Introduction
Intrahepatic cholangiocarcinoma (ICC) is the second
most frequently occurring primary liver cancer with a high
mortality rate (1), which affects
1–2 per 100,000 patients (2).
Various factors have previously been suggested to contribute to the
progression of ICC, including sclerosing cholangitis and
hepatobiliary flukes (3).
Ultrasound scans may provide diagnostic information (4); however, specific diagnostic criteria
for patients with ICC remain to be elucidated. Therefore, numerous
patients present with ICC for diagnosis at an advanced stage, which
may contribute to its poor prognosis (5). It is therefore important to identify
potential biomarkers of ICC to aid prevention and identification of
therapeutic strategies.
The progression of ICC is associated with numerous
genetic factors, including gene mutations and dysregulation of gene
expression (6). Various
technological advances have identified molecular targets. Weber
et al (7) demonstrated that
low frequency alterations of phosphatase and tensin homolog,
cyclin-dependent kinase inhibitor 2A, and breast cancer 1/2 may
explain the mutation spectrum in ICC, based on clustered regularly
interspaced short palindromic repeats (CRISPR)/CRISPR-associated
protein 9-based targeted somatic multiplex-mutagenesis. Lee et
al (8) profiled the expression
of 315 genes and screened 400 alterations in 84 genes, via
hybridization capture, which may act as therapeutic targets of ICC
(8). Various drugs or therapeutic
methods have been developed based on the aforementioned identified
biomarkers. Lovastatin, which is a
3-hydroxy-3-methylglutaryl-coenzyme-CoA reductase inhibitor, may
inhibit the proliferation, migration and adhesive activities of ICC
cells, by inhibiting the expression of transforming growth
factor-β1, cyclooxygenase-2, and intracellular adhesion molecule-2
(9). Furthermore, various genes
have previously been demonstrated to affect drug sensitivity and
resistance in ICC. Tepsiri et al (10) demonstrated that the expression of
multidrug resistance-associated protein 3 was significantly
associated with the half maximal inhibitory concentration of
etoposide in ICC. Various biomarkers have been identified; however,
the underlying mechanism remains to be fully elucidated, to improve
the prognosis and therapy of ICC.
Gene microarrays profile the expression of thousands
of genes simultaneously and have been incorporated in numerous
cancer and ICC associated investigations (11–13).
The present study compared gene and microRNA (miRNA/miR) expression
data in ICC samples with healthy liver tissues, and differentially
expressed genes (DEGs) and miRNAs (DEMs) were identified.
Functional and pathway analyses were conducted, and subsequently, a
miRNA-gene regulation network was constructed to explore potential
biomarkers of ICC to improve its prognosis.
Materials and methods
Microarray data
The miRNA expression dataset GSE32957 and mRNA
expression dataset GSE32879 (14)
were downloaded from the Gene Expression Omnibus database
(http://www.ncbi.nlm.nih.gov/geo/). A
total of 16 ICC, 7 combined hepatocellular-cholangiocarcinoma (CHC)
and 2 hepatic adenoma samples were included in the mRNA and miRNA
expression datasets. In addition, the miRNA expression dataset
contained 3 focal nodular hyperplasia (FNH) and 5 healthy liver
tissue, and 2 cholangiocarcinoma cell lines, whereas the mRNA
expression dataset contained 5 FNH and 7 healthy liver tissue
samples. Hybridizations of miRNA and mRNA were performed on
GPL14732 Nanostring nCounter Human microRNA Expression Platform
(NanoString Technologies, Inc., Seattle, WA, USA) and GPL6244
[HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript
(gene) version; Affymetrix, Inc., Santa Clara, CA, USA],
respectively.
Differential expression analysis
The free R package NanoStringNorm (15) was used for the data preprocessing
of GSE32957, whereas the affy package (https://bioconductor.org/packages/release/bioc/html/affy.html)
was selected for analysis of GSE32879, due to the background
correction and quartile normalization qualities it exhibits. DEGs
and DEMs were identified in ICC samples compared with healthy liver
tissues, via the limma package (http://bioconductor.org/packages/release/bioc/html/limma.html)
in R. DEGs and DEMs were screened with the thresholds of
Benjamini-Hochberg adjusted P<0.05 and |log2(fold
change)|>1.
Alternative splicing (AS)
analysis
AS, which is prevalent in the mammalian genome, may
generate a complex transcriptome from a finite genome and thus,
result in a diverse range of proteins with various functions
(16,17). The present study used AltAnalyze
(18) software to infer AS via the
interpretation of alternative exon inclusion, in ICC samples.
