Introduction
Choroideremia (CHM, OMIM: 303,100) is an X-linked
recessive chorioretinal dystrophy characterized by progressive
degeneration of the choroids, the retinal pigment epithelium, and
the retinal photoreceptors. Its incidence is estimated to be
between 1 in 50,000 and 1 in 100,000 people (1). However, it is likely that
choroideremia is under-diagnosed due to its similarities to other
eye disorders, such as retinitis pigmentosa (RP) (1,2).
Affected males initially exhibit symptoms of night blindness by
10–20 years of age, followed by progressive peripheral visual field
loss, with complete blindness occurring by age 40 in certain
patients (3–5). Carrier females are usually
asymptomatic; however, certain individuals do develop night
blindness and/or visual field loss (6).
Choroideremia is caused by mutations in the
CHM gene (OMIM: 300,390), which consists of 15 exons
spanning at least 150 kb and is located at Xq21.2-q21.3 (7). The CHM gene encodes Rab escort
protein 1 (REP1), a subunit of the dimeric holoenzyme, Rab
geranylgeranyl transferase, which attaches 20-carbon isoprenoid
groups to the cysteine residues of Rab proteins, a family of
GTP-binding proteins that regulate vesicular traffic. Rab
geranylgeranyl transferase is an enzyme complex that mediates
correct intracellular vesicular transport (8). REP1 is composed of 653 amino acids
and a single 95-kDa polypeptide (9). Cremers et al (10,11)
first identified that the CHM gene is the disease-causing
gene that, when partially deleted or disrupted in male patients
with tapetochoroidal dystrophy (choroideremia, OMIM: 303,100),
leads to choroideremia. Thus far, there are at least 195 CHM
gene mutations reported in the Human Genome Mutation database
(HGMD), and various methods were used to detect these CHM
gene variants. Direct sequence analysis for identifying
choroideremia-causing variants results in a detection rate of
~60–95%; however, this is recommended as the first investigative
option (6,12,13),
even though next-generation sequencing (NGS) will result in an
almost 100% detection rate (14).
Human gene replacement clinical trials for
choroideremia are currently under way. Initially, these phase 1/2
trials used an adeno-associated viral vector (AAV2) to deliver the
REP1 gene directly to the retina, which improved visual outcomes in
six individuals (15). With the
increased likelihood that individuals affected with choroideremia
may benefit from gene therapy trials or gene-specific treatments in
the near future, the genetic confirmation of this disease, and
identification of the specific variants in clinically prenatal
diagnosed individuals, is increasingly a high priority for
reproductive health in China and other developing countries.
Prenatal diagnosis is used to detect whether the
fetus will suffer from a genetic disease after birth. Traditional
invasive methods of sampling fetal materials include amniocentesis,
chorionic villus sampling, umbilical cord blood sampling, and so
forth (16–18), whereas non-invasive prenatal
testing (NIPT) involves testing fetal genetic material through
isolation of cell-free fetal DNA (cffDNA) from maternal peripheral
blood, plasma, or serum. Using cffDNA for prenatal diagnosis to
replace or complement existing invasive methods may greatly reduce
the pain and the risk of a miscarriage; therefore, there is
increased interest in using NIPT.
Herzenberg et al (19) first isolated fetal nucleated red
blood cells using flow cytometry, but no efficient prenatal
diagnosis was available at the time due to the limited amount of
usable sample recovered. The use of NIPT did not become promising
until Lo et al (20)
confirmed that cffDNA exists in maternal peripheral blood. Although
the concentration of cffDNA is extremely low in maternal peripheral
blood (3–6%) (21), the goal of
the present authors was to establish an effective method to measure
the concentrations of cffDNA at different stages of gestation, and
to optimize NIPT (22).
The cffDNA in NIPT is mainly used in the clinic for
prenatal identification of X-linked disorders, including
hemophilia, Huntington's disease, Duchenne muscular dystrophy,
congenital adrenal cortical hyperplasia prenatal screening of fetal
chromosomal aneuploidy, and detection of the Rhesus (Rh) factor
(21,23–27).
Maternal plasma cell-free DNA is mixed with maternal and fetal DNA,
of which fetal DNA represents a minor portion of the total. The
SRY gene is a sex-determining gene on the Y chromosome,
which only exists in males. Using cffDNA from maternal plasma for
fetal gender determination is mainly limited to Y chromosome
sequences absent in the maternal genome, such as the SRY.
Thus, the only way to identify those sequences is through
male-bearing pregnancies (21).
Prenatal genetic diagnosis is often used when there
is a family history of sex-linked diseases. Most sex-linked
diseases are recessive X-linked and caused by a particular gene
mutation on the X chromosome, such as choroideremia. The disease is
normally manifested only in males, who carry a single X chromosome,
whereas, in females, the normal allele on the second X chromosome
compensates for the diseased allele (28). Using cffDNA for the combined use of
NGS for detecting choroideremia mutations and NIPT for Y chromosome
determination is novel.
The aim of the present study was to genetically
identify variants of the choroideremia-causing gene by NGS, and to
analyze the SRY gene using NIPT for the presence of X-linked
recessive disease in a large family cohort diagnosed with
choroideremia.
Materials and methods
Collection for samples
The present research study was approved by the
Southwest Medical University review board, and was conducted
according to the principles of the Declaration of Helsinki.
Informed consent was obtained from all participants. Full medical
and family histories were collected, a pedigree was formed, and an
ophthalmologic examination that included wide-field fundus
photography, wide-field autofluorescence, spectral domain optical
coherence tomography, fundus photography, and eye/orbit ultrasound
was performed.
Pregnant women signed informed consent forms for
approval of the prenatal diagnosis. Methods for DNA extraction from
the peripheral blood of associated family members and the amniotic
fluid from the fetus were described previously (22,29,30).
Briefly, 1X Blood Lysis buffer was added into peripheral blood on
ice for 30 min, and centrifuged for 10 min at 1,600 × g. The
filtrate was discarded, 1 ml Nucleic Lysis Buffer, 100 µl 20% SDS
and 10 µl Protease K (20 mg/ml; Roche Diagnostics, Indianapolis,
IN, USA) were added, and the mixture was incubated at 56°C for 4 h.
