatured at 95°C for 3 min, followed by 35 cycles of
95°C for 15 sec and 60°C for 60 sec. The data were quantified using
the 2-ΔΔCq method (24). The quantities of mRNA were
normalized to that of U6 RNA.
3′UTR luciferase reporter assay
The cDNA fragment corresponding to the 3′UTR of NFIA
mRNA, containing the binding site of miR-191 with XhoI and
NotI cutting sites, was constructed and confirmed by
sequencing. The fragments were cloned into psiCHECK™-2 vectors
(Promega, Madison, WI, USA) at the 3′-end of the Renilla gene. The
HEK293T cells (20×103) were seeded into 96-well plates for 24 h,
following which transfection was performed using 1.0 µl
Lipofectamine 3000 transfection reagent (Roche, Mannheim, Germany),
100 ng of the vector constructs, and 50 nM of either the miR-191
mimic or NC per well. The cells were harvested 48 h following
transfection. Luciferase activity was measured using a DualGlo
Luciferase Assay system (Promega). Renilla luciferase activity was
measured and normalized to the corresponding firefly luciferase
activity.
Western blot analysis
The total proteins were extracted using a Total
Protein Extraction kit (BestBio, Shanghai, China) and quantified
using a BCA Protein assay kit (Merck, Whitehouse Station, NJ, USA),
according to the manufacturer's protocols. Equal quantities of
protein (20 µg) from each sample were separated by 12% SDS-PAGE and
electro-transferred onto polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA) for immunoblotting analysis. The
membranse were blocked using blocking buffer (Abcam, Cambridge, UK)
overnight at 4°C. The following primary antibodies were used:
Anti-CCAAT-enhancer-binding protein α (C/EBPα; 1:200; cat. no.
ab8227; Abcam), anti-NFIA (1:300; cat. no. ab41851; Abcam) and
anti-β-actin (1:200; cat. no. ab32358; Abcam), which was used as
the internal reference. The antibodies were diluted in 5% non-fat
milk in tris-buffered saline containing Tween-20 (TBST) and the
incubation was for 2 h at 37°C. Following washing three times (8
