Introduction
Articular cartilage has a poor regenerative capacity
which often results in joint osteoarthritis, with cartilage
degradation as the primary characteristic. Currently available
treatments for osteoarthritis yield variable outcomes (1) and for this reason novel treatments
are required. One promising alternative is regenerative medicine
using stem cells, producing cells that it is possible to
differentiate into articular chondrocytes to replenish the damaged
cartilage. Initially, researchers focused on somatic stem cells,
including bone marrow mesenchymal stem cells, which were considered
a potent cell source for cartilage repair (2,3).
However, their limited proliferative potential made them unsuitable
for cartilage regeneration. The search for alternative approaches
led to the development of human induced pluripotent stem cells
(hiPSCs), which have unrestricted proliferative activity and
pluripotency. As hiPSCs are developed from adult human cells, they
are free from the ethical concerns associated with human embryonic
stem cells (hESCs) (4). HiPSCs are
formed by inducing a pluripotent state, usually achieved by
overexpression of transcription factors (known as Yamanaka factors)
and proteins with varied cellular functions (including RNA-binding
protein Lin28) (5,6).
There are numerous techniques used for chondrogenic
differentiation of hiPSCs, including micromass culture, directed
differentiation, pellet culture and formation of embryoid bodies
(EBs) (7–10). Of these methods, the most common
and efficient method of obtaining chondrocyte-like cells from
hiPSCs is EB formation. However, multiple aspects of this process
remain poorly understood, including how the specific medium used
for chondrogenic differentiation affects gene expression (11). Likewise, although a wide range of
markers are used to assess cell differentiation during the
differentiation process, the relative utility of these markers is
not well understood (12). As a
result, it is difficult to select the optimal medium and markers to
achieve optimal cell yield.
It was within this context that the present study
was conducted. The present study has two main aims: To determine
the gene expression profile of chondrogenic-like cells derived from
hiPSCs cultured in medium conditioned with HC-402-05a cells or
supplemented with transforming growth factor β3 (TGF-β3), and to
determine the relative value of the most commonly used chondrogenic
markers as indicators of cell differentiation. The cells were
differentiated in chondrogenic mediums supplemented with either
TGF-β3, the member of the TGF-β superfamily with the most
chondrogenic potential (13) or
conditioned with growth factors from the human primary chondrocyte
cell line (HC-402-05a). The gene expression profiles of the
chondrogenic-like cells derived from the hiPSCs cultured in the
TGF-β-supplemented medium (TGF-β3 medium) was then assessed and
compared with the cells cultured in the HC-402-05a-conditioned
medium (conditioned medium). The type of medium was demonstrated to
have a large impact on the gene expression profiles. A total of 22
different markers of chondrogenic differentiation were also
evaluated, and the most promising gene markers of hiPSC
differentiation during late stage chondrogenesis were runt-related
transcription factor 2 (RUNX2), matrix
metalloproteinase-13 (MMP-13), and vascular endothelial
growth factor (VEGF), which engaged in the formation of
hypertrophic chondrocytes during skeletal development. Useful,
however less valuable markers included collagens and members of the
TGF-β superfamily, which serve functions in several stages of
chondrogenesis. Furthermore, the shared mesodermal origin of
fibroblasts and chondrocytes should be taken into consideration, as
several genes are common between stem cell-derived chondrocytes and
human fibroblasts, including SMAD family member 3 (SMAD3)
and bone morphogenetic protein-2 (BMP-2), decreasing their
utility in the evaluation of chondrogenic process in
vitro.
Cells differentiated in the conditioned medium were
demonstrated to present features that were characteristic of mature
chondrocytes. In contrast, cells cultured in the presence of TGF-β3
presented characteristics of hypertrophic chondrocytes, which may
result in a decreased capacity to repair and regenerate articular
cartilage and impaired viscoelastic properties compared with normal
chondrocytes. Consequently, the HC-402-05a-conditioned medium
offers greater potential for in vitro chondrogenesis. The
present study contributes to an improved understanding of the
changes in gene expression during the in vitro chondrogenic
process and the short-term culture of stem-derived chondrocytes, in
addition to clarifying the relative value of a wide range of
chondrogenic differentiation markers.
This is a two-part study. The first part of the
study (14) described markers
characteristic for the pluripotent state and early and advanced
stage chondrogenesis. Part B, presented here, focuses on markers
that are characteristic of late stage chondrogenesis, hypertrophy,
and ossification (Table I).
 | Table I.Analysis of the usefulness of
selected markers for advanced hiPSC chondrogenic differentiation
in vitro. |
Table I.
Analysis of the usefulness of
selected markers for advanced hiPSC chondrogenic differentiation
in vitro.
| Marker | Function of marker
(stage of presentation) | Influence on
chondrogenesis: Positive (+) or negative (−) | The usefulness of
marker in evaluation of chondrogenic progression (+, ++, +++) |
|---|
| TGF-βIR,
TGF-βIIR, | Pluripotency,
chondrogenesis, ossification, osteoarthritis | −/+ | + |
| TGF-βIIIR, |
|
|
|
| TGF-β2, -β3 |
Chondrocytes/hypertrophic
chondrocytes/osteoblasts | + | ++ |
| BMP-2, BMP-4 |
Chondrocytes/hypertrophic
chondrocytes/osteoblasts | + | ++ |
| GDF-5 |
Chondrocytes/hypertrophic
chondrocytes/osteoblasts | + | ++ |
| SMAD3 | Chondrocytes | + | ++ |
| TYPE I
COLLAGEN | Dedifferentiated
chondrocytes | − | ++ |
| TYPE II
COLLAGEN |
Chondroprogenitors/mature
chondrocytes | + | ++ |
| TYPE X
COLLAGEN | Hypertrophic
chondrocytes/endochondral ossification | − | ++ |
| TYPE XI
COLLAGEN | Mature
chondrocytes | + | ++ |
| IHH | Hypertrophic
chondrocytes | + | ++ |
| PTHLH |
Chondrocytes-prevents from
hypertrophy | − | ++ |
| PTCH1 |
Proliferating/hypertrophic
chondrocytes | −/+ | ++ |
| RUNX2 |
Chondrocytes/hypertrophic
chondrocytes/osteoblasts | − | ++ |
| CH13L1 | OA
chondrocytes | − | +++ |
| MMP-2 | Chondrocytes | + | ++ |
| MMP-13 | Hypertrophic
chondrocytes/OA chondrocytes/endochondral bone formation | − | ++ |
| ALPL | Maintenance of
pluripotency/hypertrophic chondrocytes/OA chondrocytes | − | + |
| VEGF | Angiogenesis | − | +++ |
Materials and methods
Culturing human induced pluripotent
stem cells
The hiPSCs obtained during the reprogramming process
as previously described (15) were
seeded on 10 cm Petri dishes in Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA) which had previously been coated with inactivated
murine embryonic fibroblasts as a feeder layer (1×106).