An equivalent volume of phenol was added, mixed, and centrifuged
for 10 min at 1,600 × g. Subsequently, a repeat step was
performed using an equivalent volume of phenol and chloroform. The
supernatant was retained, and 2.5 vols absolute ethyl alcohol was
added to precipitate the DNA. Finally, the precipitated DNA was
dissolved in 1X TE buffer. For DNA extraction from amniotic fluid,
it was necessary to move directly to the Nucleic Lysis Buffer
treatment step following centrifugation of the amniotic fluid, and
then the identical procedure was followed with respect to DNA
extraction from the peripheral blood.
cffDNA isolation from plasma and
nested polymerase chain reaction (PCR)
Blood samples from the peripheral blood of pregnant
women were collected in 2–3 ml EDTA-K2 anticoagulative
tubes. Plasma separation from these blood samples was performed
using a two-step centrifugation method, as previously described
(22). Then, 20 µl Protease K (20
mg/ml) and 200 µl plasma were mixed, and centrifuged briefly after
adding 200 µl Buffer AL (Qiagen, Dusseldorf, Germany). The solution
was incubated at 56°C for 10 min, centrifuged briefly, 200 µl
absolute ethyl alcohol was added, mixed, and subsequently
transferred to the spin columns (Qiagen). The spin columns were
centrifuged for 1 min at 6,000 × g, 500 µl Buffer AW2
(Qiagen) was added prior to centrifugation for 3 min at 20,000 ×
g, and then the filtrate was discarded. Next, the spin
columns were centrifuged for 1 min at 20,000 × g, and the DNA was
eluted with 60 µl Buffer AE (Qiagen), separated into aliquots and
stored at −80°C prior to use.
The amplification of the SRY gene (Y
chromosomal material) using nested PCR was performed as previously
described (22). The primers used
for this nested PCR were as follows: SRY-138 forward (F)
(5′-TACAGGCCATGCACAGAGAG-3′) and SRY-138 reverse (R)
(5′-TGTTGTCCAGTTGCACTTCG-3′) (first round); and SRY-116F
(5′-GCACAGAGAGAAATACCCGAAT-3′) and SRY-116R
(5′-GCACTTCGCTGCAGAGTACC-3′) (second round).
Amplification of DNA of the amniotic
fluid
Amplification reactions were set up in a reaction
volume of 10 µl (29,30). Each reaction contained 50 ng DNA,
300 nmol each gene specific primer for SRY or CHM, 5
µl 2X PCR MasterMix (Tiangen Biotech Co., Ltd., Beijing, China),
and doubly distilled (dd)H2O. The following primer sets
were used for these reactions: XES7
(5′-CCCGAATTCGACAATGCAATCATATGCCCC0CCC1-3′) and XES2
(5′-CTGTAGCGGTCCCTGTGCTGCGGTG-3′), or CHM7L (TGG GAG AAA AGG ATT
TGT GTG) and CHM7R (ATG GAT CAG GTT TTG CTG CT), which produced a
609 or 637 base pair (bp) amplification product for the SRY
or CHM gene, respectively. The following PCR program was
used: An initial denaturation step at 95°C for 90 sec, followed by
34 cycles, each consisting of a denaturation step at 94°C for 30
sec, an annealing step at 65 or 62°C for 30 sec, and an extension
step at 72°C for 25 sec, followed by an extension step at 72°C for
2 min.
Electrophoresis and silver
staining
The PCR products were resolved on a 2% agarose gel
or an 8% PAGE gel and detected using ultraviolet (UV) light or
silver staining, respectively.
Real-time quantitative PCR
(RT-qPCR)
RT-qPCR for the SRY gene and the control
gene, β-ACTIN, were performed according to previously
reported methods and using cffDNA as the sample template (22). Sequence-specific
fluorescence-labeled probes and primers for TaqMan qPCR were
matched by the Universal Probe Library Center software (Roche
Applied Science, Penzberg, Germany) (31,32).
The β-ACTIN gene was set up as a internal control using
genomic DNA as the template. The amplification primers for the
SRY gene were: SRY-71F (5′-CCAGCTAGGCCACTTACCG-3′), SRY-71R
(5′-AGCTTTGTCCAGTGGCTGTAG-3′), and the Taqman probe was no. 71; and
the primers for the β-ACTIN gene were: Q-β-actin55GF
(5′-AAGTCCCTTGCCATCCTAAA-3′), and Q-β-actin55GR
(5′-ATGCTATCACCTCCCCTGTG-3′), and the Taqman probe was no. 55. The
following PCR program was used: An initial denaturation step at
95°C for 10 min, followed by 45 cycles, each consisting of a
denaturation step at 94°C for 15 sec and an annealing step at 60°C
for 1 min.
Capture panel design, library
preparation, and targeted sequencing
A capture panel of retinal disease genes was used,
as previously described (33,34).
The probes on the panel covered 4,405 exons and corresponded to the
splice junctions of 163 known retinal disease genes within a span
of 1,176 Mbp.
Illumina paired-end libraries (Illumina, Inc., San
Diego, CA, USA) were generated according to the manufacturer's
sample preparation protocol for genomic DNA. Briefly, 1 µg each
patient's genomic DNA (proband) was sheared into fragments of
~300-500 bp. The DNA fragments were end-repaired using
polynucleotide kinase and Klenow fragment (large protein fragment).
The 5′ ends of the DNA fragments were phosphorylated, and a single
adenine base was added to the 3′ end. Illumina Y-shaped index
adaptors were ligated to the repaired ends, and subsequently the
DNA fragments were amplified by PCR for eight cycles. Fragments of
300–500 bp were isolated using bead purification. The pre-capture
libraries were quantified, and their size distributions were
determined using a commercial bio-analytical system (Agilent 2100
BioAnalyzer; Agilent Technologies, Santa Clara, CA, USA). For each
capture reaction, 50 pre-capture libraries (60 ng/library) were
pooled together. Hybridization and wash kits (Agilent Technologies,
Inc., Santa Clara, CA, USA) were used for the washing and recovery
of captured DNA, and performed according to the standard
manufacturer's protocol. Captured libraries were quantified and
sequenced (Illumina HiSeq 2000; Illumina, Inc.) as 100 bp
paired-end reads, according to the manufacturer's protocol.
Bioinformatic analysis of sequenced
data
Sequence reads were aligned to human genome
reference version hg19 using the Burrows-Wheeler Aligner version
0.5.9 (35). After recalibration
and local realignment using the Genome Analysis Toolkit (GATK,
version 1.0.5974) (36), the
refined sequencing results were subjected to variant calling using
the Atlas2 toolkit (37). Several
common variant databases, such as the 1,000 genome database [Build
20110521 and 20101123], the single nucleotide polymorphism database
(dbSNP)137, the National Heart, Lung, and Blood Institute (NHLBI)
GO Exome Sequencing Database, the National Institute of
Environmental Health Sciences (NIEHS) Exome Sequencing Database,
the YanHuang Project Database, and an internal control database of
997 exomes, were used to filter out common polymorphisms at an
allele frequency >0.5% in all the above databases (35,38).