min each) with TBST, the membranes were incubated with goat
anti-rabbit horseradish-peroxidase-conjugated secondary antibody
(cat. no. ab6721; 1:2,000; Abcam) for 1 h at 37°C. The membranes
were washed as before and the the proteins were detected using a
ChemiDoc XRS imaging system and Quantity One version 4.62 analysis
software (Bio-Rad Laboratories, Inc.).
Prediction software
The targeting association between NFIA and miR-191
was predicted using the online server, TargetScan (http://www.targetscan.org/cgi-bin/targetscan/vert_61/view_gene.cgi?taxid=9606&rs=NM_001134673&members=miR-191&showcnc=0&shownc=0).
Statistical analysis
All data were obtained from at least three
independent experiments. Values are expressed as the mean ±
standard error of the mean. Statistical analyses were performed
using SPSS 19.0 (IBM SPSS, Armonk, NY, USA). Multiple comparisons
were assessed using one-way analysis of variance followed by
Dunnett's tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
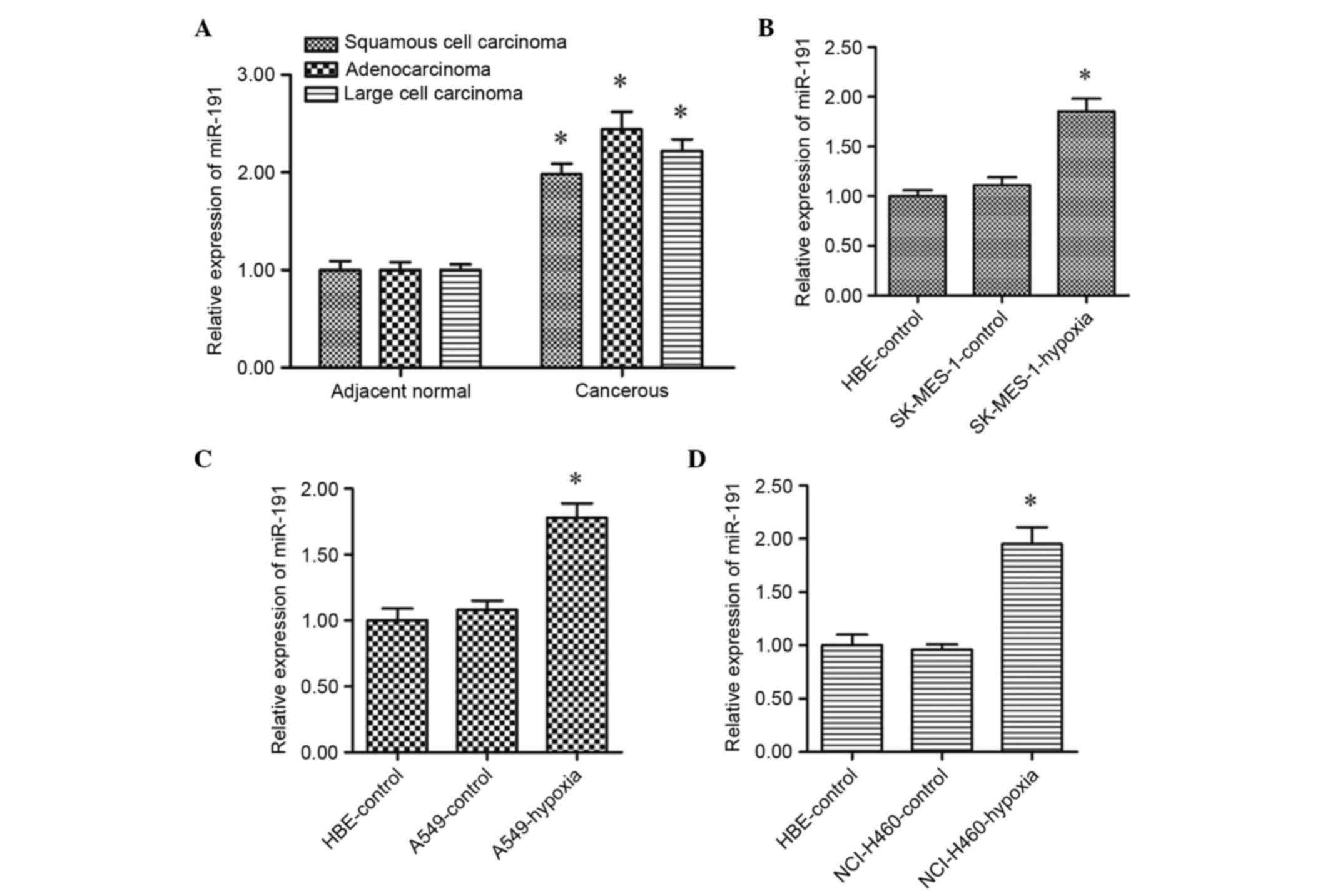
Expression of miR-191 is significantly
upregulated in NSCLC cells under chronic hypoxic conditions
The expression levels of mature miR-191 in cancerous
tissues and adjacent non-cancerous tissues from the patients with
NSCLC were detected using RT-qPCR analysis. The results
demonstrated that the levels of miR-191 were significantly
upregulated in the cancerous tissues, compared with the adjacent
tissues (Fig. 1A). However, the
data obtained from the lung cancer cell lines showed that there
were no significant differences in the levels of miR-191 between
the normal lung cell line and the NSCLC cell lines (Fig. 1B-D). Subsequently, the NSCLC cells
were incubated in a chronic hypoxic environment (15%
O2). It was found that the levels of miR-191 were
markedly upregulated in response to chronic hypoxia, compared with
the cells under normal conditions (Fig. 1B-D). These data suggested that
miR-191 may be involve in the progression of NSCLC cells under
chronic hypoxic conditions.