Following 24 h preparation of the feeder layer, hiPSCs were seeded
at 2×106 in hiPSC growth medium: Dulbecco's modified
Eagle's medium (DMEM) F12 with L-glutamine (Merck Millipore,
Darmstadt, Germany), 20% knockout serum replacement (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 1% non-essential amino acids
(Merck Millipore), 0.1 mM β-mercaptoethanol (Merck Millipore), 1%
penicillin/streptomycin (P/S; Merck Millipore). Prior to use, the
medium was supplemented with fibroblast growth factor 2 (FGF-2; 10
ng/ml; Merck Millipore). The complete hiPSC growth medium was
supplemented with ciprofloxacin (0.5 µg/ml; Sigma Aldrich; Merck
Millipore) to avoid Mycoplasma spp. contamination for the first 7
days of culture. The culture medium was changed daily.
EB formation
At 80% confluency, hiPSC colonies were passaged and
dissociated into clumps with 0.1% collagenase IV solution (Thermo
Fisher Scientific, Inc.). The cells were centrifuged (300 × g, 5
min, room temperature) in order to remove the collagenase and
transferred into non-adherent 96-well plates (1,000 cells per well;
Brand GmbH, Wertheim, Germany) in EB growth medium, which is a
hiPSC growth medium without FGF-2. EBs formed within 24 h and were
observed as free-floating aggregates. The culture medium was
changed every 48 h. On day 7 the EBs were used for chondrogenic
differentiation.
Chondrogenesis in vitro
A standard chondrogenic medium was used: DMEM F12
with L-glutamine (Merck Millipore), 10% fetal bovine serum (FBS;
Biowest, Nuaillé, France), 50 µM L-proline (Sigma Aldrich; Merck
Millipore), 50 µM ascorbic acid (Sigma Aldrich; Merck Millipore), 1
mM sodium pyruvate (Biowest), 1% ITS + Premix (Corning Life
Sciences, Big Flats, NY, USA), 1% P/S (Merck Millipore) and
10−7 M dexamethasone (Sigma Aldrich; Merck
Millipore).
Medium conditioning
Standard chondrogenic medium was used: DMEM F12 with
L-glutamine (Merck Millipore), 10% FBS (Biowest), 50 µM L-proline
(Sigma Aldrich; Merck Millipore), 50 µM ascorbic acid (Sigma
Aldrich; Merck Millipore), 1 mM sodium pyruvate (Biowest), 1% ITS +
Premix (Corning, Life Sciences), 1% P/S (Merck Millipore) and
10−7 M dexamethasone (Sigma Aldrich; Merck Millipore),
which was conditioned on the HC-402-05a cell line (up to 3
passages). Medium was collected following 24 h conditioning and
administered to the differentiated EBs.
Chondrogenesis using EBs
The mature EBs were transferred onto 6-well plates
(10 EBs per well) previously coated with 0.1% gelatin (Merck
Millipore) and allowed to adhere for the next 24 h, following which
the medium was replaced with a chondrogenic medium This was either
supplemented with TGF-β3 (10 ng/ml; ImmunoTools GmbH, Friesoythe,
Germany), as a growth factor with the most chondrogenic potential,
or conditioned with the HC-402-05a cell line as above. The positive
influence of standard chondrogenic medium with the addition of
exogenous TGF-β3 (10 ng/ml) on pluripotent stem cells was
previously tested and confirmed by our group (16). The chondrogenic medium was changed
every 48 h. The culture period lasted 21 days. In order to confirm
that chondrocyte-like cells had been obtained, immunofluorescence
analysis was performed on passage 0 (p0). Subsequently, to evaluate
the expression profile of chondrogenic markers (p3), reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis was performed (14). In
all analyses, the stable adult human articular chondrocyte cell
line (HC-402-05a) served as a positive control, as the European
Collection of Authenticated Cell Cultures recommended it for the
evaluation of the differentiation process in in vitro model
systems.
Culture of differentiated cells
The derived stem cells were cultured in 0.1% gelatin
(Merck Millipore) in DMEM F12 with L-glutamine (Merck Millipore),
10% FBS (Biowest), and 1% P/S (Merck Millipore) up to 3
passages.
