Introduction
microRNAs (miRNAs) are endogenous single-stranded
noncoding RNAs that negatively regulate diverse biological and
pathological processes, including apoptosis, myogenesis and
tumorigenesis. miRNAs post-transcriptionally regulate gene
expression via inhibiting mRNA translation or promoting mRNA
degradation following binding to the 3′ untranslated region of
target mRNA (1–4). Accumulating evidence suggests that
miRNAs are involved in osteoblast differentiation. Previous studies
have revealed that miRNA (miR)-20a is upregulated in
naringin-induced osteogenesis in bone marrow stromal cells (BMSCs)
(5) and that inhibition of
miR-103a increased runt-related transcription factor 2 (Runx2)
protein expression levels in osteoblasts (6). A recent study has demonstrated that
miR-21 promotes osteogenesis and mineralization in MC3T3-E1 cells
by inhibiting the translation of mothers against decapentaplegic 7
(7). Additionally, it has been
demonstrated that miR-188 is a key regulator of the age-associated
switch from osteogenic to adipogenic differentiation of BMSCs
(8). Therefore, miRNAs may be
potential therapeutic targets for the treatment of bone-associated
diseases, including osteoporosis and osteonecrosis.
miR-133a is a highly conserved miRNA. Previous
studies have revealed that mir-133a is specifically expressed in
cardiac and skeletal muscle and acts as a key regulator during the
development of these tissues (3,9).
Studies have demonstrated the expression of miR-133a in mesenchymal
stem cells, where it acts as a negative regulator of osteoblast
differentiation (10–12). Furthermore, Wang et al
(13) suggested that miR-133a
expression levels in circulating monocytes were upregulated in
patients with osteoporosis and Wu et al (14) discovered that mir-133a was
significantly downregulated in non-traumatic osteonecrosis of the
femoral head samples compared with femoral neck fracture samples,
indicating that miR-133a may be a key regulator of bone
metabolism.
In humans, miR-133a appears to have a direct
regulatory effect on the expression of vitamin K epoxide reductase
complex subunit 1 (VKORC1), which is crucial in the vitamin K
(VK)-epoxide cycle (15). The
VK-epoxide cycle is closely associated with the γ-carboxylation of
VK-dependent proteins during bone metabolism, including osteocalcin
(OCN), matrix Gla protein and protein S. OCN is synthesized by
osteoblasts during the mineralization of bone; γ-carboxylation of
OCN confers greater affinity for calcium ions, whereas the failure
of carboxylation results in the accumulation of uncarboxylated OCN,
which has decreased affinity for calcium ions, reducing the bone
quality (16,17). Due to the effect of vitamin
K2 (VK2) on the γ-carboxylation of OCN, and
other bone metabolism effects, including the stimulation of
osteogenesis and inhibition of adipogenesis (18,19),
VK2 has been used as a treatment for osteoporosis.
As miR-133a is a regulator of osteogenesis, the
present study hypothesized that miR-133a additionally mediates the
effect of VK2 on bone metabolism, functioning in the
following two ways: Direct suppression of mir-133a by
VK2, which further increases Runx2 and promotes
osteogenesis via a positive feedback mechanism, and suppression of
the expression of mir-133a by VK2, resulting in the
upregulation of VKORC1 and γ-carboxylation of OCN.
Materials and methods
Cell culture and treatment
Human BMSCs (hBMSCs) were harvested from the femoral
heads of patients undergoing total hip arthroplasty, as reported in
a previous study (18). Ethical
approval was obtained by the Office of Research Ethics at the
Shanghai Sixth People's Hospital of Shanghai Jiao Tong University
(Shanghai, China). hBMSCs were cultured in complete medium
consisting of alpha minimal essential medium (MEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS, Invitrogen; Thermo Fisher Scientific,
Inc.), 100 units/ml penicillin and 10 µg/ml streptomycin. The
culture medium was replaced with fresh medium every 3 days. Cells
were passaged at ~90% confluence, and cells at passages 3–6 were
used in the experiments. To induce osteogenic differentiation,
hBMSCs were cultured in osteogenic induction medium, which
consisted of complete medium, 10 mM β-sodium glycerophosphate, 50
µg/ml L-ascorbic acid and 10 nM dexamethasone. The following graded
concentrations of VK2: 10−5 M
(VK−5), 10−6 M (VK−6) and
10−7 M (VK−7), were used to treat hBMSCs
(18).
Alizarin red S staining
Following culture in osteogenic medium for 21 days,
hBMSCs were rinsed with PBS, fixed with 4% paraformaldehyde for 20
min and subsequently stained with 40 mM alizarin red working
solution for 10 min. Following this, cells were rinsed twice with
PBS, and images were captured using an inverted phase contrast
microscope.