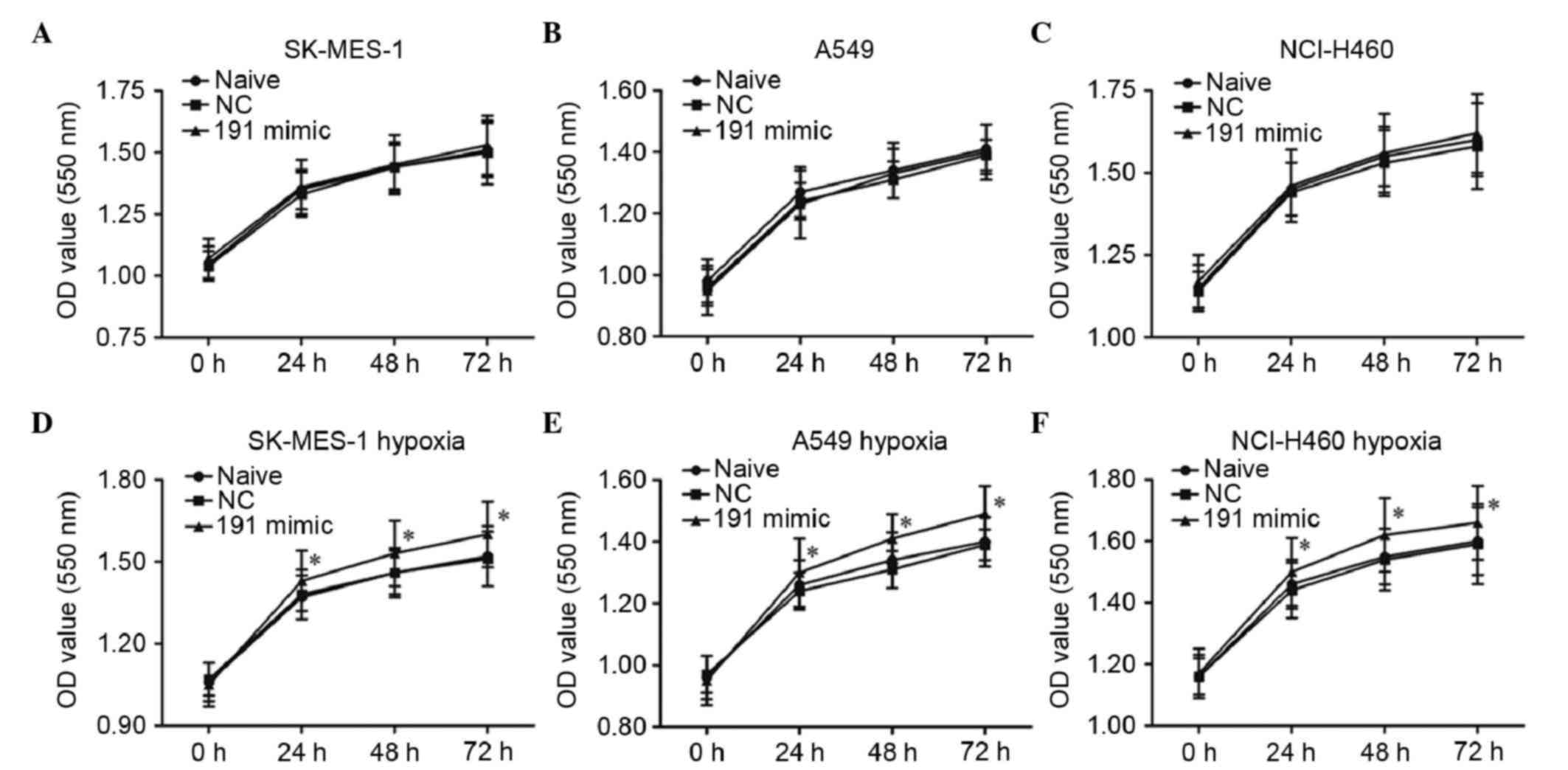
Overexpression of miR-191 has no
effect on the proliferation of NSCLC cell lines under normal
conditions, but promotes their proliferation under chronic hypoxic
conditions
To investigate the role of miR-191 in the
progression of NSCLC in vitro, the NC mimic or miR-191 mimic
were transfected into the SK-MES-1, A549 and NCI-H460 human NSCLC
cell lines under normal conditions and chronic hypoxic conditions.