Briefly, raw CEL microarray data were normalized using the FIRMA
method implemented in AltAnalyze, followed by the identification of
alternative splicing genes (ASGs) with all of the default
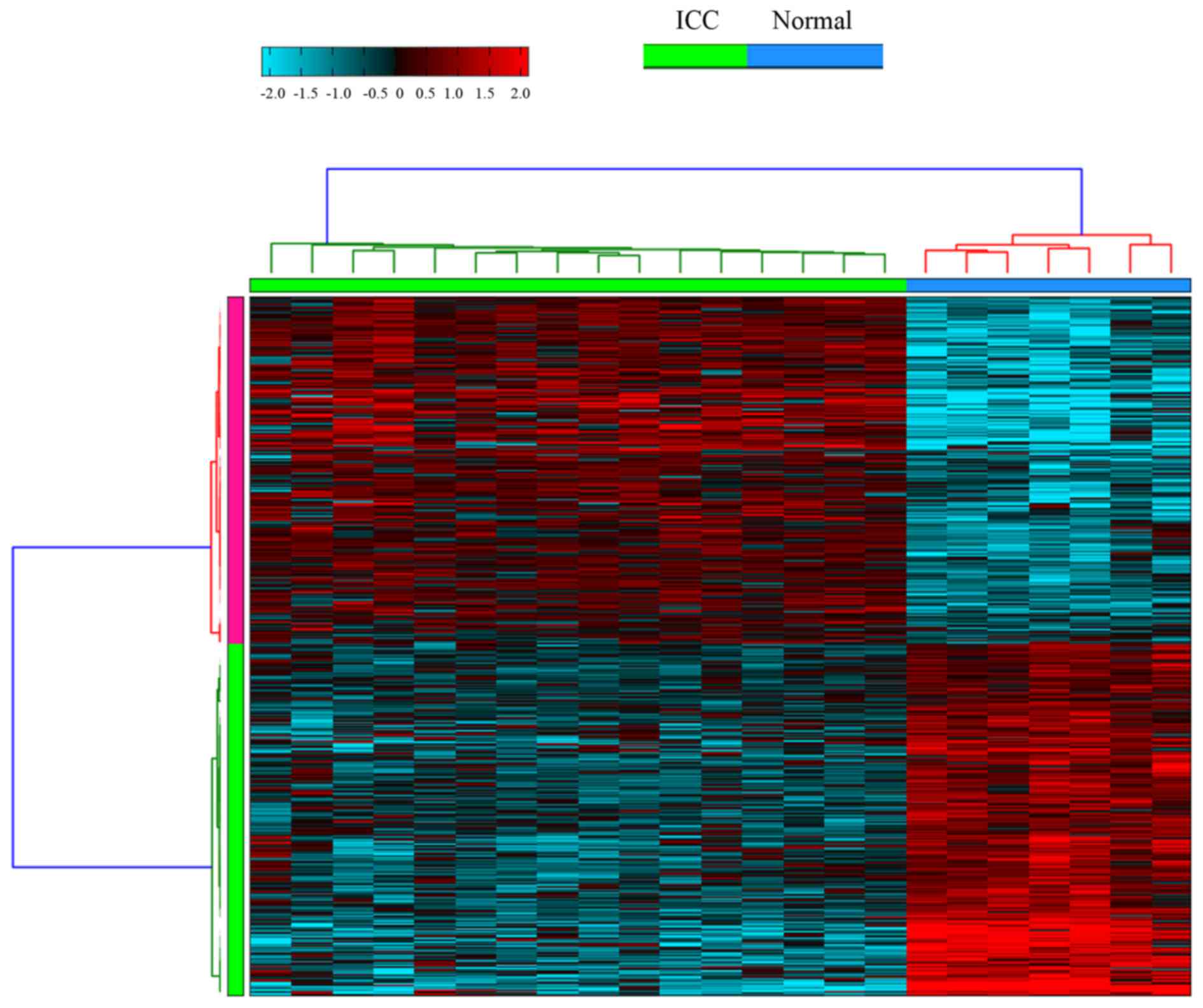
parameters of AltAnalyze. In addition, hierarchical clustering of
ASGs in ICC and healthy liver tissue samples were conducted.
Functional and pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Analysis (DAVID; https://david.ncifcrf.gov/) is a widely used web-based
tool for functional and pathway enrichment analysis (19). DAVID was used to conduct functional
and pathway enrichment analysis of DEGs. P<0.05 was used as the
criterion to identify significantly enriched Gene Ontology (GO;
http://geneontology.org/) terms and Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathways.