To remove synonymous mutations, variant annotation was performed
using ANNOVAR (39), and RefSeq
genes were used as references to coordinate the mutations. SIFT,
Polyphen2, LRT, MutationTaster, and MutationAssessor were used to
make functional predictions of missense variants (40–42).
The pathogenicity of novel missense mutations was predicted using
the database, dbNSFP (43). The
HGMD was used to search for known pathogenic mutations.
Mutation validation and segregation
tests
The putative NGS mutations were validated using
Sanger sequencing. Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) was used to
design primers at least 100 bp upstream and downstream from the
mutation, and the assigned names of the primers used for the
CHM gene causative variation were CHM7L and CHM7R. After PCR
amplification, the amplicons were sequenced on an ABI3500
sequencer. The DNA materials of other family members were also
sequenced using Sanger sequencing in order to perform segregation
tests.
The DNA from fetal amniotic fluid was PCR-amplified
using the previously mentioned primer pairs, CHM7L and CHM7R, and
Sanger sequencing was performed using primer CHM7 L for the
prenatal gene diagnosis.
Results
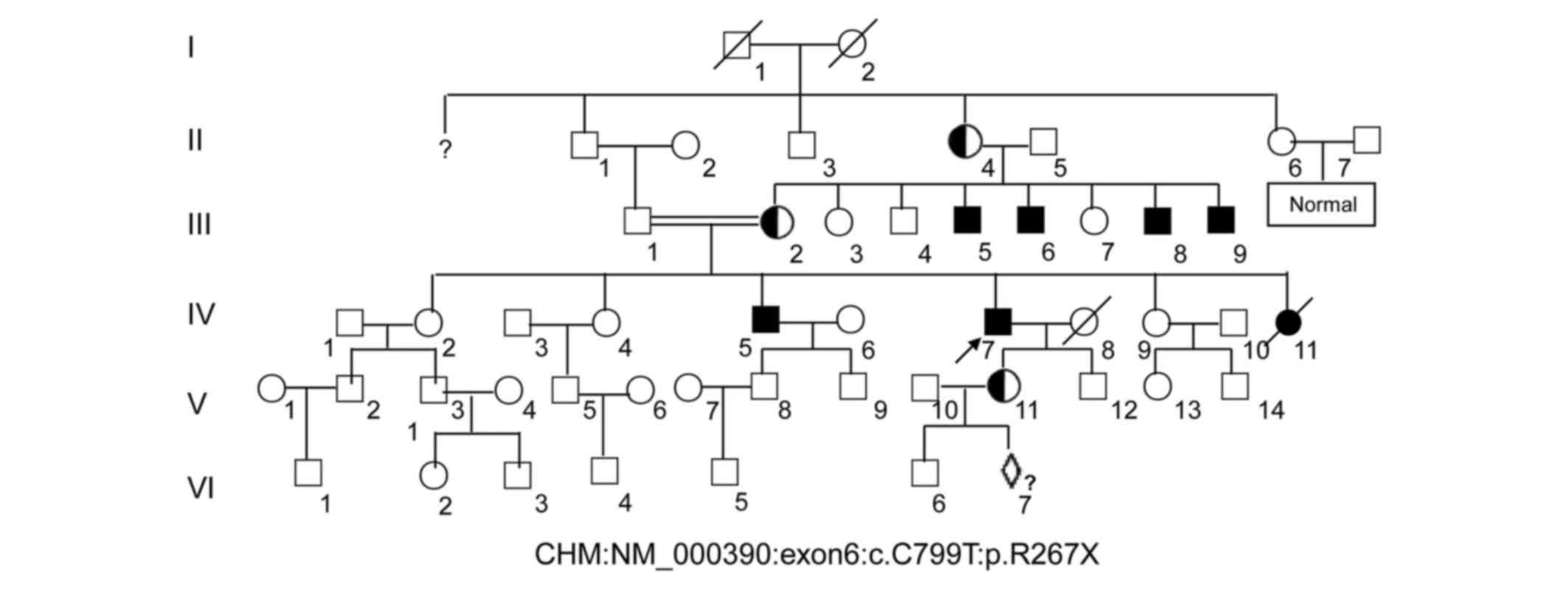
Pedigree collection and clinical
phenotypes
A six-generation pedigree of Chinese descent with 30
years of follow-up was recruited to the present study. The
60-year-old proband (IV:7, Fig. 1)
presented with early clinical signs of nyctalopia or night
blindness at around 1 year of age. This patient now presents with
extreme ametropia (both eyes >1,000 degrees), severe retinal
pigment degeneration, and coloboma of the choroid (Fig. 2). A progressive narrowing of the
field of vision, or tunnel vision, and decreased visual acuity was
also observed. The fundus photographs shown in Fig. 2 reveal that the retinal pigment
epithelial layer was patchy, with shaded areas of scattered
pigmentation; however, the layer appeared transparent in the
central area called the macula (Fig.
2A and B, denoted by the black arrows). The pigment layer was
extremely thin, possibly absent, or lacking pigment (Fig. 2A and B, denoted by the blue
arrows), leading to its transparent nature, which allowed the
sclera and deep choroidal blood vessels (Fig. 2A and B, blue arrows) in the areas
outside the central macula to be visible (Fig. 2A and B, black arrows). The observed
blood vessels were markedly smaller in size, and the boundary of
the optic-papilla was clear. This observation was further confirmed
in optical coherence tomography images, which revealed atrophy of
the retina at the macular fovea, thinning of the fovea, and loss of
choroids.