The proliferation of the cells were then detected using an MTT
assay. The results showed that miR-191 had no effect on the
proliferation of these three cell lines (Fig. 2A-C), whereas transfection with the
miR-191 mimic led to significant promotion of the proliferation of
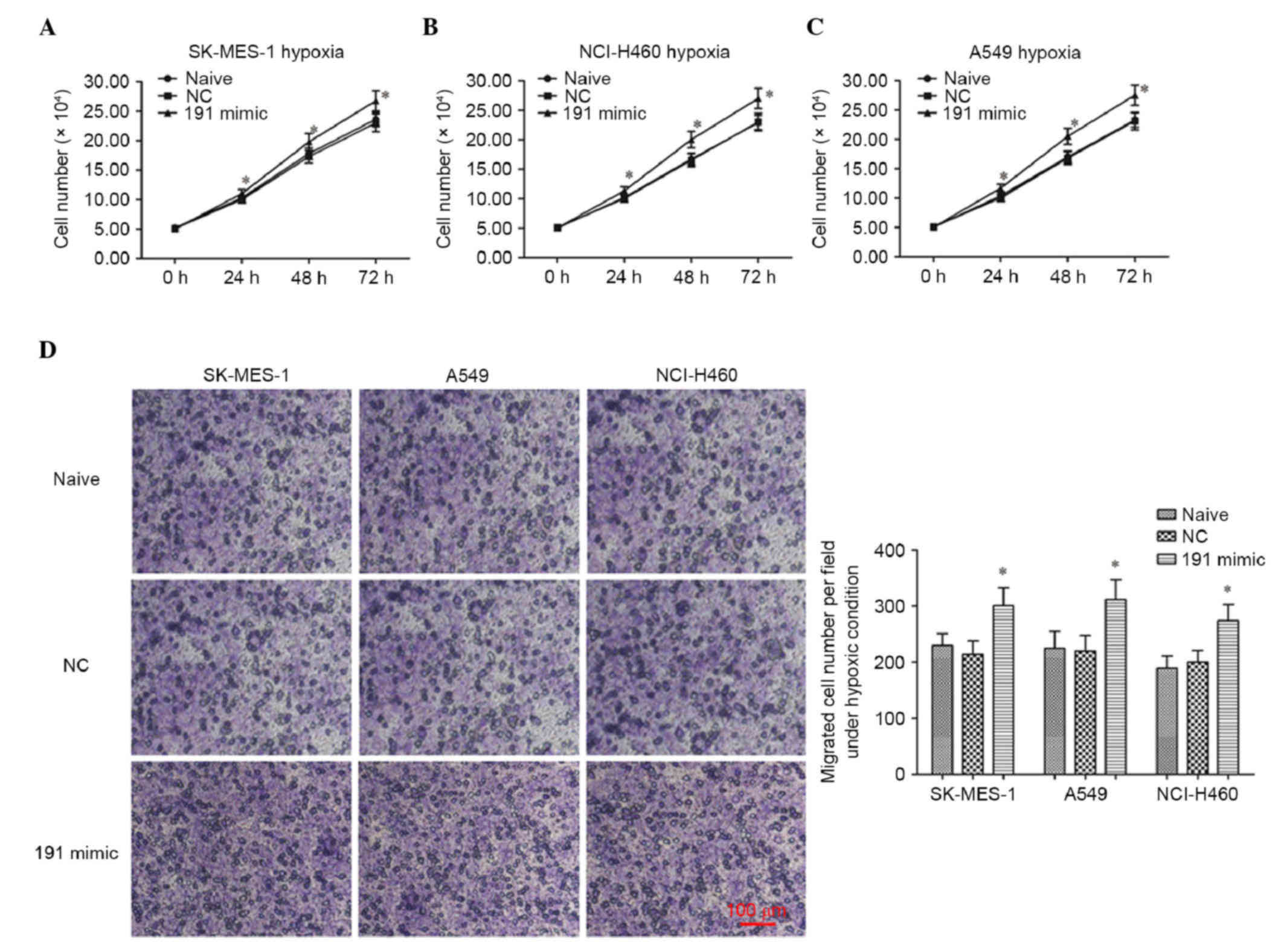
the cells under chronic hypoxic conditions (Fig. 2D-F). The data obtained from the
CCK-8 assay also indicated that the overexpression of miR-191
promoted the proliferation of the cells under chronic hypoxic
conditions (Fig. 3A-C).