Screening of target genes of DEMs
TargetScan (20)
(http://www.targetscan.org/) is a
web-based software that was used to predict the target genes of
DEMs. Overlaps among these target genes, DEGs and ASGs were
obtained, and regulatory associations between the overlaps and DEMs
were retrieved to construct the miRNA-gene regulation network,
which was visualized using Cytoscape software (21).
Results
Identification of DEGs, DEMs and
ASGs
A total of 2,327 DEGs; 1,214 of which were
downregulated and 1,113 of which were upregulated, and 70 DEMs,
including 5 down- and 65 upregulated, were identified in ICC
samples compared with healthy liver tissues. A total of 623 genes
exhibited alternative splicing in ICC samples compared with healthy
liver tissue samples. Hierarchical clustering of ASGs in ICC and
healthy liver tissue samples was conducted, as presented in
Fig. 1.
Functional and pathway enrichment
analysis of DEGs
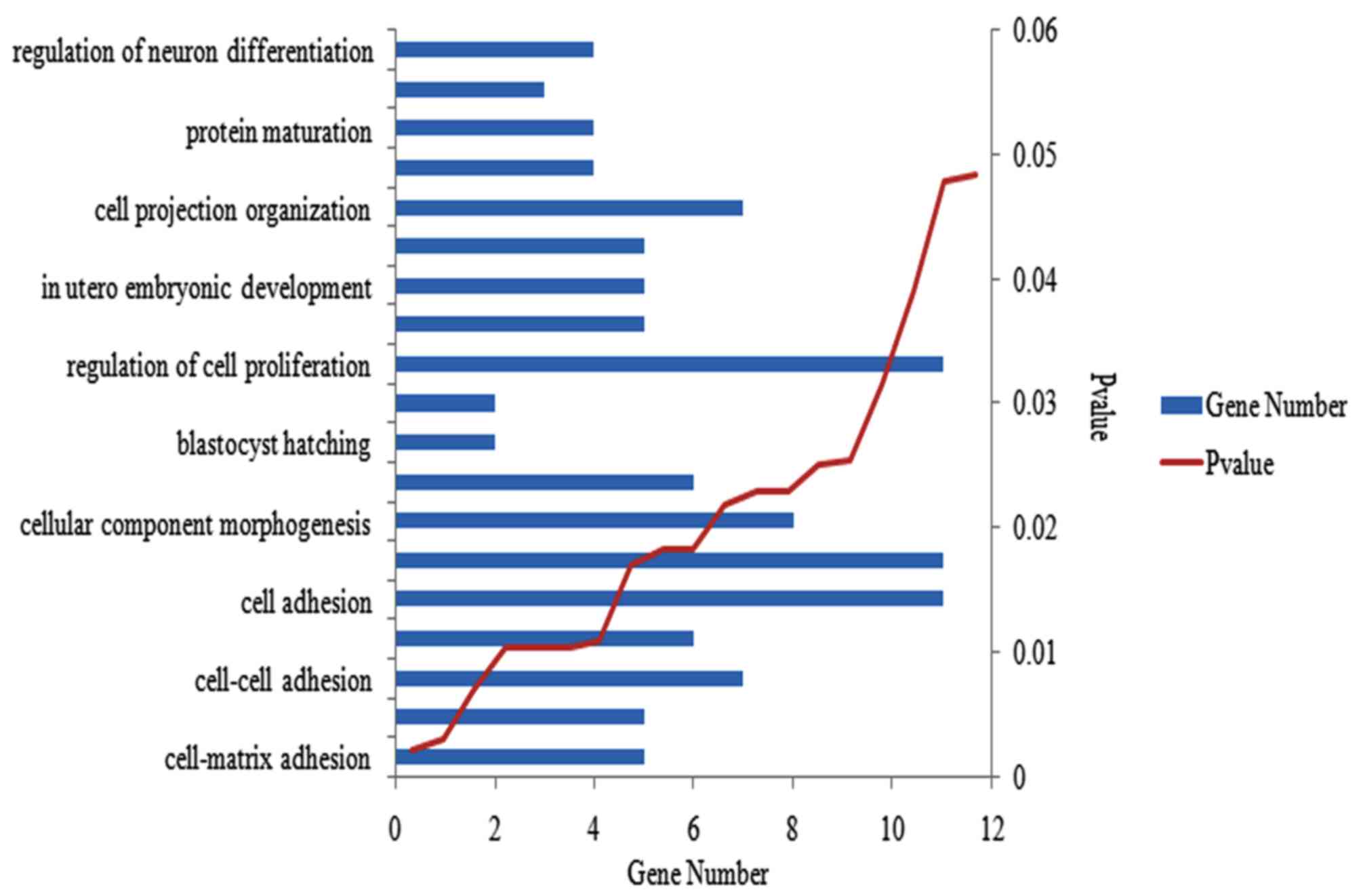
Functional and pathway enrichment analysis was
performed for the 2,327 DEGs. The majority of enriched GO terms
were involved in the activity of the cell, and the most
significantly enriched were presented in cell-matrix adhesion. Ten
of the GO terms that were randomly selected at P<0.05, are
presented in Fig. 2. A total of 51
KEGG pathways associated with metabolism and degradation were
revealed to be enriched in DEGs; 10 randomly selected pathways are
presented in Table I.
