Introduction
Gastric cancer is the second leading cause of
cancer-associated mortality worldwide, and is the most common
gastrointestinal tumor in East Asia (1,2).
Gastrectomy remains the primary treatment for gastric cancer.
Although the majority of patients at an early stage of gastric
carcinoma may be cured by surgery, >50% of patients do not
survive due to carcinoma recurrence at an advanced stage of the
disease, despite undergoing a curative gastrectomy (3). Therefore, an improved understanding
of the pathogenesis and identification of the molecular alterations
is essential for the development of diagnostic markers that may aid
novel effective therapies for gastric cancer (4,5).
Long non-coding RNA (lncRNAs), which are transcripts
>200 bp in size with no protein-encoding function, represent a
less investigated class of non-coding RNAs (6). Previous studies have reported that
lncRNAs are crucial for the regulation of chromatin structure, gene
expression and translational control (7–9). A
significant number of studies have investigated small interfering
RNAs and microRNAs, and their functions and molecular mechanisms
have been illustrated in recent years (10,11).
However, our understanding of the function of lncRNAs in different
diseases remains limited.
An increasing number of studies are investigating
the role of lncRNAs in the origin and development of cancers,
particularly digestive system cancers. High expression levels of
PVT1 was demonstrated to be a potential candidate biomarker for
gastric cancer, and promoted cell proliferation through the
epigenetic regulation of p15 and p16 (12). In addition, upregulation of
LINC00152 is reportedly correlated with depth of tumor invasion in
patients with gastric cancer (13). The lncRNA CCAT2 is highly expressed
and associated with poor prognosis in gastric cancer (14). Although several lncRNAs have been
reported to function in the development of gastric cancer, the
function of <1% of the 30,000 lncRNAs identified to date has
been reported (15). Therefore,
identification of gastric cancer-associated lncRNAs and
investigation of their molecular mechanisms and biological
functions, is essential for understanding the molecular biology of
gastric cancer development.
A previous study demonstrated that LET was
downregulated in gastric cancer and was associated with poor
prognosis (16). The aim of the
present study was to verify the expression of LET in gastric cancer
tissues and assess its impact on gastric cancer cell proliferation,
migration and apoptosis. The results demonstrated that LET may
function as a tumor suppressor lncRNA in gastric cancer.
Materials and methods
Patients and specimens
A total of 37 cases of paired gastric cancer
specimens used for the purposes of the present study, were
collected from gastrectomy procedures at the Department of
Gastroenterology, NanKai Hospital (Tianjin, China) between June
2008 and June 2014. Samples were collected from 17 male and 20
female patients. The average weight of male patients was 63.4±3.4
kg and the average weight (mean ± standard deviation) of female
patients was 52.6±3.8 kg. The average age for male and female
patients (mean ± standard deviation) was 48.5±5.3 years and
57.6±6.2 years, respectively. All tissue samples were reviewed and
diagnosed as gastric cancer according to the American Joint
Committee on Cancer staging manual based on histopathological
evaluation (17). None of the
patients received percutaneous ablation, chemoembolization or
radiotherapy prior to the gastrectomy operation. Written informed
consent was provided by all patients, and the clinical research was
approved by the Institutional Review Board of NanKai Hospital. All
specimens were immediately frozen in liquid nitrogen for downstream
analysis.
Cell culture and transfection
The human gastric cancer cell lines, SGC-7901 and
MGC-803, were purchased from the American Type Culture Collection
(Manassas, VA, USA). The normal human gastric epithelial cell line,
GES-1, was obtained from the Cell Line Resource Center, Shanghai
Institute of Biochemistry and Cell Biology, the Chinese Academy of
Sciences. (Shanghai, China). Cells were maintained in Dulbecco's
Modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum at 37°C in a humidified
atmosphere with 5% CO2.