RT-qPCR
Total RNA was extracted from cells (p3;
2×106 cells) with TRIzol (Sigma Aldrich; Merck
Millipore). Total RNA (1 µg per 20 µl reaction volume) free of
genomic DNA contamination was reverse-transcribed using the
iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) according to the manufacturer's protocol (25°C for 5 min,
42°C for 30 min, 85°C for 5 min). qPCR reactions were performed
using the LightCycler 480 Probes Master mix and appropriate probes
labeled with fluorescein for each primer (Roche Diagnostics, Basel,
Switzerland). The reaction conditions for all amplicons were as
follows: Initially 95°C for 10 min, followed by 45 cycles at 94°C
for 10 sec, 60°C for 15 sec and 72°C for 1 sec. All reactions were
performed in the presence of 3.2 mM MgCl2. cDNA samples
(2.5 µl for a total volume of 10 µl) were analyzed for genes of
interest and for the reference gene glyceraldehyde 3-phosphate
dehydrogenase, which were selected based on the latest literature
data concerning chondrogenic differentiation of hiPSCs (17). The level of expression of each
target gene was calculated as −2ΔΔCq (18). The reaction was performed in
triplicate for genes of interest: TGF-β receptor 1 (TGF-βIR),
TGF-βIIR, TGF-βIIIR, TGF-β2, TGF-β3, BMP-2, BMP-4, growth
differentiation factor 5 (GDF-5), SMAD3, type I collagen, type II
collagen, type XI collagen, Indian hedgehog (IHH), parathyroid
hormone-like hormone (PTHLH), patched 1 (PTCH1), RUNX2,
chitanise-3-like protein (CH13L1), matrix metalloproteinase 2
(MMP-2), MMP-13, alkaline phosphatase (ALPL), VEGF. Primer
information is available upon request.
Statistical analysis
All experiments were performed a minimum of three
times. The results are reported as the mean ± standard deviation.
Comparisons between the study groups and controls were performed
using one-way analysis of variance. Where the analysis of variance
results were significant, post hoc analysis was performed via
Tukey's multiple comparison test with a single pooled variance.
Statistical tests were performed with GraphPad Prism (version 5.0a;
GraphPad Software, Inc., San Diego, CA, USA). *P<0.05 was
considered to indicate a statistically significant difference.
Results
Gene expression profiles of stem
cell-derived chondrocytes revealed the presence of receptors and
members of TGF-β superfamily
Immunofluorescence analysis confirmed that
chondrocyte-like cells were obtained (14). The presence of the following TGF-β
receptors in the cultured cells was determined: TGF-βRI, TGF-βRII,
and TGF-βRIII. The presence of members of the TGF-β superfamily was
also observed, as follows: TGF-β2, TGF-β3, BMP-2, BMP-4, and
GDF-5.
TGF-βRI was expressed by all the cells, but
most prominently by cells differentiated in TGF-β3 medium and by
PHDFs (Fig. 1). TGF-βRII
was also expressed by all cells, but was most prominent in cells
differentiated in in conditioned medium, HC-402-05a cells and PHDFs
(Fig. 1). TGF-βRIII was
highly expressed by HC-402-05a cells and PHDFs (Fig. 1). TGF-β2 expression was also
observed at relatively low levels in HC-402-05a cells and in cells
differentiated in the conditioned medium, but cells differentiated
in the TGF-β3 medium demonstrated relatively high expression
(Fig. 2). TGF-β3 was only present,
at similar levels, in the two types of chondrocyte-like cells
differentiated in vitro (Fig.
2). BMP-2 was expressed by all cells, but its expression
level was highest in PHDFs (Fig.
2). BMP-4 was also expressed by all cells, with the
highest levels of expression observed in the positive-control
HC-402-05a cells. Among the differentiated cells, BMP-4
expression was higher in those cultured in the presence of TGF-β3
(Fig. 2).
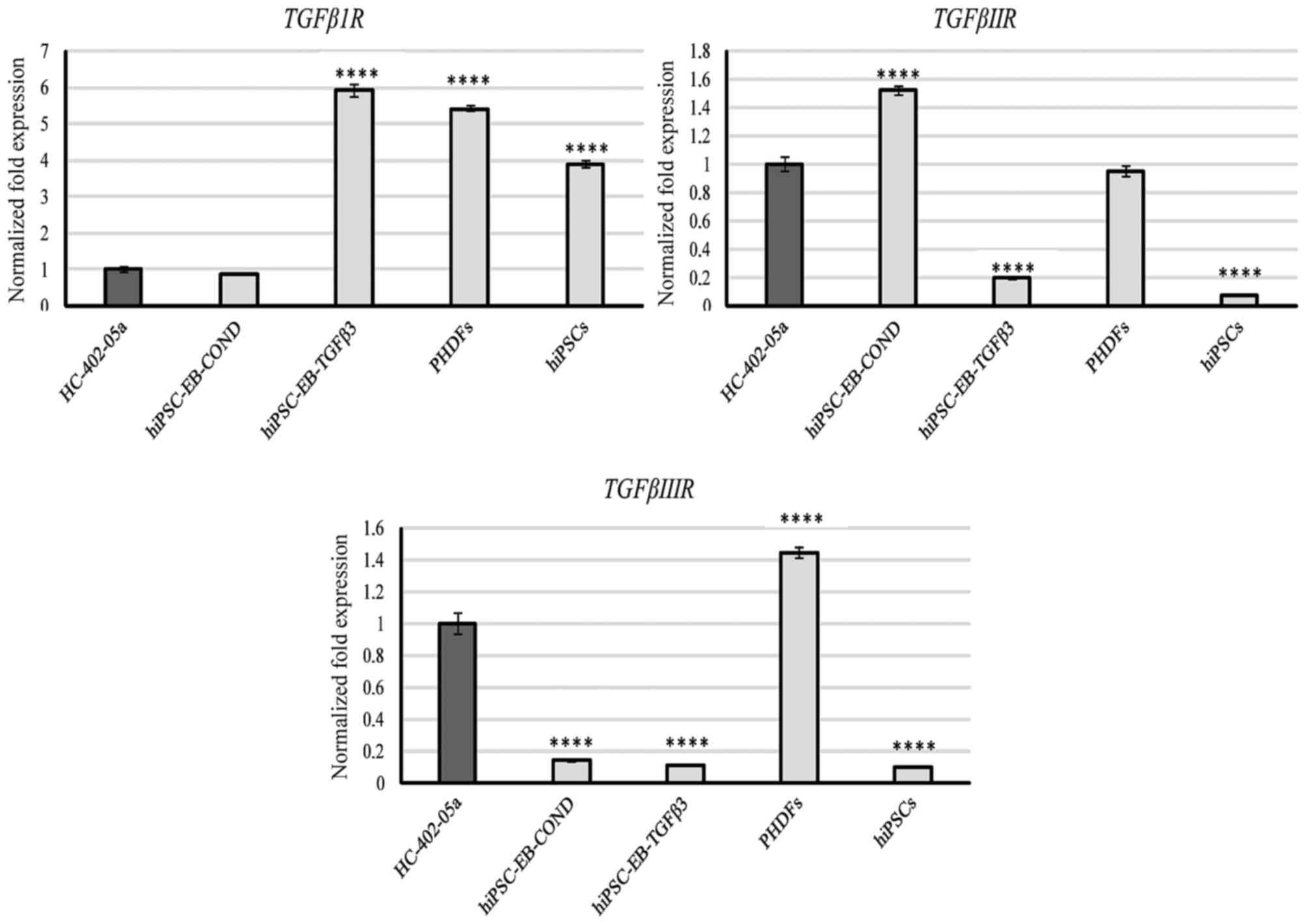 | Figure 1.The hiPSC-derived chondrocytes
differentiated in chondrogenic medium with TGF-β3 (10 ng/ml) or
following conditioning on HC-402-05a cells indicated the presence
of mRNA characteristic for TGF-βIR, TGF-βIIR and TGF-βIIIR. The
HC-402-05a cell line served as a positive control. PHDFs and hiPSCs
were used as negative controls. ****P<0.0001 vs. HC-402-05a.