 | Table I.A total of 10 Kyoto Encyclopedia of
Genes and Genomes pathways enriched in differentially expressed
genes. |
Table I.
A total of 10 Kyoto Encyclopedia of
Genes and Genomes pathways enriched in differentially expressed
genes.
| Pathway name | P-value | Gene number |
|---|
| hsa04610: Complement
and coagulation cascades | 5.92E-24 | 49 |
| hsa00071: Fatty acid
metabolism | 1.89E-13 | 28 |
| hsa00260: Glycine,
serine and threonine metabolism | 5.08E-13 | 24 |
| hsa00280: Valine,
leucine and isoleucine degradation | 5.72E-12 | 28 |
| hsa00380: Tryptophan
metabolism | 1.99E-11 | 26 |
| hsa03320: PPAR
signaling pathway | 2.63E-10 | 34 |
| hsa00982: Drug
metabolism | 1.22E-09 | 31 |
| hsa00330: Arginine
and proline metabolism | 1.10E-08 | 27 |
| hsa00980: Metabolism
of xenobiotics by cytochrome P450 | 1.22E-08 | 29 |
| hsa00250: Alanine,
aspartate and glutamate metabolism | 7.26E-08 | 19 |
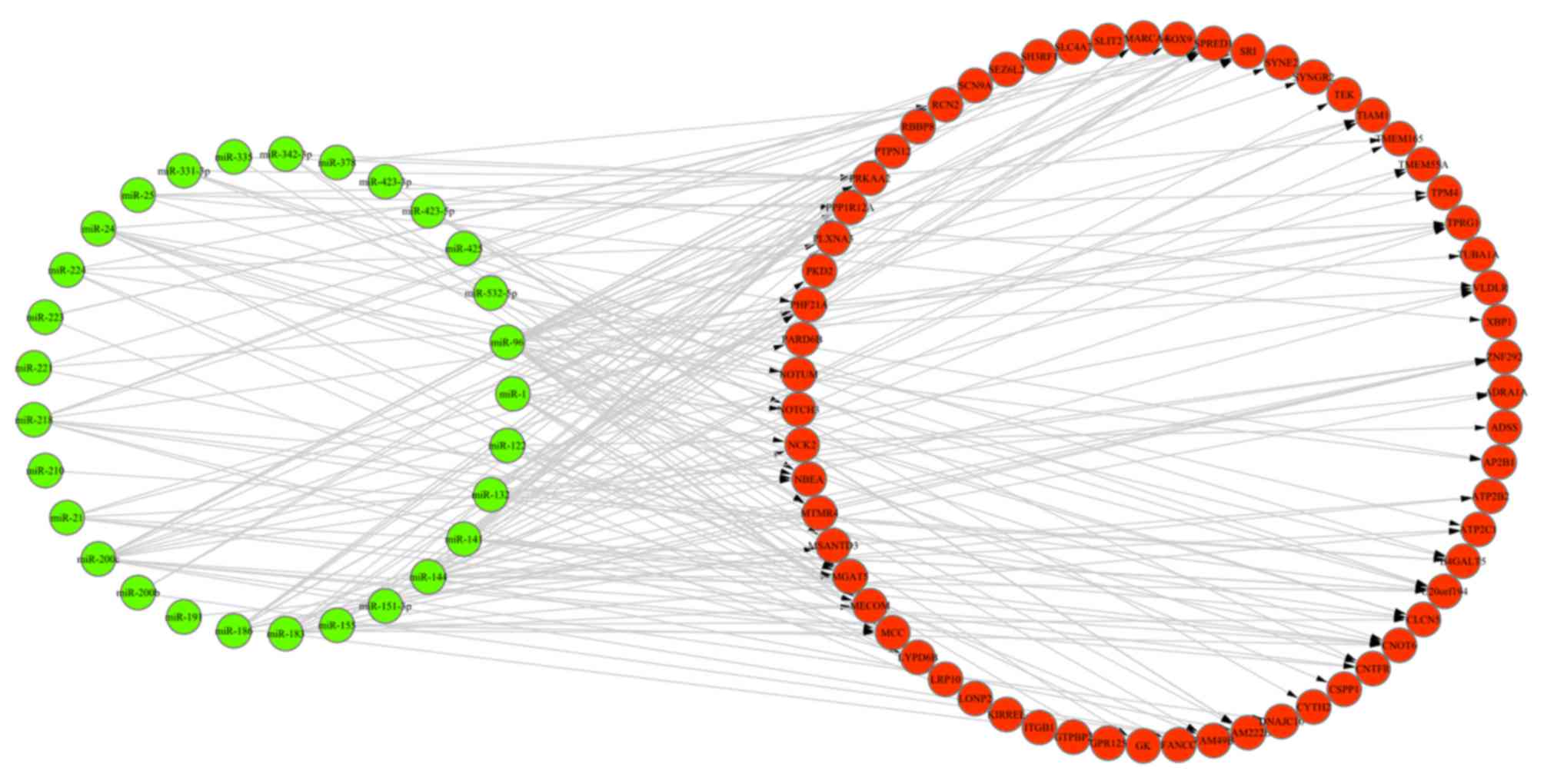
Target genes of DEMs
A total of 956 target genes of DEMs were obtained
via TargetScan software (data not shown). In addition, 63 overlaps
were identified among these target genes, DEGs and ASGs, and 243
miRNA-gene regulation pairs were retrieved between these overlaps
and DEMs. The miRNA-gene regulation network is presented in
Fig. 3. Furthermore, 52 miRNA-gene
pairs (Table II) exhibited an
opposite trend in the alteration of miRNA and gene expression
values in ICC samples, compared with healthy liver tissues.
 | Table II.A total of 52 miRNA-gene pairs
presenting opposite trends in the alterations of miRNA and gene
expression values in intrahepatic cholangiocarcinoma samples
compared with those in healthy liver tissues. |
Table II.
A total of 52 miRNA-gene pairs
presenting opposite trends in the alterations of miRNA and gene
expression values in intrahepatic cholangiocarcinoma samples
compared with those in healthy liver tissues.