To investigate the function of LET in gastric cancer
cells, an overexpression plasmid containing the LET lncRNA sequence
was generated using gene recombination technology. The sequence of
human LET was amplified by polymerase chain reaction (PCR) using
the following primers: Forward,
5′-CGCGGATCCCTCACAGACAAAGGAGAGTCTGATG-3′, and reverse,
5′-CCGGAATTCTGGGTGTTTTCATGTAGGAAATGGT-3′. All primers were obtained
from Invitrogen (Invitrogen, CA, USA). PCR products were
double-digested using BamHI and EcoRI restriction endonucleases
(Takara Biotechnology Co., Ltd., Otsu, Japan) according to the
manufacturer's instructions. The digested products were sub-cloned
into the pcDNA3.1(+)vector (Invitrogen; Thermo Fisher Scientific,
Inc.) to produce a pcDNA3.1(+)-LET fusion overexpression vector,
and the sequence of pcDNA3.1(+)-LET were confirmed by Sanger DNA
sequencing (Invitrogen; Thermo Fisher Scientific, Inc.). SGC-7901
and MGC-803 cells were transfected with 4 µg pcDNA3.1(+)-LET
recombinant overexpression vector or pcDNA3.1(+)empty vector using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at
a ratio of 1.25:1.00 Lipofectamine:vector DNA.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from gastric cancer tissues and cells
(1×106) was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. cDNA was synthesized from 1 µg RNA using the
PrimeScript RT kit (Takara Biotechnology Co., Ltd.). RT-qPCR
reactions were performed using the SYBR® Premix EX Taq™
II PCR kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions, with the Roche LightCycler®
480 Instrument II (Roche Diagnostics, Indianapolis, IN, USA). All
primers were obtained from Invitrogen (Thermo Fisher Scientific,
Inc.). The primer sequences were as follows: LET, forward,
5′-GGGAGTAAAGGGAAAGAGTTGC-3′, and reverse,
5′-AGGCTGAGGAAGGTGGTATTGG-3′; GAPDH, forward,
5′-CATCAAGAAGGTGGTGAAGCAGG-3′, and reverse,
5′-AAAGGTGGAGGAGTGGGTGTCG-3′. Data were collected and analyzed
using Roche LightCycler® 480 software (version 1.5;
Roche Diagnostics). The expression of LET was normalized internally
using the quantification cycle of the GAPDH housekeeping gene. The
relative quantitative value was calculated using the
2-ΔΔCq method (18).
Each experiment was performed in quintuplicate and repeated five
times.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was detected using the CCK-8
assay kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
according to the manufacturer's instructions. SGC-7901 and MGC-803
cells were seeded in 96-well plates at a density of 4×103
cells/well and cultured in normal medium for 24 h, before they were
transfected with the pcDNA3.1(+)-LET vector or the pcDNA3.1(+)empty
vector. At 0, 24, 48 and 72 h following transfection, the
absorbance of each well at 450 nm (with 630 nm as the reference
wavelength) was measured using an ELISA microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All assays were repeated at
least three times.
Cell migration assay
The migratory ability of gastric cancer cells
(SGC-7901 and MGC-803) was analyzed using an in vitro wound
scratch assay. Cells (~5×107) were cultured in a 6-well dish for 24
h, before they were transfected with the pcDNA3.1(+)-LET vector or
the pcDNA3.1(+)empty vector using Lipofectamine 2000. Vertical
horizontal wounds were generated with a sterile 10 µl pipette tip
at 6 h following transfection. The cells were then washed with
phosphate-buffered saline and maintained at 37°C in the incubator.
The wound images were acquired with a digital camera system at 0
and 24 h after the wounds were generated. The width of wounds was
measured using a standard caliper. All experiments were performed
in triplicate and repeated at least three times.
Cell apoptosis assay
The extent of apoptosis was evaluated using a
caspase 3 ELISA assay. SGC-7901 and MGC-803 gastric cancer cells
were seeded in 6-well plates at a density of 2×106 cells/well and
transfected with pcDNA3.1(+)-LET vector or pcDNA3.1(+)empty vector.