hiPSCs, human induced pluripotent stem cells; TGF-β3, transforming
growth factor β3; TGF-βIR, TGF-β type I receptor; TGF-βIIR, TGF-β
type II receptor; TGF-βIIIR, TGF-β type III receptor; PHDFs,
primary human dermal fibroblasts; EB, embryoid bodies, COND,
conditioned medium. |
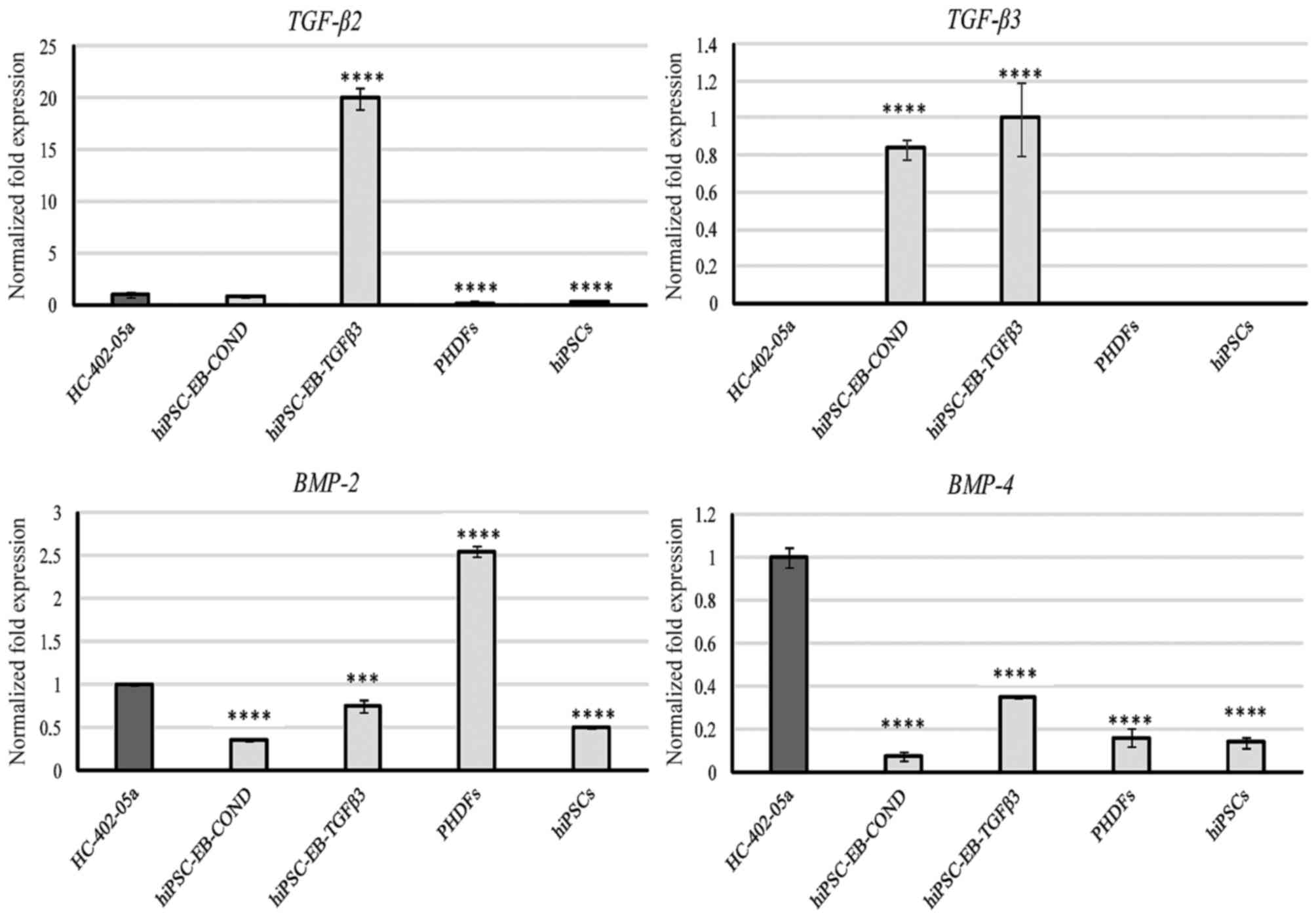 | Figure 2.The chondrocyte-like cells obtained
following differentiation in chondrogenic medium with TGF-β3 (10
ng/ml) or on medium conditioned with HC-402-05a cells demonstrated
expression of members of the TGF-β superfamily: TGF-β2, TGF-β3,
BMP-2 and BMP-4. The HC-402-05a cell line served as a positive
control. PHDFs and hiPSCs were used as negative controls.