| miRNA | Gene |
|---|
| hsa-miR-186 | ADRA1A |
| hsa-miR-122 | MGAT5 |
| hsa-miR-132 | ADRA1A |
| hsa-miR-96 | TPRG1 |
| hsa-miR-96 | SCN9A |
| hsa-miR-122 | LRP10 |
| hsa-miR-96 | LONP2 |
| hsa-miR-122 | ADSS |
| hsa-miR-96 | TIAM1 |
| hsa-miR-96 | CLCN5 |
| hsa-miR-96 | GK |
| hsa-miR-200c | TPRG1 |
| hsa-miR-21 | TIAM1 |
| hsa-miR-141 | SCN9A |
| hsa-miR-21 | CNTFR |
| hsa-miR-144 | MGAT5 |
| hsa-miR-218 | TPRG1 |
| hsa-miR-144 | PTPN12 |
| hsa-miR-144 | TMEM165 |
| hsa-miR-200c | CNTFR |
| hsa-miR-141 | TIAM1 |
| hsa-miR-224 | TPRG1 |
| hsa-miR-183 | TIAM1 |
| hsa-miR-141 | CLCN5 |
| hsa-miR-200c | MCC |
| hsa-miR-335 | MTMR4 |
| hsa-miR-144 | ZNF292 |
| hsa-miR-144 | SPRED1 |
| hsa-miR-144 |
C20orf194 |
| hsa-miR-218 | CLCN5 |
| hsa-miR-1 | GPR125 |
| hsa-miR-223 | LONP2 |
| hsa-miR-144 | TMEM55A |
| hsa-miR-186 | TPRG1 |
| hsa-miR-155 | CLCN5 |
| hsa-miR-186 | SCN9A |
| hsa-miR-191 | ATP2B2 |
| hsa-miR-144 | VLDLR |
| hsa-miR-144 | SMARCA4 |
| hsa-miR-144 | FAM222B |
| hsa-miR-423-5p | CNTFR |
| hsa-miR-218 | MCC |
| hsa-miR-144 | RCN2 |
| hsa-miR-144 | ATP2C1 |
| hsa-miR-24 | FANCC |
| hsa-miR-24 | CNTFR |
| hsa-miR-24 | CLCN5 |
| hsa-miR-224 | NOTUM |
| hsa-miR-25 | XBP1 |
| hsa-miR-24 | MCC |
| hsa-miR-532-5p | MTMR4 |
| hsa-miR-186 | MCC |
Discussion
In recent years, the emergence of novel diagnostics
and treatments, including B-mode ultrasound and chemotherapy, has
improved the prognosis of ICC. However, limitations are present in
terms of its early diagnosis and cure. The present study conducted
a statistical comparison of mRNA and miRNA expression profiles
between ICC and healthy liver tissues samples, in order to identify
DEGs, DEMs and ASGs. Functional and pathway analyses of DEGs were
conducted to explore associated functions and pathways. The
miRNA-gene regulation network was constructed to identify the
potential biomarkers of ICC.