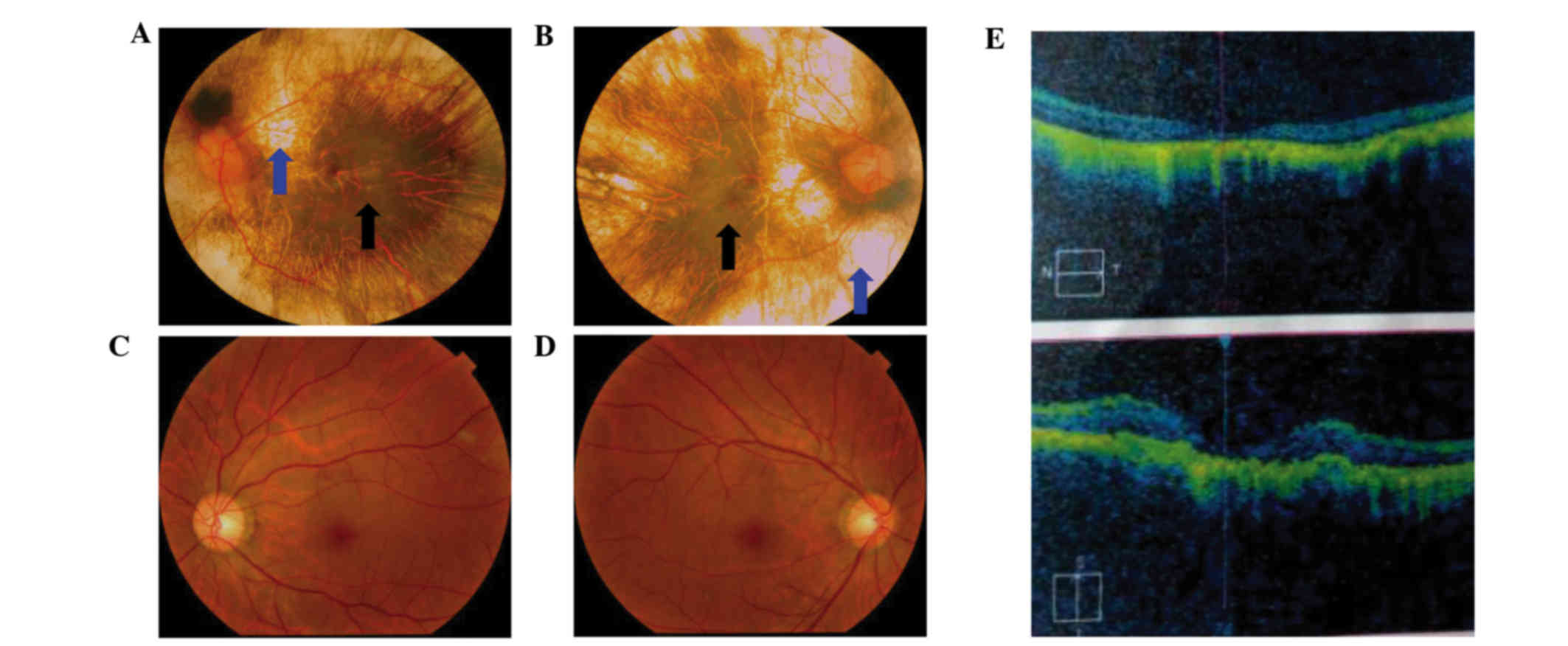 | Figure 2.Fundus photograph and OCT images of
choroideremia proband. (A) The left eye fundus photograph. (B) The
right eye fundus photograph. Fundus photographs revealed that the
retinal pigment epithelial layer was patchy with shaded areas of
scattered pigmentation; however, the layer appeared transparent in
the central area, called the macula (black arrow). The pigment
layer was extremely thin, possibly absent, or lacking pigment (blue
arrow), leading to its transparent nature, which allowed the sclera
and deep choroidal blood vessels (blue arrow) in the areas outside
the central macula to be visible (black arrow). The observed blood
vessels were markedly smaller in size, and the boundary of the
optic-papilla was clear. (C and D) Normal fundus photographs
(control) are shown. (E) The OCT images reveal atrophy of the
retina at the macular fovea, thinning of the fovea, and loss of
choroids. OCT, optical coherence tomography. |
A further 5 males were identified as having night
blindness and severely restricted peripheral visual fields within
the first decade of life, as is classically expected. The female
carriers disclosed no noteworthy visual defects. The patients for
III:5, III:8 and III:9 in Fig. 1
did not marry and produced no offspring, whereas the male of III:6
married and assisted in producing 3 girls, who were normal.
The pregnant woman (V:11, Fig. 1) with G2P1001 was recruited and
signed the informed consents for prenatal diagnosis. DNAs from the
peripheral blood of this pregnant woman and the amniotic fluid from
the fetus (VI:7, Fig. 1) at 17
weeks of pregnancy were extracted for further analysis.
Capture sequencing, data processing,
mutation screening and validation
To identify causative mutations in this family,
targeted capture sequencing was performed. DNA from the proband
(IV:7) was captured, and sequenced with at least ×20 depth
coverage. An automatic variant calling, filtering, and annotation
pipeline was used to process the capture sequencing data from the
sample. The common polymorphisms with >0.5% frequency were
filtered out, and non-pathogenic variations in any of the variant
or internal control databases were queried. Sequence variants that
were not annotated in any of the above public databases were
prioritized for further analysis (data not shown).
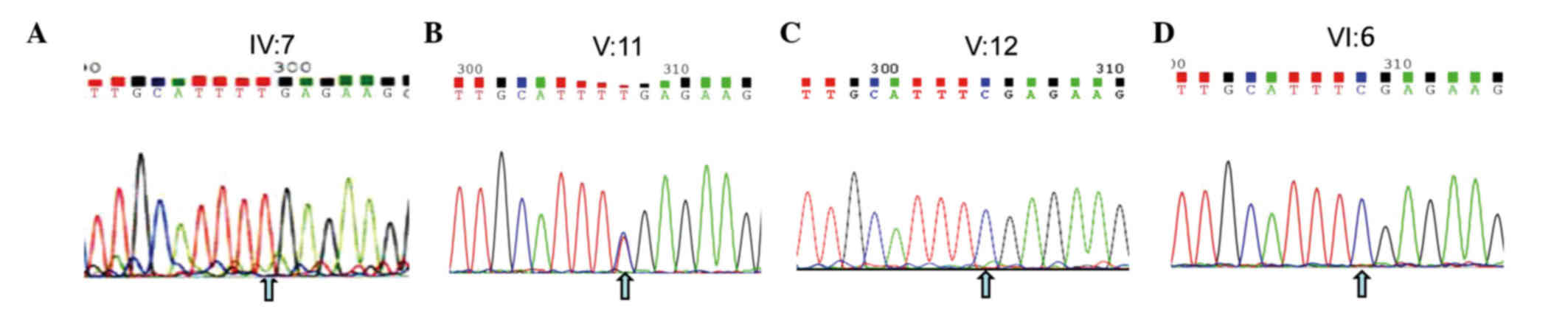
A nonsense mutation (c.C799T:p.R267X) located in
exon 6 of the CHM gene (GenBank accession number:
NM_000,390, NP_000,381.1) on the X chromosome from the proband was
detected. This nonsense mutation was subsequently confirmed using
Sanger sequencing (Figs. 1 and
3A), while other known retinal
disease-causing gene mutations, such as RP, were excluded. This
mutation was not identified in 100 healthy controls, and introduced
a termination codon into the open reading frame (ORF) of the
CHM gene. This ORF disruption was predicted to result in a
distinct truncated protein product, whereby 60% of the C-terminal
amino acids of the REP1 protein were lost, leading to a functional
defect in its protein escort capabilities. The identical mutation
was subsequently identified in 5 males (III:5, 6, 8, 9, and IV:5)
of this family (data not shown). The identical heterozygous
mutation was also subsequently identified in two female carriers
(III:2 and V:11) of this family (Fig.