At 48 h following transfection, cell apoptosis was determined by
calculating the activity of caspase 3 using a caspase 3 ELISA kit
(cat. no. KHO1091; Thermo Fisher Scientific, Inc.) and a Hoechst
33258 staining assay (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The value of optical
density at 405 nm was measured using an ELISA microplate reader
(Bio-Rad Laboratories, Inc.). The ratio of cell apoptosis was
calculated according to the formula: (ODxdilution
factor)/(10.5xvolume of sample in mlxreaction time in min). The
apoptosis of cell was also detected by a Zeiss LSM 710 laser
scanning confocal microscopy (Carl Zeiss AG, Oberkochen, Germany)
and analyzed using the Image-Pro Plus software program (version,
5.10; Media Cybernetics, Inc., Rockville, MD, USA). All experiments
were performed in triplicate.
Statistical analysis
Statistical analysis of data was performed using
SPSS software (version, 18.0; SPSS, Inc., Chicago, IL, USA). The
paired t-test was used to compare LET expression in gastric cancer
tissues with matched non-tumor tissues. The independent samples
t-test was used to analyze the remaining data. P<0.05 was
considered to indicate a statistically significant difference.
Results
LET expression is downregulated in
gastric cancer tissues and cell lines
A previous study reported that LET is downregulated
in gastric cancer tissues (17).
To assess the function of LET in gastric cancer in the present
study, the expression level of LET in 37 gastric cancer tissues and
matched adjacent non-tumor tissues was examined by RT-qPCR
analysis. As showed in Fig. 1A,
the expression of LET in gastric cancer tissues was significantly
lower when compared with matched adjacent non-tumor tissues
(P<0.01). LET expression in gastric cancer cell lines and the