Transfection assay
hBMSCs were seeded at 105 cells/well in a
6-well plate. After 24 h, the culture medium was replaced with
fresh MEM containing 10% FBS, and 5 µg/ml polybrene (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany), the cells were transduced
with 5×106 TU/ml lentiviral vector (Shanghai GenePharma
Co., Ltd., Shanghai, China) carrying miR-133a (miR-133a-control;
sequence, TTT GGT CCC CTT CAA CCA GCTG) or a miRNA negative control
(miRNA negative; sequence, TTC TCC GAA CGT GTC ACGT) using the four
plasmid system. Briefly, this system involves the co-transfection
of a recombinant virus plasmid encoding for a transfer vector with
three kinds of auxiliary packaging vector into 293T cells. The
supernatant containing the virus particles become concentrated
following 72 h, and then the high-titer lentivirus concentrate is
obtained within 293T cells. The medium was replaced with complete
culture medium 24 h after transduction. The transduction efficiency
was assessed 96 h following transduction and cells were selected
with 2 µg/ml puromycin until colonies of stably transduced cells
were formed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Following culture in osteogenic medium, total RNA
was isolated from cells using TRIzol® reagent; 1 µg RNA
was reverse-transcribed with the EasyScript One-step gDNA Removal
and cDNA Synthesis supermix (Beijing Transgen Biotech Co., Ltd.,
Beijing, China) in a total volume of 20 µl, according to the
manufacturer's protocol. Following this, qPCR was performed using
SYBR® Green reagents (Beijing Transgen Biotech Co.,
Ltd.) for 40 cycles under the following conditions: Denaturation at
95°C for 30 sec, annealing at 95°C for 5 sec and extension at 60°C
for 30 sec. A 65–95°C solubility curve was constructed with ABI
Prism 7900 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
All values were normalized to those of GAPDH, and the fold change
method (2−ΔΔCq) was used for analysis (19). The primers used for the
amplification of target mRNAs are listed in Table I.
 | Table I.The reverse
transcription-quantitative polymerase chain reaction primer
sequences. |
Table I.
The reverse
transcription-quantitative polymerase chain reaction primer
sequences.
| Gene | Forward
sequence | Reverse
sequence |
|---|
| Runx2 |
GCGGTGCAAACTTTCTCCAG |
TGCTTGCAGCCTTAAATGACTC |
| ALP |
GAGAAGCCGGGACACAGTTC |
CCTCCTCAACTGGGATGATGC |
| OCN |
CTCACACTCCTCGCCCTATTG |
GCTTGGACACAAAGGCTGCAC |
| GAPDH |
CCTCGCCTTTGCCGATCC |
ATCATCATCCATGGTGAGCTGG |
For the detection of mir-133a, miRNAs were isolated
using the EasyPure miRNA kit (Beijing Transgen Biotech Co., Ltd.)
according to the manufacturer's protocol. qPCR analysis of mir-133a
was performed using the SYBR Green RT-qPCR kit (BioTNT Co., Ltd.,
Shanghai, China). The stem-loop RT primer for miR-133a was
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGCTGGT-3′, and qPCR
primers were: Forward, 5′-TTTGGTCCCCTTCAAC-3′ and reverse,
5′-TCAACTGGTGTCGTGG-3′. U6 served as a control with primers as
described previously (20). The
thermocycling conditions were as follows: Denaturation at 95°C for
5 min, annealing at 95°C for 5 sec and extension at 60°C for 30
sec, for a total of 40 cycles. A 65–95°C solubility curve was
constructed, and the value of mir-133a expression was normalized to
that of U6 using the 2−ΔΔCq method.
Western blotting
Total protein extracted with Cell Lysis buffer
(#9803; Cell Signaling Technology, Inc., Danvers, MA, USA) from
osteogenic-induced hBMSCs was quantified using a Bicinchoninic Acid
(BCA) kit (Thermo Fisher Scientific, Inc.). Following this, total
protein was denatured by heating at 95°C for 5 min, mixed with
loading buffer, and 30 µg proteins were separated by SDS-PAGE on
10% acrylamide gels. Proteins were subsequently transferred onto
polyvinylidene difluoride membranes. The membranes were blocked
with 5% dried skimmed milk, and incubated with rabbit anti-human
GAPDH (1:2,000; #2118; Cell Signaling Technology, Inc., Danvers,
MA, USA) and rabbit anti-human Runx2 (1:1,000; ab48811; Abcam,
Cambridge, MA, USA) antibodies at 4°C overnight and subsequently
incubated with a horseradish peroxidase-conjugated anti-rabbit
secondary antibody (1:2,000; #7074; Cell Signaling Technology,
Inc.) at 37°C for 1 h. Following chemiluminescence with a
commercial assay (Pierce™ ECL Western Blotting substrate, #32106,
Thermo Fisher Scientific, Inc.), protein bands were visualized by
Odyssey scanner (LI-COR Biosciences, Lincoln, NE, USA). The bands
were quantified using Quantity One software (version 4.52; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and normalized to GAPDH.
Alkaline phosphatase (ALP) activity
and staining
hBMSCs were lysed with 0.2% Triton-X 100 and
centrifuged at 14,000 × g for 10 min at 4°C,, and the supernatant
was detected with an ALP assay kit (Nanjing Jiancheng
Bioengineering Institute, Nanjing, China) according to the
manufacturer's protocol. Absorbance values were measured at a
wavelength of 520 nm using the iMark Microplate Absorbance reader
(Bio-Rad Laboratories, Inc.) and normalized against the protein
concentration determined with the use of a BCA kit.