3, and data not shown), which indicated that the mutation of
the proband (IV:7) was inherited from the mother (III:2), leading
to the pathogenic mutation in the male offspring. Ultimately, this
mutation demonstrated perfect co-segregation with the disease in
the family. The father, son, and grandson of the proband (III:1,
V:12 and VI:6, respectively) and other members of the family were
normal, with wild-type alleles of the CHM gene (Fig. 3 and data not shown).
NIPT
cffDNA isolation from maternal peripheral blood
plasma (V:11) was performed. PCR-based amplification of the
SRY gene by nested PCR and subsequent silver staining
revealed that the plasma DNA from both a pregnant woman with a
known female fetus and no template were negative, whereas the
plasma DNA from a pregnant woman carrying a male fetus were
positive (Fig. 4A). The results of
the SRY gene amplification from this woman's plasma DNA were
verified by extracting the DNA from fetal amniocytes and using the
amplification primer set, XES7 and XES2 (Fig. 4B). These results confirmed the
successful identification of the fetus as female (VI: 7) using the
cffDNA from the maternal peripheral blood plasma (V: 11).
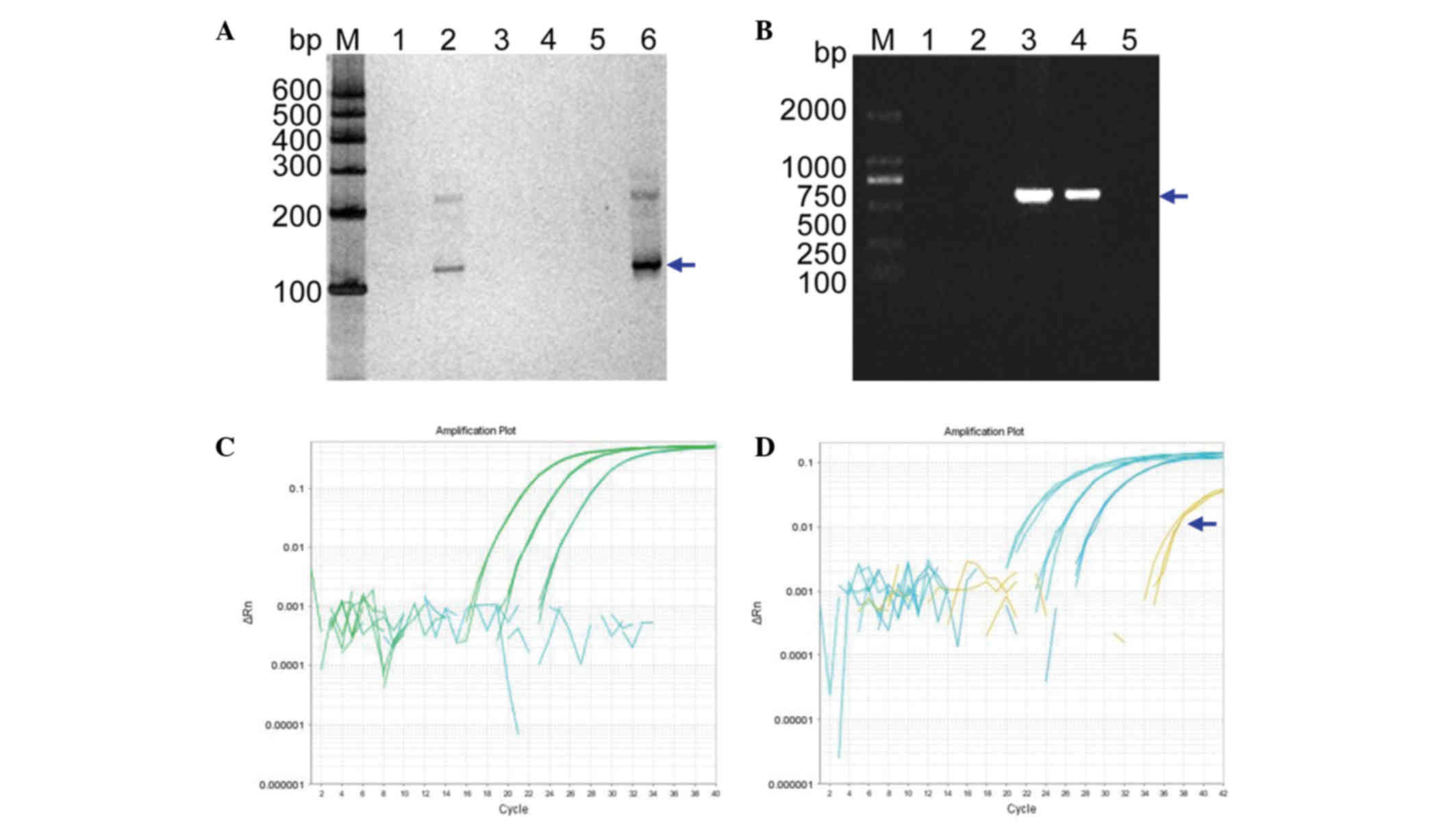 | Figure 4.NIPT. (A) NIPT using cffDNA from the
mother's blood (V:11 in Fig. 1).
Lanes 1–6 are DNA PCR products determined using nested PCR for the
mutational diagnosis from the maternal blood plasma (V: 11 in
Fig. 1), a known male genomic DNA
sample, a known female genomic DNA sample, a blank control without
any DNA, a plasma DNA sample from a pregnant woman with a known
female fetus, and a plasma DNA sample from a pregnant woman with a
known male fetus, respectively. Lanes 2 and 6 show the positive
amplification. Lane M indicates the DNA molecular-weight marker
DL600, with fragment sizes of 600, 500, 400, 300, 200, and 100 bp.