****P<0.0001 vs. HC-402-05a. TGF-β, transforming growth factor
β; BMP-2, bone morphogenetic protein-2; BMP-4, bone morphogenetic
protein-4; PHDFs, primary human dermal fibroblasts; hiPSCs, human
induced pluripotent stem cells; EB, embryoid bodies, COND,
conditioned medium. |
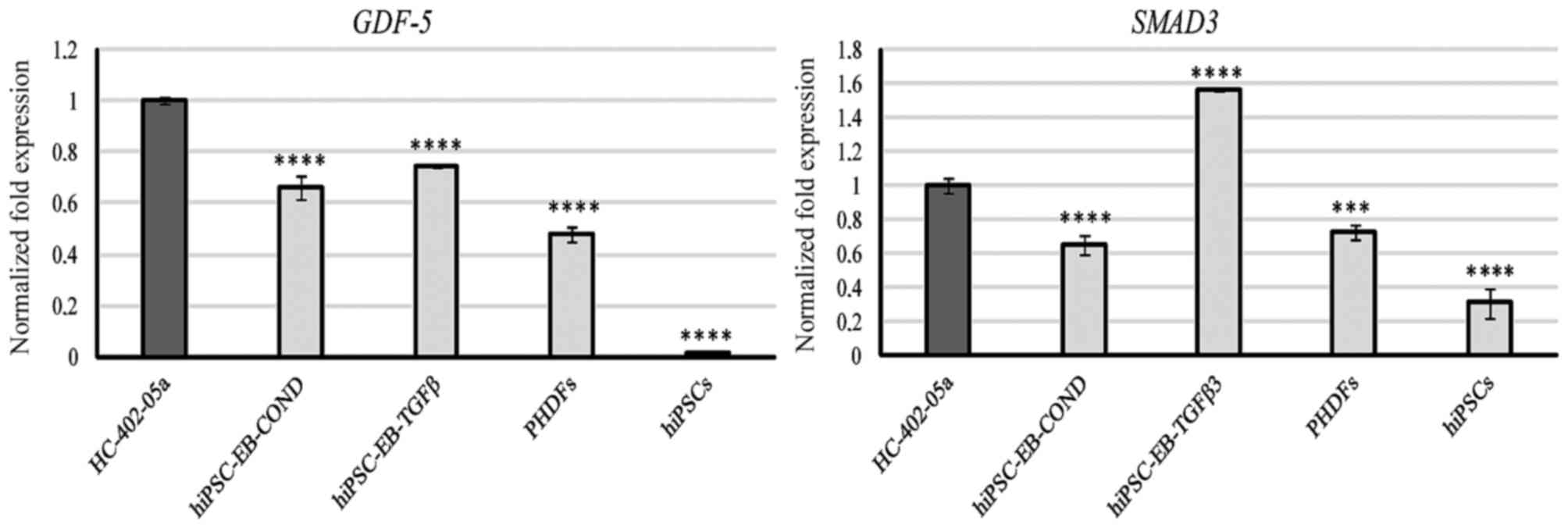
GDF-5 mRNA was present in all cells, with the
highest levels of expression in HC-402-05a cells and similar levels
of expression in the two differentiated cell groups (Fig. 3). SMAD3 expression was
observed in all cells, with the highest level of expression in
cells differentiated in the presence of TGF-β3 (Fig. 3).
Collagen expression
Type I collagen was expressed primarily by PHDFs,
but was also detectable in HC-402-05a cells, the two hiPSC-derived
chondrocyte groups, and at very low levels in hiPSCs (Fig. 4). No expression of type II collagen
was observed. Expression of type X collagen in PHDFs was relatively
high compared with the other cell groups (Fig. 4). Cells differentiated in
HC-402-05a or TGF-β3 medium presented with a higher production of
type X collagen at the mRNA level compared with positive control
HC-402-05a cells (Fig. 4). Type XI
collagen was most highly expressed by cells obtained by
TGF-β3-mediated chondrogenesis and, to a lesser extent, by
HC-402-05a (Fig. 4).