ALP staining was performed using a 5-Bromo-
4-chloro-3-indolyl Phosphate/Nitro Blue Tetrazolium ALP Color
Development kit (Beyotime Institute of Biotechnology, Shanghai,
China) according to the manufacturer's protocol. Briefly, cells
were rinsed with PBS, fixed with 4% paraformaldehyde for 20 min and
stained with the ALP Color Development kit for 30 min. Images were
captured using an inverted phase contrast microscope.
OCN detection by ELISA
Following incubation of cells for 1 week, the
osteogenic medium was removed and the cultures were incubated with
MEM for 24 h. Concentrations of OCN in the medium were determined
using an ELISA kit (#MK128, Takara Bio, Inc., Otsu, Japan). The
values were normalized against the total protein concentration
determined using a BCA kit.
Statistical analysis
SPSS software (version, 20.0; IBM SPSS, Armonk, NY,
USA) was used to analyze the values in each group. All experiments
were performed in triplicate, and data are expressed as the mean ±
standard deviation. One-way analysis of variance followed by
Tukey's post hoc test was performed to determine statistical
significance. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-133a expression levels are reduced
during the osteogenic differentiation of hBMSCs
To detect miR-133a expression during osteogenic
differentiation, RT-qPCR was performed on samples collected at days
0, 1, 3, 7, 14 of differentiation. Osteogenic differentiation was
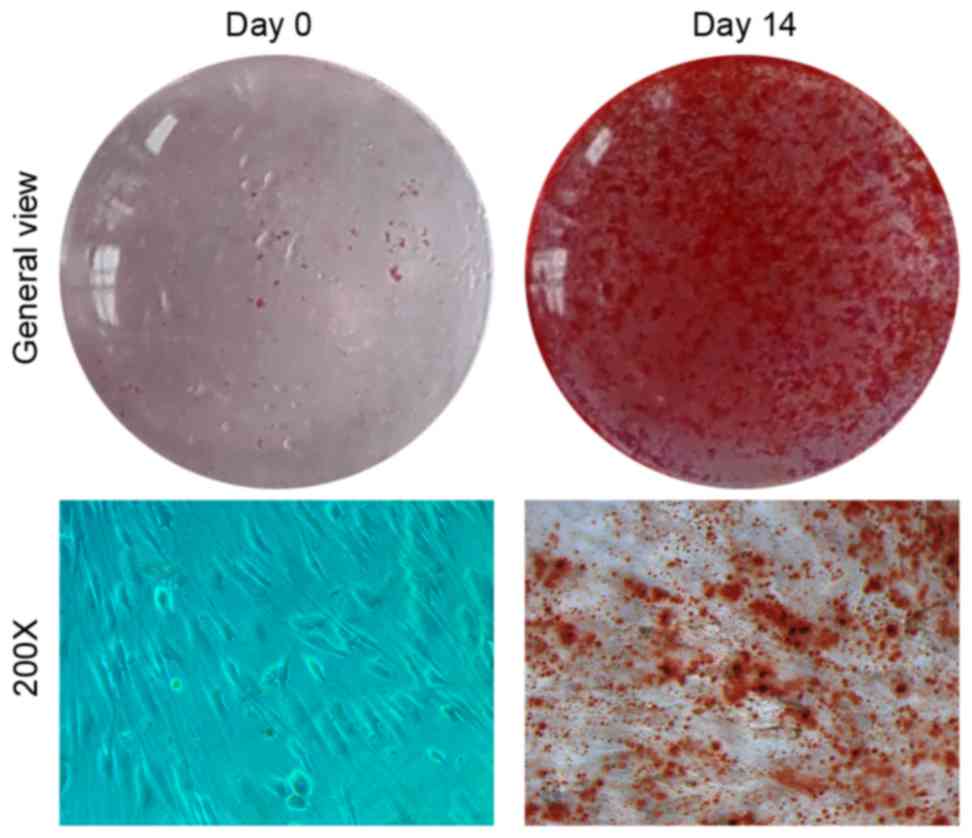
confirmed by Alizarin red S staining (Fig. 1). Compared with undifferentiated
cells at day 0, miR-133a expression levels rapidly declined during
osteogenic induction to 0.05-fold after 14 days (P<0.05;
Fig. 2A). By contrast, the
expression levels of Runx2 (Fig.
2B), ALP (Fig. 2C) and OCN
(Fig. 2D) mRNA steadily increased
in a time-dependent manner, exhibiting a negative correlation with
miR-133a expression levels. In addition, the expression of VKORC1
mRNA (Fig. 2E) was inversely
correlated with miR-133a expression, as previously reported
(14), increasing 13-fold in
osteogenic-induced hBMSCs at day 14. These results suggested that
the osteogenic differentiation of BMSCs was associated with a
reduction in miR-133a expression levels, indicating that miR-133a
may be associated with the osteoblast differentiation of
mesenchymal cells.