(B) Verification of the SRY gene from the fetal DNA of
amniotic fluid in the pregnant woman. Lanes 1–5 represent the
diagnosis from fetal amniotic fluid DNA (VI:7 in Fig. 1), a female genomic DNA sample
(fetus's mother; V:11 in Fig. 1),
a male genomic DNA sample (fetus's father; V:10 in Fig. 1), a male genomic DNA sample
(fetus's brother; VI:6 in Fig. 1)
and a blank control without any DNA, respectively. Lane M indicates
the DNA molecular weight marker DL2000, with fragment sizes of
2000, 1000, 750, 500, 250 and 100 bp. (C and D) The real-time
quantitative PCR profiles. (C) The profile for a plasma DNA sample
from a pregnant woman and β-ACTIN as control using different
dilution of genomic DNA. (D) The profile for a plasma DNA sample
from a pregnant woman with the known baby boy and β-ACTIN as
control using different dilution of genomic DNA. NIPT, non-invasive
prenatal testing; bp, base pairs. |
To further validate these cffDNA results, RT-qPCR
was performed to determine the quantity of cffDNA in the maternal
peripheral blood plasma. The results did not show any curve of
amplification (Fig. 4C), whereas a
positive control using male cffDNA did (Fig. 4D, denoted by the blue arrow). Thus,
these data further supported the results obtained from the nested
PCR experiment.
Verification and prenatal gene
diagnosis by amniotic fluid DNA
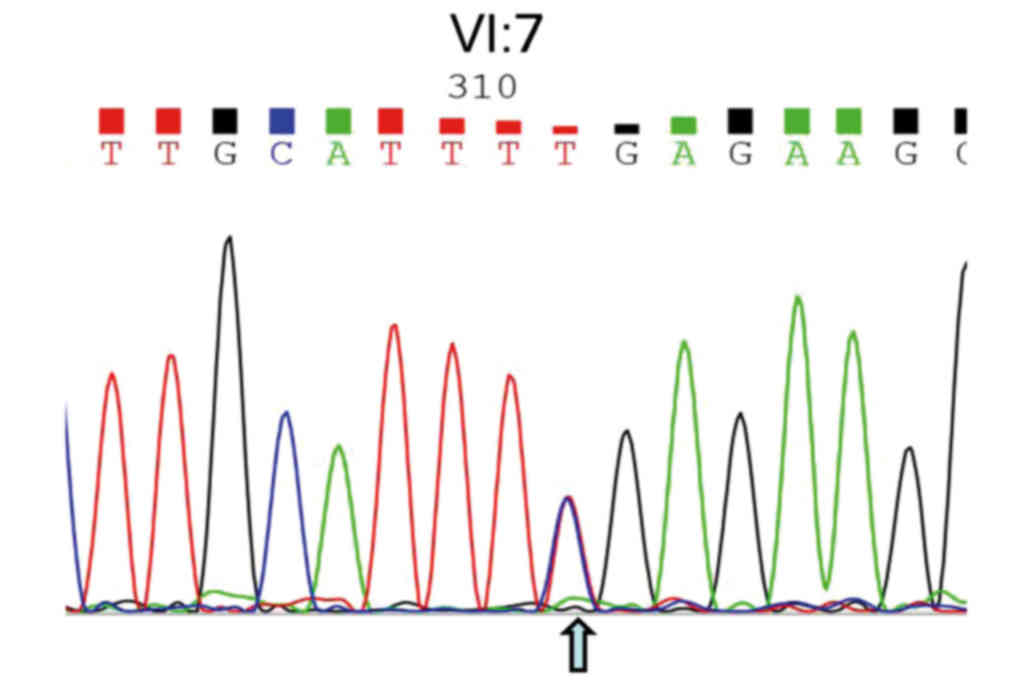
DNA isolated from the amniotic fluid of the pregnant
woman (V:11, Fig. 1) with the
fetus (VI:7) was amplified, and Sanger sequencing was performed for
prenatal gene diagnosis of the CHM gene mutation. The
results shown in Fig. 5 illustrate
that the heterozygous nonsense mutation (c.C799T:p.R267X) has been
identified, indicating that this baby will inherit the pathogenic
DNA mutation from her mother, since her father exhibited both a
normal phenotype and genotype (data not shown). At the age of one
and a half, this female baby exhibited no symptoms associated with
this disease mutation.
Discussion
Choroideremia is an X-linked recessive chorioretinal
dystrophy caused by mutations in the CHM gene and
characterized by progressive degeneration of the choroids, the
retinal pigment epithelium, and the retinal photoreceptors
(1,2,10,11).
Detection of variants in the CHM gene were previously
identified using a range of methods (6,12–14).
Human gene replacement clinical trials are currently under way for
treatment of choroideremia (15).
With the increased likelihood that individuals affected with
choroideremia may benefit from gene therapy or gene-specific
treatments in the near future, the genetic confirmation of this
disease and the identification of specific variants present
prenatally are now a high priority for reproductive health in China
and other developing countries.
In 1997, scientists identified cffDNA in maternal
peripheral blood during pregnancy, opening the possibility for
early NIPT for a variety of genetic conditions (20). Using cffDNA for prenatal testing
may be able to replace or complement existing invasive methods, and
greatly reduce the pain and risk of miscarriage compared with
current methods. cffDNA has only been used for NIPT with a select
number of single gene inheritance diseases and aneuploidy in
chromosomes (23–25,44,45)
due to the mixture of maternal DNA.
Due to the risk of miscarriage associated with
traditional prenatal diagnostic methods or invasive methods, there
is a growing interest for using NIPT (23–25,44,45).
Currently, NIPT is mainly used to detect aneuploidy, the Rh antigen
D (RhD) group, X-linked genetic disease, and certain single gene
inheritance diseases (21,26,27).
Although the gender of the fetus can often be determined using an
ultrasound scan of the fetus in the second or third trimester,
isolation of cffDNA in the first trimester would open up novel
fields of NIPT with an earlier diagnosis and earlier, more precise
management of medical care. Using isolated cffDNA present in the
maternal plasma from women 6–10 weeks into their pregnancies will
provide the required sensitivity, specificity, and accuracy
required for NIPT. The present study has successfully managed to
isolate cffDNA from maternal peripheral blood, and laid the
foundation for a novel use of non-invasive prenatal diagnosis
(NIPD) in medicine in the future.
In the present study, cffDNA was successfully used
for the combined use of NGS for diagnosing choroideremia and NIPT
for Y chromosome determination. First, a six generation pedigree of
Chinese descent was recruited. The 60-year-old proband (IV:7)
presented with early clinical signs of nyctalopia or night
blindness at around 1 year of age. The proband's daughter became
pregnant, and was included in our study. Secondly, a nonsense
mutation (c.C799T:p.R267X) in the CHM gene on the X
chromosome from the proband was detected by NGS, and confirmed by
Sanger sequencing (Fig. 3). This
mutation introduces a termination codon into the ORF of the
CHM gene, thereby producing a distinct truncated protein
product. The reduced quantity of fully functional REP1 prevents Rab
proteins from reaching and attaching to targeted organelle
membranes, which ultimately leads to premature cell death (8,46,47).