Overexpression of miR-191 promotes the
migration of NSCLC cell lines under chronic hypoxic conditions
Subsequently, the role of miR-191 in the migration
of NSCLC cell lines under hypoxic conditions was investigated.
Following transfection, the migration of the NSCLC cells was
detected using a Transwell migration assay. The results of the
crystal violet staining and cell counting revealed that miR-191
promoted the migration of the cells under chronic hypoxic
conditions (Fig. 3D and E).
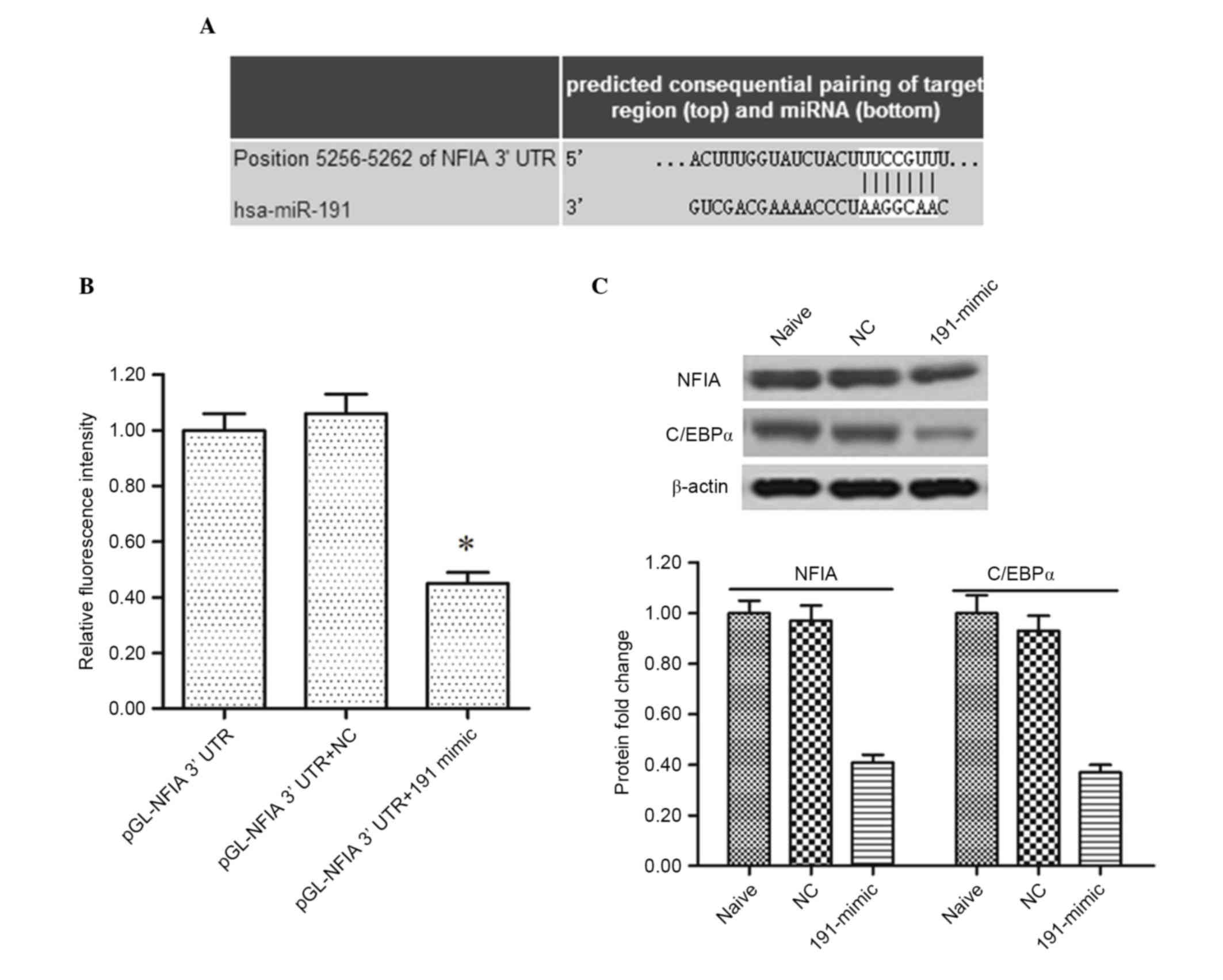
miR-191 targets NFIA and negatively
regulates the tumor-suppressing NFIA-C/EBPα axis
To examine the mechanism underlying the
miR-191-induced promotion of proliferation and migration in the
NSCLC cell lines, the online server, TargetScan, was accessed to
identify its potential target genes. The output of TargetScan
showed that the NFIA mRNA was perfectly matched by the miR-191 seed
sequence at the 3′UTR region (Fig.
4A). The 3′UTR luciferase reporter assay was then applied to
validate their targeting association. The data showed that miR-191
markedly reduced the relative fluorescence intensity (Fig. 4B), which supported the results
described above. In addition, western blot analysis of the A549
cells showed that the protein expression of NFIA was markedly
reduced by miR-191mimic transfection (Fig. 4C). Its downstream tumor suppressing
gene, C/EBPα was also suppressed (Fig.
4C).
Discussion
The role of miR-191 in lung cancer has been
disputed. Certain studies have reported that the expression of
miR-191 is stable in NSCLC, and that overexpression of miR-191 does
not lead to changes in cell cycle, proliferation, xenograft
formation or the chemosensitivity of lung adenocarcinoma cells
in vitro (23,25). However, in other studies, miR-191
has been found to be upregulated in patients with lung cancer,
suggesting that it may be involved in the progression of lung
cancer. A study by Nadal et al involved profiling 61 lung