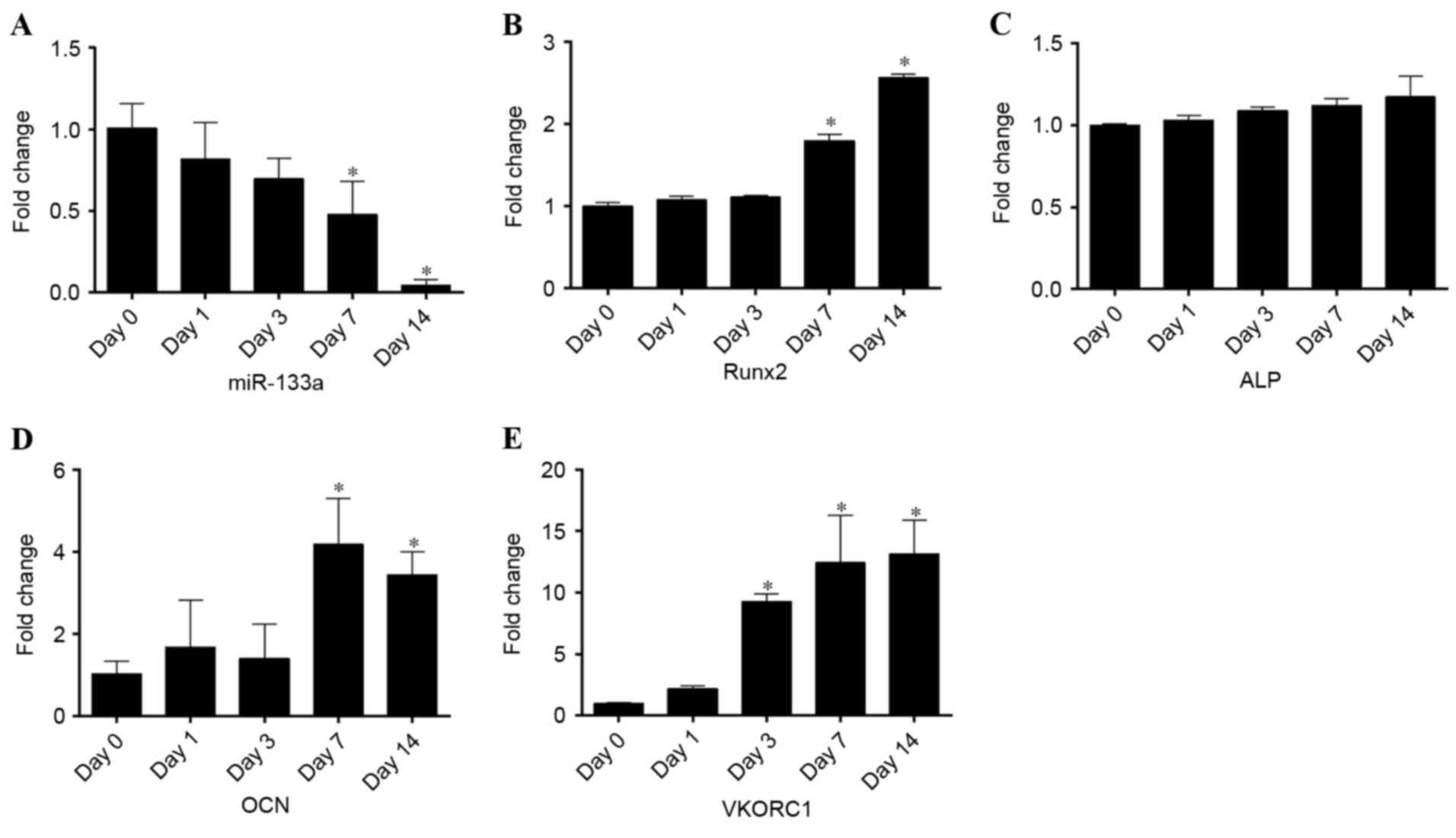 | Figure 2.Expression levels of miR-133a and
osteoblastic markers during the osteogenic differentiation of human
bone marrow stromal cells, as assessed by reverse
transcription-quantitative polymerase chain reaction. Expression
levels of (A) miR-133a, (B) Runx2, (C) ALP, (D) OCN and (E) VKORC1
at days 0, 1, 3, 7 and 14 of osteogenic differentiation. Data are
expressed as the mean ± standard deviation (n=3). *P<0.05 vs.