The GeneChip Human Gene 1.0 ST array
(HuGene-1_0-st-v1) is a transcript-based array, used for the
detection of gene expression values, and estimation of AS based on
the well-annotated exon probes (22). The present study identified the
DEGs and ASGs in ICC samples compared with healthy liver tissues,
based on the data detected by HuGene-1_0-st-v1. Functional
enrichment analysis indicated that the DEGs were primarily involved
in the biological processes associated with cell activity,
including cell adhesion and regulation of cell proliferation, which
have previously been demonstrated to be associated with cancer
progression (23). Pathway
enrichment analysis revealed that DEGs were significantly enriched
in the pathways associated with substance metabolism and
degradation, including fatty acid, glycine, serine and threonine
metabolism, and valine, leucine and isoleucine degradation. The
extracellular matrix-receptor pathway was also demonstrated to be
enriched in DEGs, consistent with the findings of Lee et al
(24). The genes that were
simultaneously presented in the extracellular matrix-receptor
pathway and were revealed to be ASGs, included chondroadherin
(CHAD), integrin subunit (ITG)-A3, ITG-B1 and laminin subunit
alpha-5 (LAMA5). Three of the aforementioned genes, (ITGA3, ITGB1
and LAMA5) have previously been demonstrated to be associated with
the progression of ICC (25,26).
CHAD encodes a cartilage matrix protein, which may mediate adhesion
of isolated chondrocytes. CHAD may be included in the
phosphoinositide 3-kinase-Akt and focal adhesion signaling
pathways, and may be regulated by p53, which is associated with the
pathogenesis and development of numerous cancers, including ICC
(http://www.genecards.org). Therefore, CHAD may be
a novel biomarker of ICC that may contribute to its progression;
however, further studies are required to confirm this.
miRNA are non-coding RNA molecules, which contain
~22 nucleotides, and are present in animals, plants and viral
genomes. miRNAs may induce RNA-silencing and regulate gene
expression at the post-transcriptional stage (27,28).
The dysregulation of miRNA may result in numerous diseases,
including ICC (29–31). The present study identified DEMs in
ICC samples compared with healthy liver tissues samples, and the
screening of DEM target genes was conducted using the TargetScan
database. A miRNA-gene regulation network was constructed based on
the DEMs and the overlapping genes among the DEGs, ASGs and target
genes of the DEMs. In the regulation network, hsa-miR-96 regulated
22 target genes and 6 out of these regulation pairs
[hsa-miR-96-tumor protein P63 regulated 1, hsa-miR-96-sodium
voltage-gated channel alpha subunit 9, hsa-miR-96-lon peptidase 2,
peroxisomal, hsa-miR-96-T-cell lymphoma invasion and metastasis 1
(TIAM1), hsa-miR-96-chloride voltage-gated channel 5,
hsa-miR-96-glycerol kinase] revealed an opposite trend in the
alterations of miRNA and mRNA expression values (upregulated miRNA
and downregulated gene expression levels). hsa-miR-96 is closely
associated with cell proliferation and growth, and Collins et
al (32) demonstrated that
hsa-miR-96 may be used to identify cholangiocarcinoma and
pancreatic adenocarcinoma. Furthermore, the dysregulation of
hsa-miR-96 was revealed to contribute to the progression of ICC
(33). TIAM1 is a target gene of
hsa-miR-96, which is important in cellular migration and remodeling
of the actin cytoskeleton, and may be involved in the proliferation
and migration of ICC via effects on RasGEF domain family member 1A
expression (34). Further target
genes of hsa-miR-96 have previously been identified, and may act as
novel biomarkers of ICC, however further experimental verification
is necessary (35).