squamous cell carcinoma tissues and 10 matched adjacent normal lung
tissues using miRNA arrays, and it was concluded that miR-191 in
the serum was a predictor of prognosis in surgically resected lung
squamous cell carcinoma (22,26).
In the present study, the results showed that
expression levels of miR-191 were significantly upregulated in the
lung tissues of patients with middle- and late-stage NSCLC.
However, the overexpression of miR-191 did not lead to changes in
the proliferation of the SK-MES-1, A549 or NCI-H460 NSCLC cell
lines under normal conditions (20% O2/5%
CO2). This suggested that there are certain important
distinctions between the in vitro and in vivo
environments of NSCLC cells. Studies have revealed that HIF-1α is
induced in the advanced stages of several types of cancer (27–30),
suggesting that NSCLC cells are adversely affected by a hypoxic
environment in patients. Therefore, the present study hypothesized
that miR-191 may be induced by hypoxia, and affect the progression
of NSCLC under hypoxic conditions. The data from the MTT and CCK-8
assays demonstrated that miR-191 promoted the proliferation of
NSCLC under mild hypoxic conditions (15% O2).
NFIA is a key regulator of gene expression in normal
and cancerous cells. A previous study showed that the miR-223/NFIA
axis suppressed tumorigenesis in a human glioma cell line (31). In the present study, the output of
TargetScan showed that NFIA mRNA was perfectly matched by the
miR-191 seed sequence at the 3′UTR region. A 3′UTR luciferase
reporter assay showed that miR-191 markedly reduced the relative
fluorescence intensity, supporting the results described above. The
western blot analysis revealed that the protein levels of NFIA and
its downstream tumor suppressing gene, C/EBPα, was markedly reduced
by miR-191 mimic transfection. These results contradicted those of
previous reports in other types of cancer (31,32).