day 0. miR, microRNA; Runx2, runt-related transcription factor 2;
ALP, alkaline phosphatase; OCN, osteocalcin; VKORC1, vitamin K
epoxide reductase complex subunit 1. |
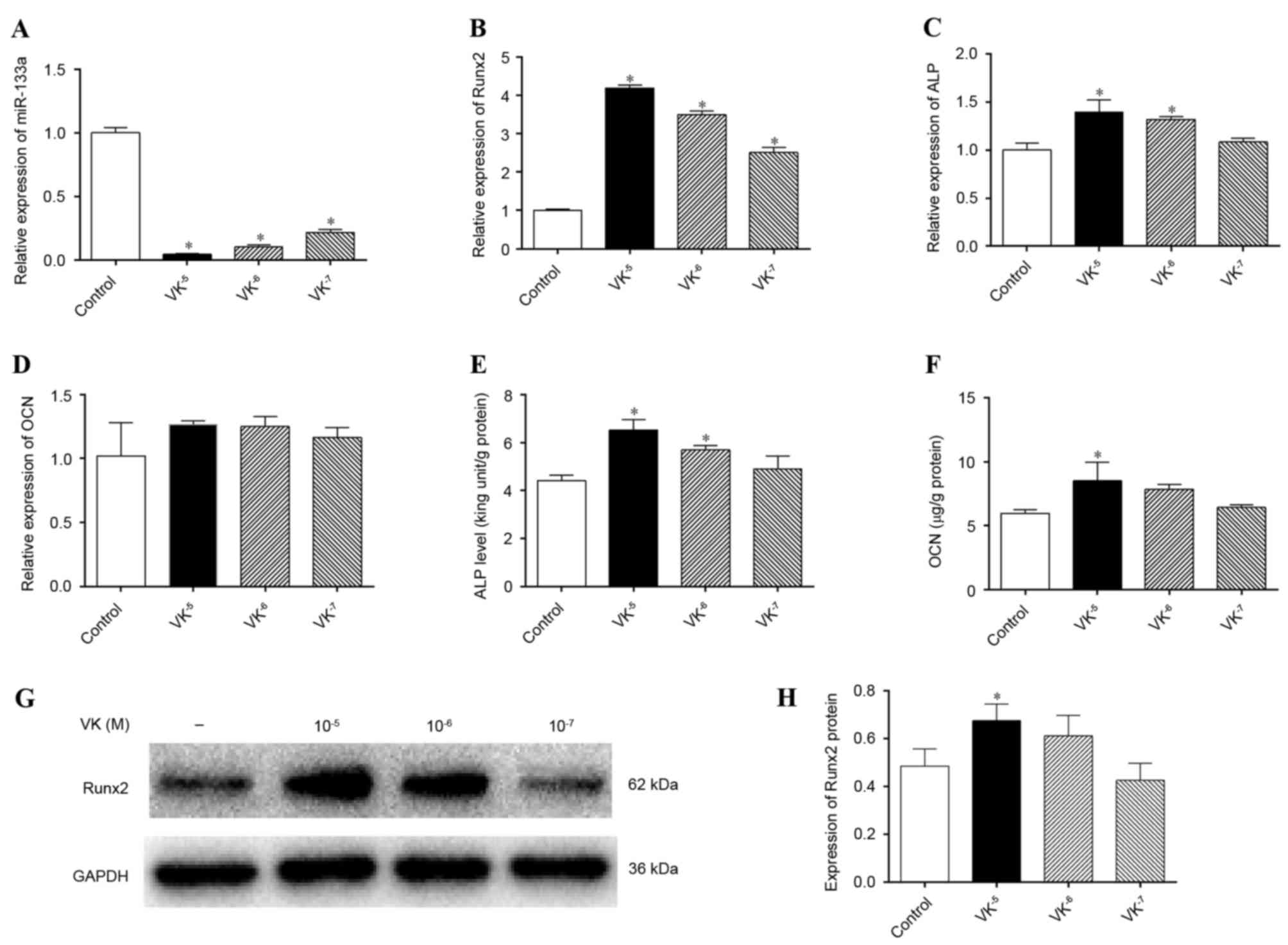
VK2 inhibits miRNA-133a expression and
promotes the osteogenic differentiation of hBMSCs
To evaluate the association between miR-133a and
VK2, osteogenic induction of hBMSCs was performed for 1
week in the presence or absence of gradient concentrations of
VK2, and miR-133a expression levels were detected by
RT-qPCR. miR-133a expression levels were markedly downregulated by
VK2 treatment compared with the control group
(P<0.05), to the greatest extent by 10−5 M
VK2 (Fig. 3A). In
addition, mRNA expression levels of Runx2 (Fig. 3B) and ALP (Fig. 3C) were significantly upregulated by
VK2 treatment (P<0.05), whereas those of OCN were not
(P>0.05) (Fig. 3D). ALP and OCN
are two late-stage osteogenic differentiation markers of
mesenchymal cells, and were therefore further analyzed. Greater ALP
activity was detected in cells treated with 10−5 M and
10−6 M VK2 compared with the control
(P<0.05; Fig. 3E). OCN
concentrations in supernatant were increased by VK2
treatment, particularly by 10−5 M VK2
(P<0.05; Fig. 3F), despite the
lack of effect on OCN mRNA expression levels. Detection of Runx2
protein expression levels by western blotting (Fig. 3G) further verified the osteogenic
stimulation effect of VK2 treatment, revealing greater
Runx2 protein expression in VK2-treated cells
(P<0.05), except at a concentration of 10−7 M
VK2, although not significant for 10−6 M
VK2 (P>0.05) (Fig.
3H). Although the western blotting results were inconsistent
with the Runx2 mRNA analyses, which may be associated with the time
interval existing between gene and protein expression, these
results indicated that VK2 treatment promoted osteogenic
differentiation of hBMSCs, as previously described (18,21),
and that the stimulation of osteogenesis may be associated with the
inhibition of miR-133a.