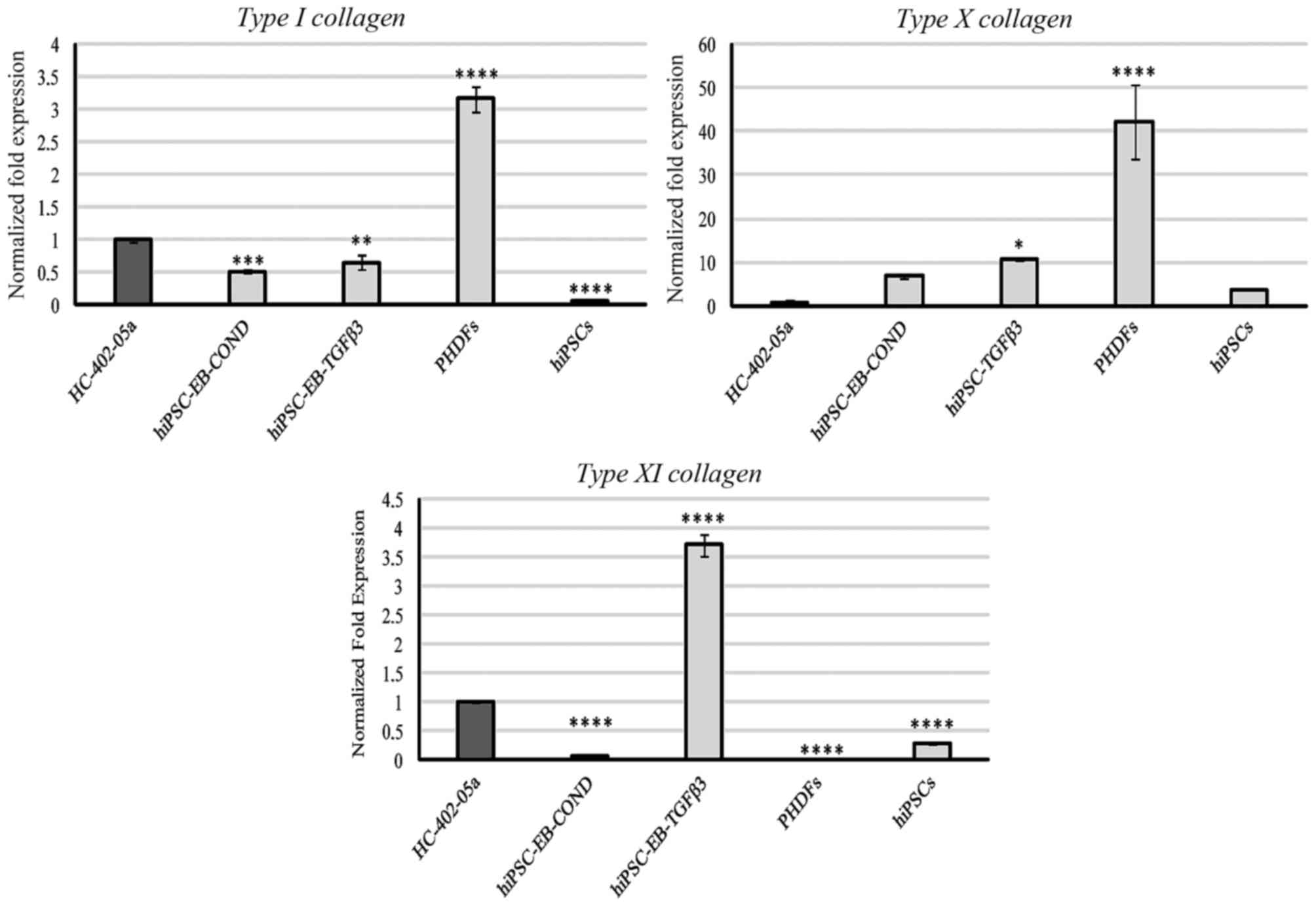 | Figure 4.Type II collagen was not expressed by
the examined cells. However, the chondrocyte-like cells obtained
following differentiation in chondrogenic medium with TGF-β3 (10
ng/ml) or on medium conditioned with HC-402-05a cells demonstrated
expression of type I, X and XI collagen characteristic of
dedifferentiated, hypertrophic and mature chondrocytes,
respectively. The HC-402-05a cell line served as a positive
control. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
vs. HC-402-05a. PHDFs and hiPSCs were used as negative controls.
TGF-β3, transforming growth factor β3; PHDFs, primary human dermal
fibroblasts; hiPSCs, human induced pluripotent stem cells; EB,
embryoid bodies; COND, conditioned medium. |
Markers of hypertrophy
Expression of IHH was detectable in all examined
cells, with the highest levels observed in the cells differentiated
with HC-402-05a-conditioned medium (Fig. 5). mRNA transcripts of PTHLH were
most abundant in PHDFs, followed by the chondrocyte-like cells
obtained in the TGF-β3 medium (Fig.
5). All cells expressed PTCH1, and in decreasing order PTCH1
expression was highest in hiPSCs, cells differentiated in the
presence of TGF-β3, PHDFs, HC-402-05a cells, and cells
differentiated in the conditioned medium (Fig. 5). RUNX2 was expressed most highly
in cells obtained via TGF-β3-mediated chondrogenesis, but was also
present in PHDFS and hiPSCs. By contrast, this marker was not
present in cells cultured in the conditioned medium (Fig. 5).
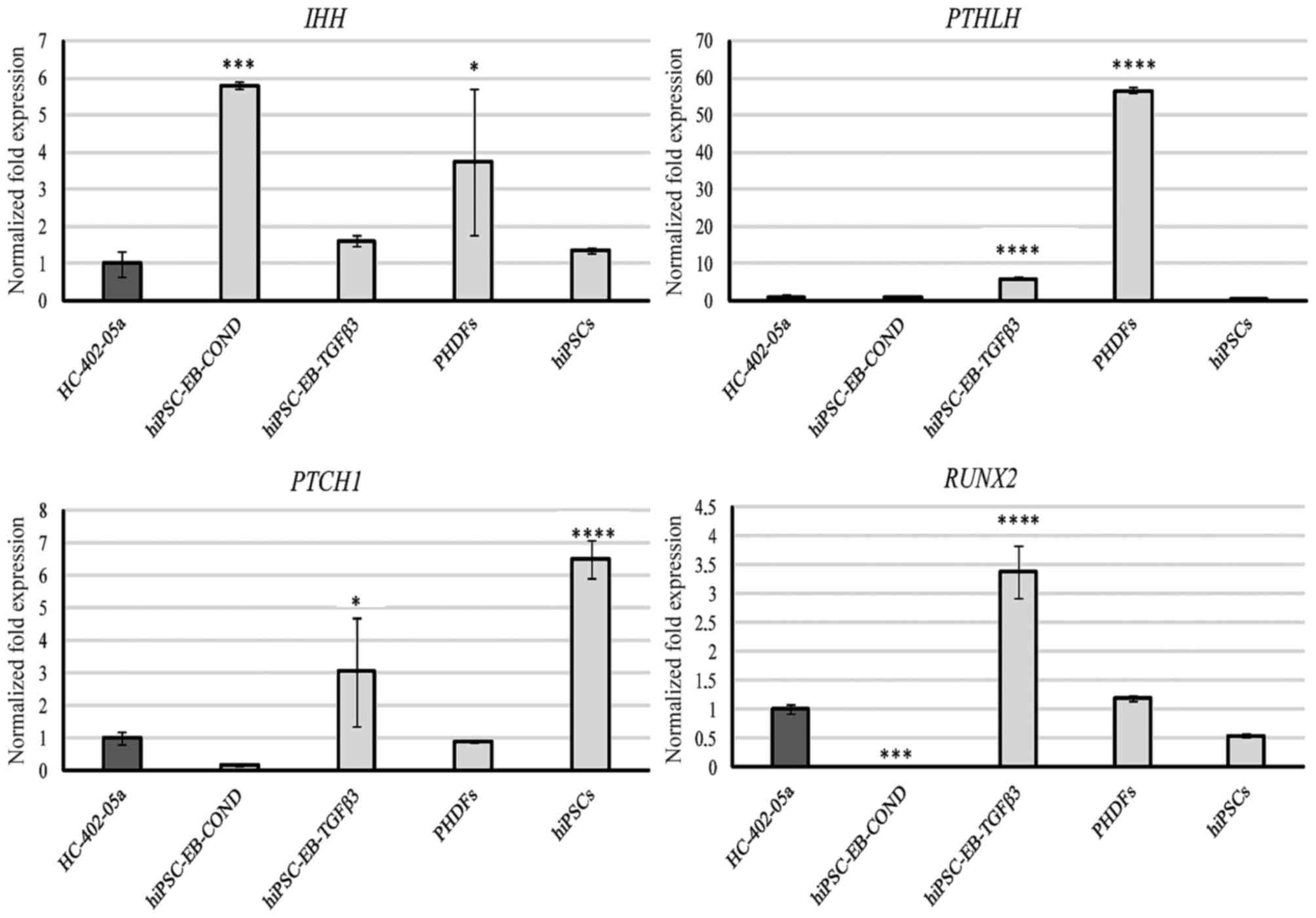 | Figure 5.The chondrocyte-like cells obtained
following differentiation in chondrogenic medium with TGF-β3 (10
ng/ml) or on medium conditioned with HC-402-05a cells expressed
genes from late chondrogenesis and hypertrophic chondrocytes: IHH,
PTHLH, PTCH1 and RUNX2. The HC-402-05a cell line served as a
positive control. PHDFs and hiPSCs were used as negative controls.
*P<0.05, ***P<0.001, ****P<0.0001 vs. HC-402-05a. TGF-β3,
transforming growth factor β3; IHH, Indian hedgehog; PTHLH,
parathyroid hormone- like hormone; PTCH1, patched 1; RUNX2,
runt-related transcription factor 2; PHDFs, primary human dermal
fibroblasts; hiPSCs, human induced pluripotent stem cells; EB,
embryoid bodies; COND, conditioned medium. |
Markers associated with osteoarthritis
and ossification
Only the HC-402-05a cell line expressed CH13L1
(Fig. 6). According to a
previously published study (19)
this marker is present in chondrocytes cultured in vitro.