human gastric epithelial mucosa cell line GES-1 was then assessed
by RT-qPCR analysis. Consistent with the results shown in Fig. 1A, LET expression was significantly
downregulated in SGC-7901 (P=0.017) and MGC-803 (P=0.032) cells
compared with GES-1 cells (Fig.
1B). These results suggest that LET expression is downregulated
in gastric cancer tissues and cell lines.
Restoration of LET expression
suppressed gastric cancer cell proliferation in vitro
To investigate the functional role of LET in gastric
cancer cell proliferation, the pcDNA3.1(+)-LET vector or
pcDNA3.1(+)empty vector was transfected into SGC-7901 and MGC-803
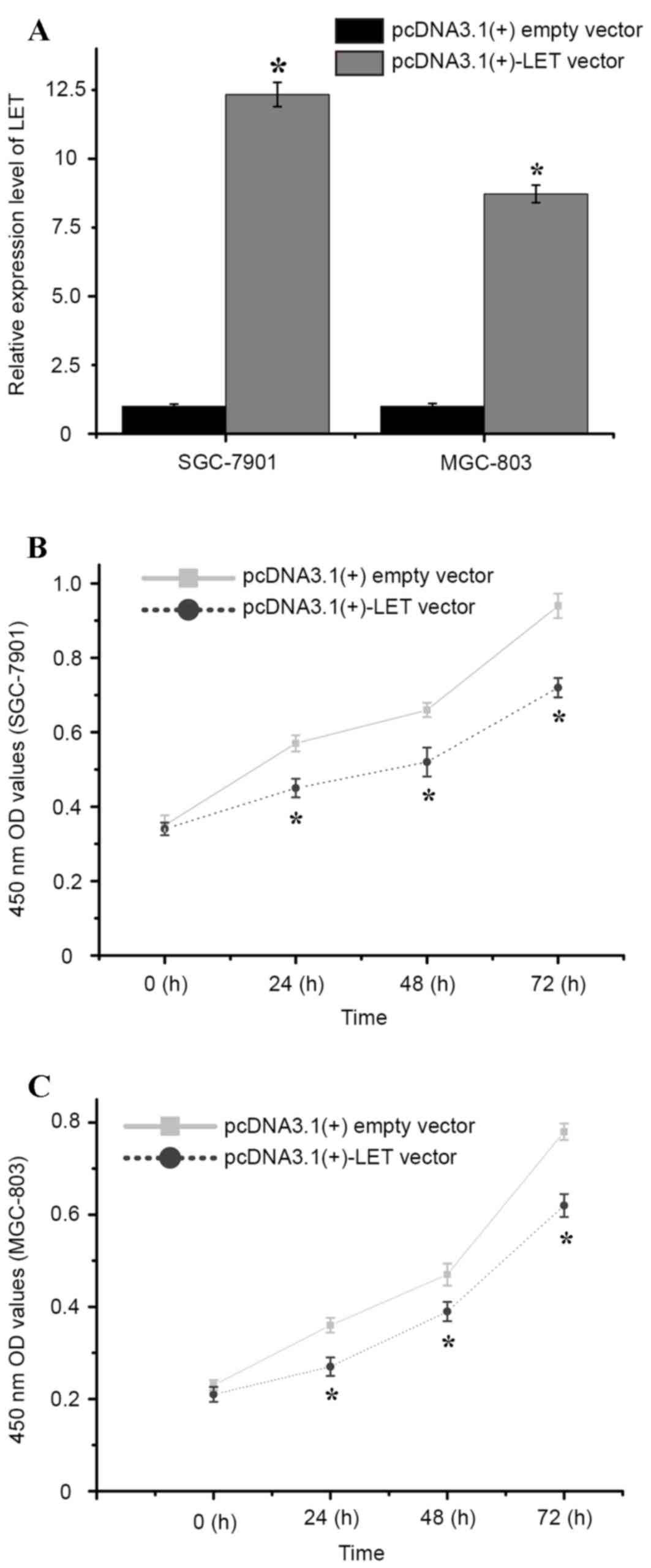
cells, and cell proliferation was examined using a CCK-8 assay. As
shown in Fig. 2A, SGC-7901 and
MGC-803 cells transfected with the pcDNA3.1(+)-LET vector
demonstrated a significant increase in LET expression when compared
with pcDNA3.1+ empty vector-transfected cells (P<0.01; Fig. 2A). The CCK-8 assay was then
performed and the optical density values of the pcDNA3.1(+)-LET
vector or pcDNA3.1(+)empty vector-transfected cells were measured
at 0, 24, 48 and 72 h following transfection. The results
demonstrated that the relative cell proliferation of
pcDNA3.1(+)-LET vector-transfected SGC-7901 cells was significantly
decreased at 24, 48 and 72 h when compared with
pcDNA3.1(+)-transfected cells (5.43, 6.07 and 9.21%, respectively;
P<0.01; Fig. 2B). Similarly,
the relative cell proliferation of pcDNA3.1(+)-LET
vector-transfected MGC-803 cells was significantly decreased at 24,
48 and 72 h following transfection when compared with
pcDNA3.1(+)-transfected cells (4.09, 3.88 and 7.47%, respectively;
P<0.01; Fig. 2C). These results
indicate that overexpression LET may significantly inhibit cell the
proliferation of gastric cancer cells in vitro.