Thirdly, NIPD was performed on cffDNA isolated from maternal
peripheral blood plasma (V:11), and the fetus was successfully
identified as a female baby (VI:7). Finally, verification of the
prenatal gene diagnosis was performed successfully on amniotic
fluid DNA. The heterozygous nonsense mutation (c.C799T:p.R267X)
from the fetus (VI:7) was identified, indicating that the mutation
in this baby was inherited from her mother (V:11). At 18 months'
follow-up, the baby revealed no symptoms of choroideremia.
Thus, cffDNA has been successfully used for the
combined use of NGS for diagnosing choroideremia and NIPD for Y
chromosome determination. This approach should result in an
increased use of prenatal diagnosis and an improved, more
sophisticated clinical management of diseases in China and other
developing countries. The establishment of a highly accurate method
for prenatal gene diagnosis will allow more reliable gene
diagnosis, improved genetic counseling, and personalized clinical
management for our patients. Earlier identification of gene
mutations responsible for diseases may aid in the diagnosis of
retinal diseases, and assist in therapeutic research approaches.
This will also afford patients the opportunity to make informed
reproductive and pregnancy management decisions based on precise
NIPD. Of course, more efficient approaches for detecting specific
point mutations from cffDNA in different pedigrees should be
developed in the future due to varied genetic mutational
spectrums.
Acknowledgements
This study was supported in part by the National
Natural Science Foundation of China (grant nos. 30371493,
81172049), the Science and Technology Innovation Team of Colleges
and Universities of Sichuan Province (grant no. 13TD0032), the
Research Foundation of the Science and Technology Department of
Sichuan Province (grant nos. 14JC0797, 2015JY0038), and the Luzhou
City Special Foundation (grant no. 2013LZLY-J10).
References
|
1
|
Coussa RG and Traboulsi EI: Choroideremia:
A review of general findings and pathogenesis. Ophthalmic Genet.
33:57–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li S, Guan L, Fang S, Jiang H, Xiao X,
Yang J, Wang P, Yin Y, Guo X, Wang J, et al: Exome sequencing
reveals CHM mutations in six families with atypical choroideremia
initially diagnosed as retinitis pigmentosa. Int J Mol Med.
34:573–577. 2014.PubMed/NCBI
|
|
3
|
Esposito G, De Falco F, Tinto N, Testa F,
Vitagliano L, Tandurella IC, Iannone L, Rossi S, Rinaldi E,
Simonelli F, et al: Comprehensive mutation analysis (20 families)
of the choroideremia gene reveals a missense variant that prevents
the binding of REP1 with Rab geranylgeranyl transferase. Hum Mutat.
32:1460–1469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roberts MF, Fishman GA, Roberts DK,
Heckenlively JR, Weleber RG, Anderson RJ and Grover S:
Retrospective, longitudinal, and cross sectional study of visual
acuity impairment in choroideremia. Br J Ophthalmol. 86:658–662.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garcia-Hoyos M, Lorda-Sanchez I,
Gómez-Garre P, Villaverde C, Cantalapiedra D, Bustamante A,
Diego-Alvarez D, Vallespin E, Gallego-Merlo J, Trujillo MJ, et al:
New type of mutations in three Spanish families with choroideremia.
Invest Ophthalmol Vis Sci. 49:1315–1321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
MacDonald IM, Smaoui N and Seabra MC:
Choroideremia. [Updated 2010] 1993–2014. In:
GeneReviews® [Internet] Seattle WA: University of
Washington, Seattle; pp. 1993–2014. 2003
|
|
7
|
van Bokhoven H, van den Hurk JA, Bogerd L,
Philippe C, Gilgenkrantz S, de Jong P, Ropers HH and Cremers FP:
Cloning and characterization of the human choroideremia gene. Hum
Mol Genet. 3:1041–1046. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Strunnikova NV, Barb J, Sergeev YV,
Thiagarajasubramanian A, Silvin C, Munson PJ and Macdonald IM:
Loss-of-function mutations in Rab escort protein 1 (REP-1) affect
intracellular transport in fibroblasts and monocytes of
choroideremia patients. PLoS One. 4:e84022009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Hurk JA, Schwartz M, van Bokhoven
H, van de Pol TJ, Bogerd L, Pinckers AJ, Bleeker-Wagemakers EM,
Pawlowitzki IH, Rüther K, Ropers HH and Cremers FP: Molecular basis
of choroideremia (CHM): Mutations involving the rab escort
protein-1 (REP-1) gene. Hum Mutat. 9:110–117. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cremers FP, Sankila EM, Brunsmann F, Jay
M, Jay B, Wright A, Pinckers AJ, Schwartz M, van de Pol DJ,
Wieringa B, et al: Deletions in patients with classical
choroideremia vary in size from 45 kb to several megabases. Am J
Hum Genet. 47:622–628. 1990.PubMed/NCBI
|
|
11
|
Cremers FP, van de Pol DJ, van Kerkhoff
LP, Wieringa B and Ropers HH: Cloning of a gene that is rearranged
in patients with choroideraemia. Nature. 347:674–677. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schwarz JM, Rödelsperger C, Schuelke M and
Seelow D: MutationTaster evaluates disease-causing potential of
sequence alterations. Nat Methods. 7:575–576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McLaren TL, De Roach JN, Montgomery H,
Hoffmann L, Kap C and Lamey TM: Genetic analysis of choroideremia
families in the Australian population. Clin Experiment Ophthalmol.
43:727–734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gregg AR, Van den Veyver IB, Gross SJ,
Madankumar R, Rink BD and Norton ME: Noninvasive prenatal screening
by next-generation sequencing. Annu Rev Genomics Hum Genet.
15:327–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
MacLaren RE, Groppe M, Barnard AR,
Cottriall CL, Tolmachova T, Seymour L, Clark KR, During MJ, Cremers
FP, Black GC, et al: Retinal gene therapy in patients with
choroideremia: Initial findings from a phase 1/2 clinical trial.
Lancet. 383:1129–1137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Simões M, Marques C, Gonçalves A, Pereira
AP, Correia J, Castela J and Guerreiro C: Amniocentesis in HIV
pregnant women: 16 years of experience. Infect Dis Obstet Gynecol.
2013:9142722013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cignini P, Dugo N, Giorlandino C, Gauci R,
Spata A, Capriglione S and Cafà EV: Prenatal diagnosis of a fetus
with a ring chromosome 20 characterized by array-CGH. J Prenat Med.