This mRNA was not observed in the differentiated cells, which
suggests a lack of inflammatory properties in stem cell-derived
chondrocyte cultures. In addition, the specificity of CH13L1 was
demonstrated. All differentiated cells expressed MMP-2 (Fig. 6). The presence of MMP-2 is likely
to be correlated with extracellular matrix (ECM) remodelling that
occurs during chondrogenesis, including the reduction of the matrix
protein content. However, cells differentiated in the presence of
TGF-β3 expressed MMP-2 and also MMP-13, and this presence in
chondrocyte-like cells differentiated from hiPSCs and chondrocytes
is undesirable (Fig. 6). This
observation demonstrated that these cells were derived from late
stage chondrogenesis. Due to the capacity of MMP-13 to degrade
chondrogenic proteins, its high mRNA expression levels and
potential high levels of its protein product may explain the loss
of proteoglycans including type II and X collagen and aggrecan.
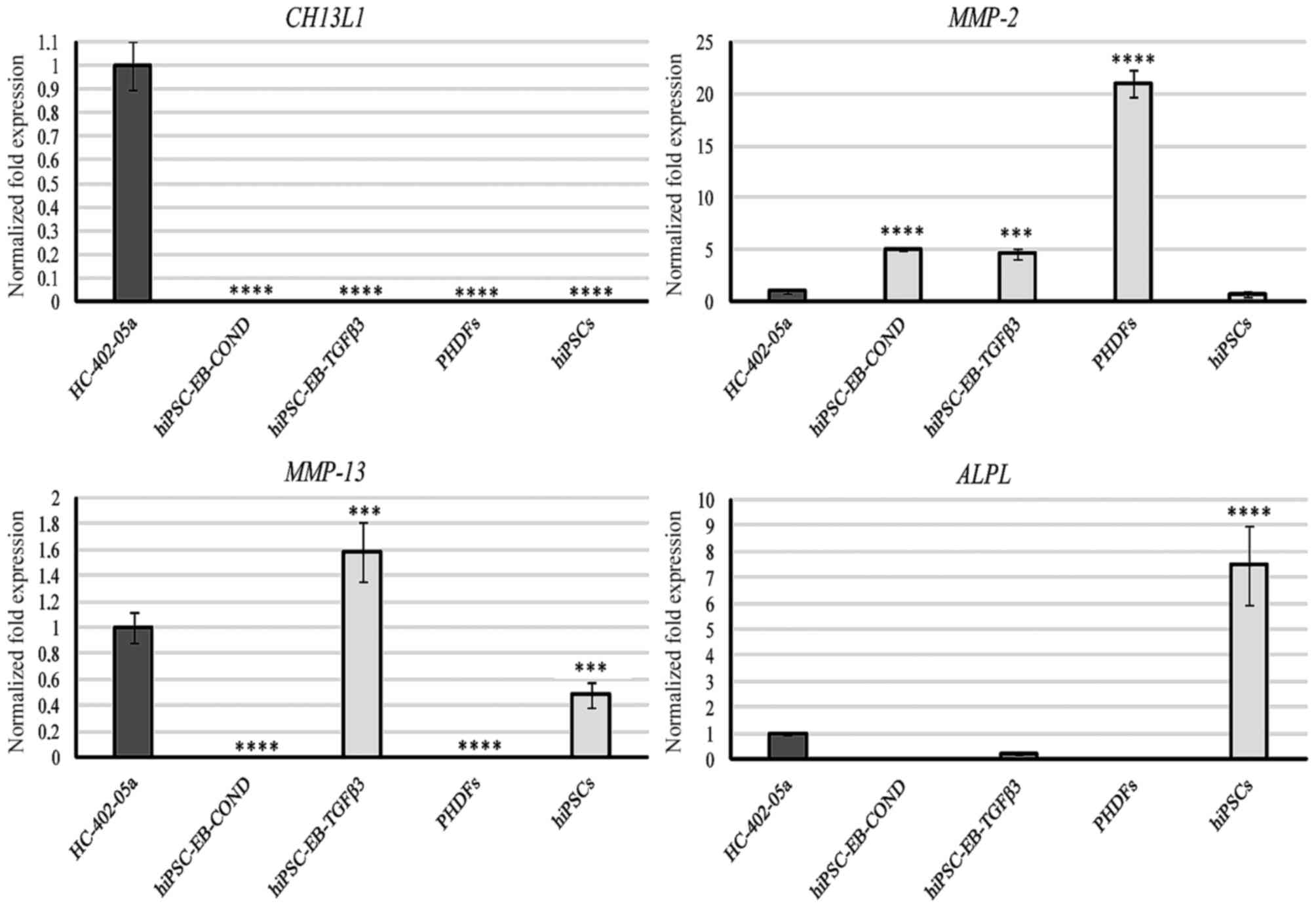 | Figure 6.The hiPSC-derived chondrocytes
obtained via differentiation in chondrogenic medium with TGF-β3 (10
ng/ml), in contrast to those differentiated in chondrogenic medium
medium conditioned with HC-402-05a cells, possessed undesirable
markers characteristic of mature and osteoarthritic chondrocytes:
Ch13L1, MMP-2, MMP-13 and ALPL. The HC-402-05a cell line served as
a positive control. PHDFs and hiPSCs were used as negative
controls. ***P<0.001, ****P<0.0001 vs. HC-402-05a. hiPSCs,
human induced pluripotent stem cells; TGF-β3, transforming growth
factor β3; Ch13L1, chitanise-3-like protein; MMP, matrix
metalloproteinase; ALPL, alkaline phosphatase; PHDFs, primary human
dermal fibroblasts; EB, embryoid bodies; COND, conditioned
medium. |
Finally, cells differentiated in the TGF-β3 medium
demonstrated relatively minor expression of ALPL (Fig. 6). These results are consistent with
other data obtained in the present study (including RUNX2;
Fig. 5), confirming that cells
differentiated in the TGF-β3 medium presented with a hypertrophic
phenotype. The highest levels of ALPL were detectable in
negative control hiPSCs, underscoring their pluripotent properties.
None of the cells in the present study expressed VEGF, a
finding that suggested that differentiated cells, regardless of
medium preparation, did not undergo vascularization following
endochondral ossification.
Discussion
In the present study, hiPSC cells were
differentiated in two different mediums: One supplemented with