Restoration of LET expression inhibits
gastric cancer cell migration in vitro
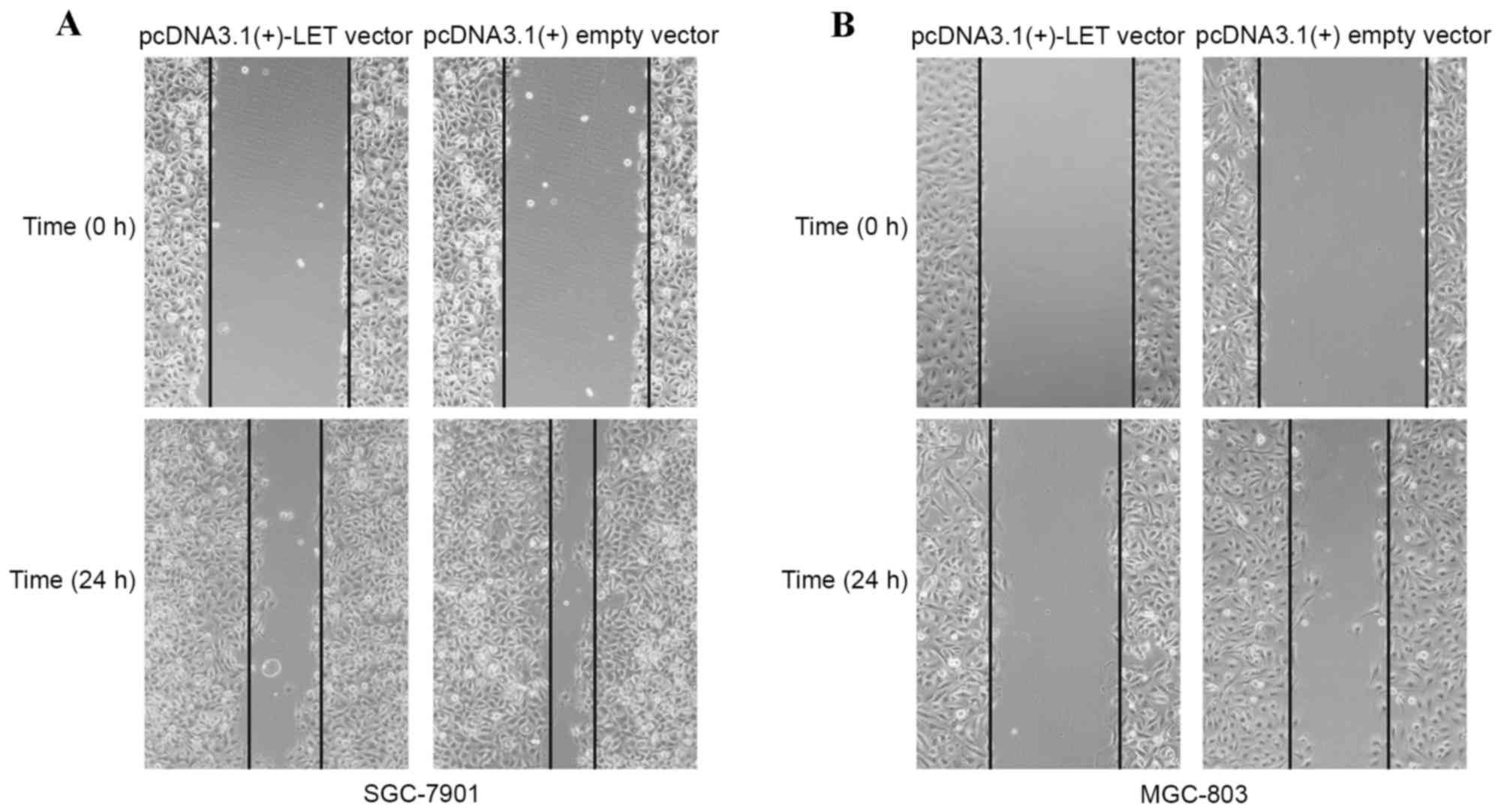
The effect of LET overexpression on gastric cancer
cell migration was determined using a wound healing assay. As shown
in Fig. 3A and B, the wound widths
of SGC-7901 and MGC-803 cells transfected with the pcDNA3.1(+)-LET
vector were markedly greater when compared with the
pcDNA3.1(+)empty vector-transfected cells. The results demonstrate
that restoration LET expression may inhibit gastric cancer cell
migration.
Restoration of LET expression induces
gastric cancer cell apoptosis in vitro
LncRNAs have been reported to serve a crucial role
in cell apoptosis, particularly in mediating escape from apoptosis
in cancer cells (19). To
determine the effect of LET on gastric cancer cell apoptosis, a
caspase 3 ELISA assay was used to measure the rate of apoptosis. As
revealed in Fig. 4, the apoptosis
rate of SGC-7901 cells transfected with pcDNA3.1(+)-LET vector was
significantly higher when compared with the pcDNA3.1(+)empty
vector-transfected cells (16.53% vs. 3.35%; P<0.01). Similarly,
the apoptosis rate of pcDNA3.1(+)-LET vector-transfected MGC-803
cells was significantly higher when compared with pcDNA3.1(+)empty
vector-transfected cells (13.30% vs. 2.19%; P<0.01; Fig. 4B). The results demonstrate that
overexpression of LET may promote apoptosis of gastric cancer
cells. The results presented thus far, identify LET lncRNA as a
novel tumor suppressor in gastric cancer.