6:72–73. 2012.PubMed/NCBI
|
|
18
|
Vora NL, Johnson KL, Peter I, Tighiouart
H, Ralston SJ, Craigo SD and Bianchi DW: Circulating cell-free DNA
levels increase variably following chorionic villus sampling.
Prenat Diagn. 30:325–328. 2010.PubMed/NCBI
|
|
19
|
Herzenberg LA, Bianchi DW, Schröder J,
Cann HM and Iverson GM: Fetal cells in the blood of pregnant women:
Detection and enrichment by fluorescence-activated cell sorting.
Proc Natl Acad Sci USA. 76:1453–1455. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW and Wainscoat JS: Presence of fetal DNA in
maternal plasma and serum. Lancet. 350:485–487. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wright CF and Burton H: The use of
cell-free fetal nucleic acids in maternal blood for non-invasive
prenatal diagnosis. Hum Reprod Update. 15:139–151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang WC, Zhu L, Qiu YM, Zhou BX, Cheng JL,
Wei CL, Chen HC, Li LY, Fu XD and Fu JJ: Isolation and analysis of
cell free fetal DNA from maternal peripheral blood in Chinese
women. Genet Mol Res. 14:18078–18089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boon EM and Faas BH: Benefits and
limitations of whole genome versus targeted approaches for
noninvasive prenatal testing for fetal aneuploidies. Prenat Diagn.
33:563–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chiu RW and Lo YM: Clinical applications
of maternal plasma fetal DNA analysis: Translating the fruits of 15
years of research. Clin Chem Lab Med. 51:197–204. 2013.PubMed/NCBI
|
|
25
|
Lv W, Wei X, Guo R, Liu Q, Zheng Y, Chang
J, Bai T, Li H, Zhang J, Song Z, et al: Noninvasive prenatal
testing for Wilson disease by use of circulating single-molecule
amplification and resequencing technology (cSMART). Clin Chem.
61:172–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van den Oever JM, Bijlsma EK, Feenstra I,
Muntjewerff N, Mathijssen IB, Bakker E, van Belzen MJ and Boon EM:
Noninvasive prenatal diagnosis of Huntington disease: Detection of
the paternally inherited expanded CAG repeat in maternal plasma.
Prenat Diagn. 35:945–949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu Y, Li X, Ge HJ, Xiao B, Zhang YY, Ying
XM, Pan XY, Wang L, Xie WW, Ni L, et al: Haplotype-based approach
for noninvasive prenatal tests of Duchenne muscular dystrophy using
cell-free fetal DNA in maternal plasma. Genet Med. 17:889–896.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baird PA, Anderson TW, Newcombe HB and
Lowry RB: Genetic disorders in children and young adults: A
population study. Am J Hum Genet. 42:677–693. 1988.PubMed/NCBI
|
|
29
|
Fu JJ, Li LY, Li XR and Lu GX: Rapid
prenatal gene diagnosis for β-thalassemia by amplification
refractory mutation system (ARMS). Chin J Obstet Gynecol.
35:359–360. 2000.
|
|
30
|
Fu JJ, Li LY and Lu GX: Prenatal gene
diagnosis of spinal muscular atrophy by combined with the technique
of PCR-SSCP, PCR followed by restriction enzyme digestion and
linkage analysis. Chin J Neurol. 34:74–78. 2001.
|
|
31
|
Fu J, Qin L, He T, Qin J, Hong J, Wong J,
Liao L and Xu J: The TWIST/Mi2/NuRD protein complex and its
essential role in cancer metastasis. Cell Res. 21:275–289. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khan MA, Tania M, Wei C, Mei Z, Fu S,
Cheng J, Xu J and Fu J: Thymoquinone inhibits cancer metastasis by
downregulating TWIST1 expression to reduce epithelial to
mesenchymal transition. Oncotarget. 6:19580–19591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang F, Wang H, Tuan HF, Nguyen DH, Sun V,
Keser V, Bowne SJ, Sullivan LS, Luo H, Zhao L, et al: Next
generation sequencing-based molecular diagnosis of retinitis
pigmentosa: Identification of a novel genotype-phenotype
correlation and clinical refinements. Hum Genet. 133:331–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Challis D, Yu J, Evani US, Jackson AR,
Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA and Yu F: An
integrative variant analysis suite for whole exome next-generation
sequencing data. BMC Bioinformatics. 13:82012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
1000 Genomes Project Consortium. Abecasis
GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME
and McVean GA: A map of human genome variation from
population-scale sequencing. Nature. 467:1061–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Q, Cheng J, Yang W, Tania M, Wang H,
Khan MA, Duan C, Zhu L, Chen R, Lv H and Fu J: Identification of a
novel heterozygous missense mutation in the CACNA1F gene in a
Chinese family with retinitis pigmentosa by next generation
sequencing. Biomed Res Int. 2015:9078272015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ng PC and Henikoff S: SIFT: Predicting
amino acid changes that affect protein function. Nucleic Acids Res.
31:3812–3814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xia H, Huang X, Guo Y, Hu P, He G, Deng X,
Xu H, Yang Z and Deng H: Identification of a novel MYO15A mutation
in a Chinese family with autosomal recessive nonsyndromic hearing
loss. PLoS One. 10:e01363062015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu X, Jian X and Boerwinkle E: dbNSFP: A
lightweight database of human nonsynonymous SNPs and their
functional predictions. Hum Mutat. 32:894–899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jensen TJ, Zwiefelhofer T, Tim RC, Džakula
Ž, Kim SK, Mazloom AR, Zhu Z, Tynan J, Lu T, McLennan G, et al:
High-throughput massively parallel sequencing for fetal aneuploidy
detection from maternal plasma. PLoS One. 8:e573812013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Srinivasan A, Bianchi DW, Huang H, Sehnert
AJ and Rava RP: Noninvasive detection of fetal subchromosome
abnormalities via deep sequencing of maternal plasma. Am J Hum
Genet. 92:167–176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bonilha VL, Trzupek KM, Li Y, Francis PJ,
Hollyfield JG, Rayborn ME, Smaoui N and Weleber RG: Choroideremia:
Analysis of the retina from a female symptomatic carrier.
Ophthalmic Genet. 29:99–110. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Krock BL, Bilotta J and Perkins BD:
Noncell-autonomous photoreceptor degeneration in a zebrafish model
of choroideremia. Proc Natl Acad Sci USA. 104:4600–4605. 2007.
View Article : Google Scholar : PubMed/NCBI
|