In conclusion, the present study identified DEGs,
ASGs and DEMs in ICC samples compared with healthy liver tissues,
via bioinformatics analysis of miRNA and mRNA expression data.
Functional and pathway analyses indicated that DEGs were primarily
involved in biological processes or pathways associated with cancer
progression or substance metabolism. A miRNA-gene regulation
network was constructed based on DEMs and overlaps among DEGs, ASGs
and target genes of DEMs. Furthermore, biomarkers that have
previously been identified, including hsa-miR-96 and TIAM1, and
numerous novel biomarkers in ICC, including anthranilate synthase
component II and sodium voltage-gated channel a subunit 9, were
identified. However, the functions of these biomarkers in ICC
remain to be elucidated.
References
|
1
|
Guglielmi A, Ruzzenente A, Campagnaro T,
Pachera S, Valdegamberi A, Nicoli P, Cappellani A, Malfermoni G and
Iacono C: Intrahepatic cholangiocarcinoma: Prognostic factors after
surgical resection. World J Surg. 33:1247–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson CD, Pinson CW, Berlin J and Chari
RS: Diagnosis and treatment of cholangiocarcinoma. Oncologist.
9:43–57. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vilana R, Forner A, Bianchi L,
García-Criado A, Rimola J, de Lope CR, Reig M, Ayuso C, Brú C and
Bruix J: Intrahepatic peripheral cholangiocarcinoma in cirrhosis
patients may display a vascular pattern similar to hepatocellular
carcinoma on contrast-enhanced ultrasound. Hepatology.
51:2020–2029. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khan SA, Thomas HC, Davidson BR and
Taylor-Robinson SD: Cholangiocarcinoma. Lancet. 366:1303–1314.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khan SA, Davidson BR, Goldin R, Pereira
SP, Rosenberg WM, Taylor-Robinson SD, Thillainayagam AV, Thomas HC,
Thursz MR and Wasan H: British Society of Gastroenterology:
Guidelines for the diagnosis and treatment of cholangiocarcinoma:
Consensus document. Gut. 51:(Suppl 6). VI1–VI9. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Isa T, Tomita S, Nakachi A, Miyazato H,
Shimoji H, Kusano T, Muto Y and Furukawa M: Analysis of
microsatellite instability, K-ras gene mutation and p53 protein
overexpression in intrahepatic cholangiocarcinoma.
Hepatogastroenterology. 49:604–608. 2002.PubMed/NCBI
|
|
7
|
Weber J, Ollinger R, Friedrich M, Ehmer U,
Barenboim M, Steiger K, Heid I, Mueller S, Maresch R, Engleitner T,
et al: CRISPR/Cas9 somatic multiplex-mutagenesis for
high-throughput functional cancer genomics in mice. Proc Natl Acad
Sci USA. 112:13982–13987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee H, Wang K, Johnson A, Jones DM, Ali
SM, Elvin JA, Yelensky R, Lipson D, Miller VA, Stephens PJ, et al:
Comprehensive genomic profiling of extrahepatic cholangiocarcinoma
reveals a long tail of therapeutic targets. J Clin Pathol.
69:403–408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang SH, Lin HY, Changou CA, Chen CH, Liu
YR, Wang J, Jiang X, Luh F and Yen Y: Integrin β3 and LKB1 are
independently involved in the inhibition of proliferation by
lovastatin in human intrahepatic cholangiocarcinoma. Oncotarget.
7:362–373. 2016.PubMed/NCBI
|
|
10
|
Tepsiri N, Chaturat L, Sripa B, Namwat W,
Wongkham S, Bhudhisawasdi V and Tassaneeyakul W: Drug sensitivity
and drug resistance profiles of human intrahepatic
cholangiocarcinoma cell lines. World J Gastroenterol. 11:2748–2753.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang MY, Li SH, Huang GL, Lin GH, Shuang
ZY, Lao XM, Xu L, Lin XJ, Wang HY and Li SP: Identification of a
novel microRNA signature associated with intrahepatic
cholangiocarcinoma (ICC) patient prognosis. BMC Cancer. 15:642015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Xie H, Ling Q, Lu D, Lv Z, Zhuang
R, Liu Z, Wei X, Zhou L, Xu X and Zheng S: Coding-noncoding gene
expression in intrahepatic cholangiocarcinoma. Transl Res.
168:107–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu YF, Ge FJ, Han B, Yang XQ, Su H, Zhao
AC, Zhao MH, Yang YB and Yang J: High-mobility group box 1
expression and lymph node metastasis in intrahepatic
cholangiocarcinoma. World J Gastroenterol. 21:3256–3265.