Discussion
Gastric cancer is one of the most common types of
digestive tumors (20). It is the
second most frequent cause of cancer-associated mortality worldwide
and presents a major public health issue (21). Despite significant advances in
cancer therapy, major limitations in the management of gastric
cancer remain. A large number of patients are diagnosed with
advanced gastric cancer and have a poor prognosis.
Emerging evidence suggests that lncRNAs serve
essential roles in the modulation of tumor behavior through various
complex mechanisms, such as modulating gene transcription and
epigenetic signaling pathways (22,23).
A recent study suggested that the HOX transcript antisense RNA
lncRNA may promote the malignant growth of human liver cancer stem
cells through downregulation of SET domain containing 2 (24). In addition, the intronic prostate
cancer antigen 3 lncRNA regulates the prune homolog 2 suppressor in
prostate cancer (25).
Furthermore, increased HOXA transcript at the distal tip lncRNA
expression is correlated with prognosis and progression in tongue
squamous cell carcinoma (26).
Silencing prostate cancer associated transcript-1 induces cell
growth arrest and apoptosis in human bladder cancer (27). However, limited data are available
regarding the expression and function of lncRNAs in gastric
cancer.
The LET lncRNA gene is located on chromosome
15q24.1. LET exhibits differential expression patterns in various
tumors (16,28–30).
Previous studies have demonstrated that LET expression is
downregulated in cervical and gastric cancer (16,28).
However, to the best of our knowledge, the precise function of LET
has not yet been reported in gastric cancer. In the present study,
the expression of LET in gastric cancer tissues and cell lines was
first examined. Consistent with previously reported results, the
expression of LET was downregulated in gastric cancer tissues when
compared with matched adjacent normal tissues. In addition, LET
expression was decreased in two gastric cancer cell lines (SGC-7901
and MGC-803) when compared with a human gastric epithelial mucosa
cell line (GES-1). These results provided a strong rationale for
subsequent functional experiments.
To further understand the biological functions of
LET in gastric cancer cells, cell proliferation, migration and
apoptosis was examined by applying a gain-of-function approach.
Transfection of SGC-7901 and MGC-803 cells with the pcDNA3.1(+)-LET
vector led to a significant reduction in cell proliferation and
migration, and increased apoptosis when compared with
pcDNA3.1(+)empty vector-transfected cells. These results suggest
that LET may function as a tumor suppressor gene in the occurrence
and development of gastric cancer.
In conclusion, the results of the present study
confirm that LET is significantly downregulated in human gastric
cancer tissues and cells. In addition, this study is the first to
demonstrate that LET serves a tumor suppressive role in gastric
cancer by influencing cellular migration, proliferation and
apoptosis. Therefore, LET presents a promising biomarker and/or a
therapeutic target for gastric cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu N, Wang Z, Song X, Wei L, Kim BS,
Freedman ND, Baek J, Burdette L, Chang J, Chung C, et al:
Genome-wide association study of gastric adenocarcinoma in Asia: A
comparison of associations between cardia and non-cardia tumours.
Gut. 65:1611–1688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakajima T: Gastric cancer treatment
guidelines in Japan. Gastric Cancer. 5:1–5. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vogiatzi P, Vindigni C, Roviello F,
Renieri A and Giordano A: Deciphering the underlying genetic and
epigenetic events leading to gastric carcinogenesis. J Cell
Physiol. 211:287–295. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa FF: Non-coding RNAs: Meet thy
masters. Bioessays. 32:599–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H
and Geng P: Forkhead box C1 promoter upstream transcript, a novel
long non-coding RNA, regulates proliferation and migration in
basal-like breast cancer. Mol Med Rep. 11:3155–3159.