Osteogenic differentiation is
regulated by miR-133a, and VK2 stimulates osteogenesis by
downregulating miR-133a expression
To examine whether miR-133a directly regulated the
osteogenic differentiation of hBMSCs, miR-133a was overexpressed in
hBMSCs via transfection with a lentiviral vector. Compared with
cells transfected with the miR-negative control, hBMSCs transfected
with miR-133a expressed significantly greater levels of miR-133a,
as demonstrated by RT-qPCR (P<0.05; Fig. 4A). To investigate the effect of
miR-133a overexpression, the mRNA expression levels of ALP, OCN and
Runx2, ALP activity, OCN secretion, and Runx2 protein expression
levels were measured in miR-133a-overexpressing hBMSCs incubated
with osteogenic induction medium for 7 days. mRNA expression levels
of ALP (P>0.05; Fig. 4B), OCN
(P>0.05; Fig. 4C) and Runx2
(P<0.05; Fig. 4D) were
decreased in miR-133a-overexpressing cells compared with control
cells, as were ALP activity (P<0.05; Fig. 4E), OCN secretion (P<0.05;
Fig. 4F), ALP staining (Fig. 4G) and Runx2 protein expression
levels (P<0.05; Fig. 4H and I),
indicating that miR-133a overexpression inhibited the osteogenic
differentiation of hBMSCs.
The effect of VK2 treatment on
miR-133a-overexpressing cells was additionally determined. After 7
days of osteogenic induction, VK2 treatment
significantly downregulated miR-133a expression levels in
miR-133a-overexpressing hBMSCs (P<0.05; Fig. 4A); the mRNA expression levels of
ALP (Fig. 4B), OCN
(Fig. 4C) and Runx2
(Fig. 4D) were upregulated by
VK2 treatment. Furthermore, greater ALP activity
(Fig. 4E) and increased
supernatant OCN concentrations (Fig.
4F), ALP-positive cells (Fig.
4G) and Runx2 protein expression levels (Fig. 4H and I) were detected in
VK2-treated miR-133a-overexpressed cells compared with
control cells.
VK2 upregulates VKORC1 expression
accompanied by the inhibition of miR-133a
VKORC1 is a key protein involved in the VK cycle,
where VK2 functions to promote the γ-carboxylation of
OCN during bone metabolism. The results of a previous (15) and the present study demonstrated
that miR-133a expression levels are inversely correlated with
VKORC1 mRNA expression levels. To investigate whether
VK2 affects the expression of VKORC1 via miR-133a,
osteogenic induction of hBMSCs was performed for 1 week in the
absence or presence of gradient concentrations of VK2,
and VKORC1 mRNA expression levels were detected by RT-qPCR. VKORC1
mRNA expression levels were significantly upregulated by
VK2 (P<0.05), particularly at a concentration of
10−5 M, where levels were 22-fold of those of the
control (Fig. 5A). An inverse
correlation between miR-133a and VKORC1 was demonstrated, even in
the presence of VK2. In addition, VKORC1 mRNA expression
levels were downregulated in miR-133a-overexpressing cells compared
with miR-negative control cells (P<0.05), and VK2
significantly increased VKORC1 mRNA expression levels in
miR-133a-overexpressing hBMSCs (P<0.05; Fig. 5B), which further demonstrated that
VK2 positively regulated VKORC1 by downregulating
miR-133a expression.
Discussion
miR-133a negatively regulates osteoblastogenensis of
mesenchymal stem cells by targeting Runx2, except in muscle
(2,11). The present study further
demonstrated that miR-133a expression in hBMSCs and overexpression
of miR-133a inhibited osteogenic differentiation in hBMSCs, using
miR-133a transfection. Furthermore, the present study revealed that
miR-133a may be a target of VK2, which promotes the
osteogenic differentiation of mesenchymal stem cells.
Human miR-133a is encoded by two miRNAs: miR-133a-1
and miR-133a-2, and the sequence of mature miR-133a-1 is identical
to that of mature miR-133a-2 (13,22).
miR-133a has been reported to serve an important role in skeletal
and cardiac muscle development; however, a role for miR-133a in
osteogenesis has additionally been described. Zhang et al
(11) reported that miR-133a
significantly inhibited osteoblast differentiation by targeting the
transcription factor Runx2. Furthermore, Liao et al
(22) demonstrated that miR-133a
inversely modulated the osteogenic differentiation of vascular
smooth muscle cells. An in vivo study has suggested miR-133a
in circulating monocytes as a potential biomarker for
postmenopausal osteoporosis, as miR-133a is upregulated in
postmenopausal women with low bone mineral density (13). A study reported that miR-133a was
significantly downregulated in the non-traumatic osteonecrosis of
femoral head samples, which failed to form bone (14). In the present study, miR-133a
expression levels declined significantly during osteogenic
differentiation and were inversely correlated with the expression
of osteogenic-associated proteins. Additionally, the overexpression
of miR-133a clearly inhibited the osteogenic differentiation of
hBMSCs. These results suggested that miR-133a acts as a negative
regulator of osteogenesis.