2015.PubMed/NCBI
|
|
14
|
Oishi N, Kumar MR, Roessler S, Ji J,
Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al:
Transcriptomic profiling reveals hepatic stem-like gene signatures
and interplay of miR-200c and epithelial-mesenchymal transition in
intrahepatic cholangiocarcinoma. Hepatology. 56:1792–1803. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Waggott D, Chu K, Yin S, Wouters BG, Liu
FF and Boutros PC: NanoStringNorm: An extensible R package for the
pre-processing of NanoString mRNA and miRNA data. Bioinformatics.
28:1546–1548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Graveley BR: Alternative splicing:
Increasing diversity in the proteomic world. Trends Genet.
17:100–107. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cieply B and Carstens RP: Functional roles
of alternative splicing factors in human disease. Wiley Interdiscip
Rev RNA. 6:311–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Emig D, Salomonis N, Baumbach J, Lengauer
T, Conklin BR and Albrecht M: AltAnalyze and DomainGraph: Analyzing
and visualizing exon expression data. Nucleic Acids Res.
38:W755–W762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pradervand S, Paillusson A, Thomas J,
Weber J, Wirapati P, Hagenbüchle O and Harshman K: Affymetrix
Whole-Transcript Human Gene 1.0 ST array is highly concordant with
standard 3′ expression arrays. Biotechniques. 44:759–762. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sawai H, Okada Y, Funahashi H, Matsuo Y,
Takahashi H, Takeyama H and Manabe T: Activation of focal adhesion
kinase enhances the adhesion and invasion of pancreatic cancer
cells via extracellular signal-regulated kinase-1/2 signaling
pathway activation. Mol Cancer. 4:372005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee JI and Campbell JS: Role of
desmoplasia in cholangiocarcinoma and hepatocellular carcinoma. J
Hepatol. 61:432–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Utispan K, Thuwajit P, Abiko Y, Charngkaew
K, Paupairoj A, Chau-in S and Thuwajit C: Gene expression profiling
of cholangiocarcinoma-derived fibroblast reveals alterations
related to tumor progression and indicates periostin as a poor
prognostic marker. Mol Cancer. 9:132010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Soejima Y, Inoue M, Takahashi Y, Uozaki H,
Sawabe M and Fukusato T: Integrins αvβ6, α6β4 and α3β1 are
down-regulated in cholangiolocellular carcinoma but not
cholangiocarcinoma. Hepatol Res. 44:E320–E334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun B, Xie C, Zheng T, Yin D, Wang J,
Liang Y, Li Y, Yang G, Shi H, Pei T, et al: Selecting molecular
therapeutic drug targets based on the expression profiles of
intrahepatic cholangiocarcinomas and miRNA-mRNA regulatory
networks. Oncol Rep. 35:382–390. 2016.PubMed/NCBI
|
|
30
|
Deng G, Teng Y, Huang F, Nie W, Zhu L,
Huang W and Xu H: MicroRNA-101 inhibits the migration and invasion
of intrahepatic cholangiocarcinoma cells via direct suppression of
vascular endothelial growth factor-C. Mol Med Rep. 12:7079–7085.
2015.PubMed/NCBI
|
|
31
|
Xiong X, Sun D, Chai H, Shan W, Yu Y, Pu L
and Cheng F: MiR-145 functions as a tumor suppressor targeting
NUAK1 in human intrahepatic cholangiocarcinoma. Biochem Biophys Res
Commun. 465:262–269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Collins AL, Wojcik S, Liu J, Frankel WL,
Alder H, Yu L, Schmittgen TD, Croce CM and Bloomston M: A
differential microRNA profile distinguishes cholangiocarcinoma from
pancreatic adenocarcinoma. Ann Surg Oncol. 21:133–138. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Papaconstantinou I, Karakatsanis A,
Gazouli M, Polymeneas G and Voros D: The role of microRNAs in liver
cancer. Eur J Gastroenterol Hepatol. 24:223–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ura K, Obama K, Satoh S, Sakai Y, Nakamura
Y and Furukawa Y: Enhanced RASGEF1A expression is involved in the
growth and migration of intrahepatic cholangiocarcinoma. Clin
Cancer Res. 12:6611–6616. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang CC, Lin CC, Hsieh WL, Lai HW, Tsai
CH and Cheng YW: MicroRNA expression profiling in PBMCs: A
potential diagnostic biomarker of chronic hepatitis C. Dis Markers.
2014:3671572014. View Article : Google Scholar : PubMed/NCBI
|