However, the results of a study on astrocytoma indicated that NFIA
is associated with improved survival rates (33). The results of the present study
suggested that NFIA has a negative effect on the progression of
NSCLC.
In conclusion, the present study demonstrated that
miR-191 was upregulated in patients with middle- and late-stage
NSCLC, and in NSCLC cell lines, under mild hypoxic conditions.
miR-191 promoted the proliferation and migration of NSCLC under
chronic hypoxic conditions, and this promotion may be associated
with its targeting of NFIA. These results indicated that miR-191
promoted NSCLC progression, which may provide novel insight for the
regulation of NSCLC progression under chronic hypoxic
conditions.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
3
|
Chen WQ, Zheng RS, Zhang SW, Zeng HM and
Zou XN: The incidences and mortalities of major cancers in China,
2010. Chin J Cancer. 33:402–405. 2014.PubMed/NCBI
|
|
4
|
McKee MD, Bondarenko I, Guclu SZ, et al:
Veliparib (ABT-888) or placebo combined with carboplatin and
paclitaxel in patients with previously untreated
advanced/metastatic squamous (Sq) non-small cell lung cancer
(NSCLC): A randomized phase 3 trial. ASCO Annual Meeting
Proceedings. pp. TPS8107Chicago, IL: 2015
|
|
5
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ettinger DS, Akerley W, Borghaei H, Chang
AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK,
Govindan R, et al: Non-small cell lung cancer. J Natl Compr Canc
Netw. 10:1236–1271. 2012.PubMed/NCBI
|
|
7
|
Li H, Chen SJ, Chen YF, Meng QC, Durand J,
Oparil S and Elton TS: Enhanced endothelin-1 and endothelin
receptor gene expression in chronic hypoxia. J Appl Physiol (1985).
77:1451–1459. 1994.PubMed/NCBI
|
|
8
|
Smith TG, Robbins PA and Ratcliffe PJ: The
human side of hypoxia-inducible factor. Br J Haematol. 141:325–334.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parvin A, Pranap R, Shalini U, Devendran
A, Baker JE and Dhanasekaran A: Erythropoietin protects
cardiomyocytes from cell death during hypoxia/reperfusion injury
through activation of survival signaling pathways. PLoS One.
9:e1074532014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang W, Liu X, Cao J, Meng F, Li M, Chen
B and Zhang J: miR-134 regulates ischemia/reperfusion
injury-induced neuronal cell death by regulating CREB signaling. J
Mol Neurosci. 55:821–829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Simon MC: Abstract IA13: A hypoxic
microenvironment influences pancreatic cancer progression. Cancer
Res. 75:IA13. 2015. View Article : Google Scholar
|
|
12
|
Shay JE, Imtiyaz HZ, Sivanand S, Durham
AC, Skuli N, Hsu S, Mucaj V, Eisinger-Mathason TS, Krock BL,
Giannoukos DN and Simon MC: Inhibition of hypoxia-inducible factors
limits tumor progression in a mouse model of colorectal cancer.
Carcinogenesis. 35:1067–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amelio I and Melino G: The p53 family and
the hypoxia-inducible factors (HIFs): Determinants of cancer
progression. Trends Biochem Sci. 40:425–434. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Flygare J, Wong P, Lim B and
Lodish HF: miR-191 regulates mouse erythroblast enucleation by
down-regulating Riok3 and Mxi1. Genes Dev. 25:119–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagpal N and Kulshreshtha R: miR-191: An
emerging player in disease biology. Front Genet. 5:992014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di Leva G and Croce CM: miRNA profiling of
cancer. Curr Opin Genet Dev. 23:3–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu R, Chen X, Du Y, Yao W, Shen L, Wang
C, Hu Z, Zhuang R, Ning G, Zhang C, et al: Serum microRNA
expression profile as a biomarker in the diagnosis and prognosis of
pancreatic cancer. Clin Chem. 58:610–618. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Collins AL, Wojcik S, Liu J, Frankel WL,
Alder H, Yu L, Schmittgen TD, Croce CM and Bloomston M: A
differential microRNA profile distinguishes cholangiocarcinoma from
pancreatic adenocarcinoma. Ann Surg Oncol. 21:133–138. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagpal N, Ahmad HM, Chameettachal S,
Sundar D, Ghosh S and Kulshreshtha R: HIF-inducible miR-191
promotes migration in breast cancer through complex regulation of
TGFβ-signaling in hypoxic microenvironment. Sci Rep. 5:96502015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He Y, Cui Y, Wang W, Gu J, Guo S, Ma K and
Luo X: Hypomethylation of the hsa-miR-191 locus causes high
expression of hsa-mir-191 and promotes the
epithelial-to-mesenchymal transition in hepatocellular carcinoma.