Vitamin K2 has been reported to stimulate
osteoblastogenesis of hBMSCs (17), except when it acts as a co-factor
of the γ-carboxylation of VK-dependent proteins. In addition,
VK2 has been revealed to activate the transcription of
extracellular matrix by activating the steroid xenobiotic receptor
(23). However, the mechanism
underlying the effects of VK2 in bone metabolism remains
to be described. The present study demonstrated that VK2
significantly inhibited the expression of miR-133a, in control and
miR-133a-overexpressing hBMSCs, suggesting that the promotion of
osteogenesis by VK2 may involve targeting miR-133a.
Runx2 is an early osteogenic marker that regulates
BMSC shape and induces osteoblast differentiation. Previous studies
have revealed that miR-133a targeted and decreased the expression
levels of Runx2 to inhibit osteoblast differentiation (11,24).
The inverse correlation between Runx2 and miR-133a, particularly in
cells overexpressing miR-133a, was additionally observed in the
present study. ALP transcription is mediated by Runx2 and acts as a
mature marker of osteogenesis (25,26);
its expression and activity is regulated by miR-133a, as confirmed
by the present study. OCN is a characteristic osteogenic lineage
maker that is preferentially expressed in mature osteoblasts. There
are Runx2 sites in the promoter of OCN, hence the involvement of
Runx2 in the transcription of OCN. (17,27).
The results of the present study revealed that OCN expression is
downregulated by miR-133a and upregulated by VK2, which
inhibited miR-133a expression. However, no alterations in OCN mRNA
expression levels were observed, whereas OCN protein expression
levels altered significantly, following VK2 treatment;
this may be due to the time interval between gene and protein
expression or detection error.
The VK-epoxide cycle, which is closely associated
with γ-carboxylation, has two activities, a VK 2,3-epoxide
reductase (VKOR) activity that converts the metabolite VK
2,3-epoxide into the native quinone form and a VKOR or nicotinamide
adenine dinucleotide (phosphate)-dependent activity that converts
VK quinone to KH2, which acts as the co-factor of carboxylase
(28). Previous studies have
revealed that VK2 stimulated osteoblastogenesis of
mesenchymal stem cells via γ-carboxylation-dependent and
-independent underlying mechanisms (18,29),
of which the γ-carboxylation of OCN was demonstrated to be
important. VKORC1 is a key enzyme involved in the VK cycle, which
is required for VK recycling and as a co-factor for γ-glutamyl
carboxylase; miR-133a has been revealed to directly downregulate
the expression of VKORC1 in humans (15). The present study observed that
VKORC1 mRNA expression levels correlated inversely with miR-133a
expression levels, which further demonstrated the regulatory effect
of miR-133a on VKORC1 expression. In addition, VK2
treatment upregulated VKORC1 expression levels; this was
accompanied by the downregulation of miR-133a expression,
indicating a possible positive feedback loop that acts via miR-133a
in the VK cycle.
In conclusion, the results of the present study
demonstrated that miR-133a inhibited osteogenic differentiation and
VKORC1 expression in hBMSCs, whereas VK2 inhibited
miR-133a expression accompanied by the stimulation of osteogenesis
and VKORC1 expression, indicating that miR-133a may participate in
the regulation of bone metabolism by VK2.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81301572) and the
SMC-Chen Xing Plan for Splendid Young Teachers of Shanghai Jiao
Tong University. The authors thank the Shanghai Institute of
Microsurgery for Extremities for providing experimental equipment
and all staff for their assistance.
References
|
1
|
Wang L, Li X, Zhou Y, Shi H, Xu C, He H,
Wang S, Xiong X, Zhang Y, Du Z, et al: Downregulation of miR-133
via MAPK/ERK signaling pathway involved in nicotine-induced
cardiomyocyte apoptosis. Naunyn Schmiedebergs Arch Pharmacol.
387:197–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Rooij E, Liu N and Olson EN: MicroRNAs
flex their muscles. Trends Genet. 24:159–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu M and Wang YZ: miR133a suppresses cell
proliferation, migration and invasion in human lung cancer by
targeting MMP-14. Oncol Rep. 30:1398–1404. 2013.PubMed/NCBI
|
|
5
|
Fan J, Li J and Fan Q: Naringin promotes
differentiation of bone marrow stem cells into osteoblasts by
upregulating the expression levels of microRNA-20a and
downregulating the expression levels of PPARγ. Mol Med Rep.
12:4759–4765. 2015.PubMed/NCBI
|
|
6
|
Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G,
Li Z, Peng J, Wang P, Shen C, et al: microRNA-103a functions as a
mechanosensitive microRNA to inhibit bone formation through
targeting Runx2. J Bone Miner Res. 30:330–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li H, Yang F, Wang Z, Fu Q and Liang A:
MicroRNA-21 promotes osteogenic differentiation by targeting small
mothers against decapentaplegic 7. Mol Med Rep. 12:1561–1567.