2015.PubMed/NCBI
|
|
8
|
Tian F, Tang Z, Song G, Pan Y, He B, Bao Q
and Wang S: Loss of imprinting of IGF2 correlates with
hypomethylation of the H19 differentially methylated region in the
tumor tissue of colorectal cancer patients. Mol Med Rep.
5:1536–1540. 2012.PubMed/NCBI
|
|
9
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rusek AM, Abba M, Eljaszewicz A, Moniuszko
M, Niklinski J and Allgayer H: MicroRNA modulators of epigenetic
regulation, the tumor microenvironment and the immune system in
lung cancer. Mol Cancer. 14:342015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia
T, Xiao B and Guo J: Increased expression of long intergenic
non-coding RNA LINC00152 in gastric cancer and its clinical
significance. Tumour Biol. 35:5441–5447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ
and Hu JH: Long non-coding RNA CCAT2 is up-regulated in gastric
cancer and associated with poor prognosis. Int J Clin Exp Pathol.
8:779–785. 2015.PubMed/NCBI
|
|
15
|
Gan L, Xu M, Zhang Y, Zhang X and Guo W:
Focusing on long noncoding RNA dysregulation in gastric cancer.
Tumour Biol. 36:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou B, Jing XY, Wu JQ, Xi HF and Lu GJ:
Down-regulation of long non-coding RNA LET is associated with poor
prognosis in gastric cancer. Int J Clin Exp Pathol. 7:8893–8898.
2014.PubMed/NCBI
|
|
17
|
Strong VE, D'Amico TA, Kleinberg L and
Ajani J: Impact of the 7th Edition AJCC staging classification on
the NCCN clinical practice guidelines in oncology for gastric and
esophageal cancers. J Natl Compr Canc Netw. 11:60–66.
2013.PubMed/NCBI
|
|
18
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Tian H, Yang J and Gong Z: Long
Noncoding RNAs regulate cell growth, proliferation, and apoptosis.
DNA Cell Biol. 35:459–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamamoto H, Watanabe Y, Maehata T, Morita
R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K,
et al: An updated review of gastric cancer in the next-generation
sequencing era: Insights from bench to bedside and vice versa.
World J Gastroenterol. 20:3927–3937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smith MG, Hold GL, Tahara E and El-Omar
EM: Cellular and molecular aspects of gastric cancer. World J
Gastroenterol. 12:2979–2990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szczesniak MW and Makalowska I: lncRNA-RNA
interactions across the human transcriptome. PLoS One.
11:e01503532016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu B, Sun L and Song E: Pinched by RNA
‘fingers’: Long noncoding RNAs hitting signal transduction
pathways. Mol Cell Oncol. 3:e10465822016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu
H and Lu D: LncRNA HOTAIR promotes human liver cancer stem cell
malignant growth through downregulation of SETD2. Oncotarget.
6:27847–27864. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salameh A, Lee AK, Cardó-Vila M, Nunes DN,
Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM,
Hosoya H, et al: PRUNE2 is a human prostate cancer suppressor
regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad
Sci USA. 112:8403–8408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Zhao L, Wang YX, Xi M, Liu SL and
Luo LL: Long non-coding RNA HOTTIP is correlated with progression
and prognosis in tongue squamous cell carcinoma. Tumour Biol.
36:8805–8809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu L, Liu Y, Zhuang C, Xu W, Fu X, Lv Z,
Wu H, Mou L, Zhao G, Cai Z and Huang W: Inducing cell growth arrest
and apoptosis by silencing long non-coding RNA PCAT-1 in human
bladder cancer. Tumour Biol. 36:7685–7689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang S, Wang HL and Yang J: Low
expression of long non-coding RNA LET inhibits carcinogenesis of
cervical cancer. Int J Clin Exp Pathol. 8:806–811. 2015.PubMed/NCBI
|
|
29
|
Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY,
Gong W, Zhang WJ and Quan ZW: Long non-coding RNA-LET is a positive
prognostic factor and exhibits tumor-suppressive activity in
gallbladder cancer. Mol Carcinog. 54:1397–1406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|