Neoplasia. 13:841–853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Z, Ren H, Gao S, Zhao X, Zhang H and
Hao J: The clinical significance and regulation mechanism of
hypoxia-inducible factor-1 and miR-191 expression in pancreatic
cancer. Tumor Biol. 35:11319–11328. 2014. View Article : Google Scholar
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Patnaik SK, Kannisto E and Yendamuri S:
Overexpression of microRNA miR-30a or miR-191 in A549 lung cancer
or BEAS-2B normal lung cell lines does not alter phenotype. PloS
One. 5:e92192010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou W, Wang G, Zhao X, Xiong F, Zhou S,
Peng J, Cheng Y, Xu S and Xu X: A multiplex qPCR gene dosage assay
for rapid genotyping and large-scale population screening for
deletional α-thalassemia. J Mol Diagn. 15:642–651. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nadal E, Chen G, Chang AC, Lin J, Reddy R,
Orringer MB and Beer DG: Ratio of miR-146b/miR-191 in serum
predicts prognosis in surgically resected lung squamous cell
carcinomas. Cancer Res. 72:41472012. View Article : Google Scholar
|
|
27
|
Puisségur MP, Mazure NM, Bertero T,
Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K,
Cardinaud B, Hofman V, et al: miR-210 is overexpressed in late
stages of lung cancer and mediates mitochondrial alterations
associated with modulation of HIF-1 activity. Cell Death Differ.
18:465–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Elson DA, Ryan HE, Snow JW, Johnson R and
Arbeit JM: Coordinate up-regulation of hypoxia inducible factor
(HIF)-1alpha and HIF-1 target genes during multi-stage epidermal
carcinogenesis and wound healing. Cancer Res. 60:6189–6195.
2000.PubMed/NCBI
|
|
29
|
Wang N, Dong CR, Jiang R, Tang C, Yang L,
Jiang QF, Chen GG and Liu ZM: Overexpression of HIF-1α,
metallothionein and SLUG is associated with high TNM stage and
lymph node metastasis in papillary thyroid carcinoma. Int J Clin
Exp Pathol. 7:322–330. 2013.PubMed/NCBI
|
|
30
|
Wu Y, Mao F, Zuo X, Moussalli MJ, Elias E,
Xu W and Shureiqi I: 15-LOX-1 suppression of hypoxia-induced
metastatic phenotype and HIF-1α expression in human colon cancer
cells. Cancer Med. 3:472–484. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu C, Duan P, Li B, Huang C, Jing Y and
Yan W: miR-29a activates Hes1 by targeting Nfia in esophageal
carcinoma cell line TE-1. Oncol Lett. 9:96–102. 2015.PubMed/NCBI
|
|
32
|
Lee JS, Xiao J, Patel P, Schade J, Wang J,
Deneen B, Erdreich-Epstein A and Song HR: A novel tumor-promoting
role for nuclear factor IA in glioblastomas is mediated through
negative regulation of p53, p21, and PAI1. Neuro Oncol. 16:191–203.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song HR, Gonzalez-Gomez I, Suh GS, Commins
DL, Sposto R, Gilles FH, Deneen B and Erdreich-Epstein A: Nuclear
factor IA is expressed in astrocytomas and is associated with
improved survival. Neuro Oncol. 12:122–132. 2010. View Article : Google Scholar : PubMed/NCBI
|