2015.PubMed/NCBI
|
|
8
|
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q,
Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, et al: MicroRNA-188
regulates age-related switch between osteoblast and adipocyte
differentiation. J Clin Invest. 125:1509–1522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu N and Olson EN: MicroRNA regulatory
networks in cardiovascular development. Dev Cell. 18:510–525. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou Q, Zhao ZN, Cheng JT, Zhang B, Xu J,
Huang F, Zhao RN and Chen YJ: Ibandronate promotes osteogenic
differentiation of periodontal ligament stem cells by regulating
the expression of microRNAs. Biochem Biophys Res Commun.
404:127–132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Xie RL, Croce CM, Stein JL, Lian
JB, van Wijnen AJ and Stein GS: A program of microRNAs controls
osteogenic lineage progression by targeting transcription factor
Runx2. Proc Natl Acad Sci USA. 108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Xie RL, Gordon J, LeBlanc K,
Stein JL, Lian JB, van Wijnen AJ and Stein GS: Control of
mesenchymal lineage progression by microRNAs targeting skeletal
gene regulators Trps1 and Runx2. J Biol Chem. 287:21926–21935.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Li L, Moore BT, Peng XH, Fang X,
Lappe JM, Recker RR and Xiao P: MiR-133a in human circulating
monocytes: A potential biomarker associated with postmenopausal
osteoporosis. PLoS One. 7:e346412012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu X, Zhang Y, Guo X, Xu H, Xu Z, Duan D
and Wang K: Identification of differentially expressed microRNAs
involved in non-traumatic osteonecrosis through microRNA expression
profiling. Gene. 565:22–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Perez-Andreu V, Teruel R, Corral J, Roldán
V, García-Barberá N, Salloum-Asfar S, Gómez-Lechón MJ, Bourgeois S,
Deloukas P, Wadelius M, et al: miR-133a regulates vitamin K
2,3-epoxide reductase complex subunit 1 (VKORC1), a key protein in
the vitamin K cycle. Mol Med. 18:1466–1472. 2013.PubMed/NCBI
|
|
16
|
Hamidi MS, Gajic-Veljanoski O and Cheung
AM: Vitamin K and bone health. J Clin Densitom. 16:409–413. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Neve A, Corrado A and Cantatore FP:
Osteocalcin: Skeletal and extra-skeletal effects. J Cell Physiol.
228:1149–1153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koshihara Y, Hoshi K, Okawara R, Ishibashi
H and Yamamoto S: Vitamin K stimulates osteoblastogenesis and
inhibits osteoclastogenesis in human bone marrow cell culture. J
Endocrinol. 176:339–348. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takeuchi Y, Suzawa M, Fukumoto S and
Fujita T: Vitamin K(2) inhibits adipogenesis, osteoclastogenesis,
and ODF/RANK ligand expression in murine bone marrow cell cultures.
Bone. 27:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koshihara Y and Hoshi K: Vitamin K2
enhances osteocalcin accumulation in the extracellular matrix of
human osteoblasts in vitro. J Bone Miner Res. 12:431–438. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao XB, Zhang ZY, Yuan K, Liu Y, Feng X,
Cui RR, Hu YR, Yuan ZS, Gu L, Li SJ, et al: MiR-133a modulates
osteogenic differentiation of vascular smooth muscle cells.
Endocrinology. 154:3344–3352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ichikawa T, Horie-Inoue K, Ikeda K,
Blumberg B and Inoue S: Steroid and xenobiotic receptor SXR
mediates vitamin K2-activated transcription of extracellular
matrix-related genes and collagen accumulation in osteoblastic
cells. J Biol Chem. 281:16927–16934. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shah A and Ahmad A: Role of microRNA in
mesenchymal stem cells differentiation into osteoblasts. Razavi Int
J Med. 1:5–10. 2013. View Article : Google Scholar
|
|
25
|
Hu N, Feng C, Jiang Y, Miao Q and Liu H:
Regulative Effect of Mir-205 on osteogenic differentiation of bone
mesenchymal stem cells (BMSCs): Possible role of SATB2/Runx2 and
ERK/MAPK pathway. Int J Mol Sci. 16:10491–10506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koromila T, Baniwal SK, Song YS, Martin A,
Xiong J and Frenkel B: Glucocorticoids antagonize RUNX2 during
osteoblast differentiation in cultures of ST2 pluripotent
mesenchymal cells. J Cell Biochem. 115:27–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma XL, Liu ZP, Ma JX, Han C and Zang JC:
Dynamic expression of Runx2, Osterix and AJ18 in the femoral head
of steroid-induced osteonecrosis in rats. Orthop Surg. 2:278–284.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shearer MJ and Newman P: Recent trends in
the metabolism and cell biology of vitamin K with special reference
to vitamin K cycling and MK-4 biosynthesis. J Lipid Res.
55:345–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Atkins GJ, Welldon KJ, Wijenayaka AR,
Bonewald LF and Findlay DM: Vitamin K promotes mineralization,
osteoblast-to-osteocyte transition, and an anticatabolic phenotype
by {gamma}-carboxylation-dependent and-independent mechanisms. Am J
Physiol Cell Physiol. 297:C1358–C1367. 2009. View Article : Google Scholar : PubMed/NCBI
|