TGF-β3 and the other conditioned with growth factors from
HC-402-05a cells. Notably, differentiation in the conditioned
medium resulted in greater chondrogenesis of the hiPSCs. On the
other hand, cells cultured in the presence of TGF-β3 presented with
characteristics shared with hypertrophic chondrocytes, which may
result in their decreased capacity to repair and regenerate
articular cartilage. Thus, the HC-402-05a-conditioned medium offers
greater potential for in vitro chondrogenesis. The
differentiated cells were then evaluated using 22 different
chondrogenic markers to assess the progression of chondrogenic
differentiation, as well as to determine the relative value of
these markers in predicting chondrogenic differentiation.
Furthermore, chondrogenic properties were demonstrated to change
even during short-term culture (passage 0 vs. 3). The aim in doing
so was to identify the best markers of late stage chondrogenesis,
hypertrophy and ossification. The most promising gene markers of
hiPSC differentiation during late stage chondrogenesis were
TGF-β2, TGF-β3, type XI collagen and
PTHLH. The best markers during hypertrophy were
RUNX2, IHH and type X collagen, and during
ossification MMP-13 and VEGF were good markers.
VEGF expression was specific to old and overgrown
chondrocytes only (data not shown). Useful however less valuable
markers included TGF-β receptors and the remaining members of the
TGF-β superfamily (Table I).
TGF-β receptors are engaged in multiple cellular
processes. The TGF-β family of receptors forms a functional complex
on the cell surface, which consists of type II and type I
transmembrane serine/threonine kinase receptors. TGF-β1 and TGF-β3
bind to their type II receptors while BMP-2 and BMP-4 bind
primarily to type I receptors. Activation of the epidermal growth
factor receptor inhibits TGF-β signaling (20). TGF-β molecules exert their effects
on cells by binding to the TGF-β receptors. The TGF-β type II
receptor (TβRII) recruits and interacts with TβRI through multiple
parallel signaling pathways, including SMAD proteins. TβRIII acts
as a co-receptor, increasing the binding rate of ligands to TβRII.
TβRII and TβRIII are present in fibroblast cells, but are
downregulated in fibroblasts and myofibroblasts in patients with
oral squamous cell carcinoma or oral carcinoma (20,21).
According to Keller et al (22), TβRI is expressed in
proliferating but not hypertrophic chondrocytes. These same authors
also demonstrated the negative regulation of BMP-2 in TGF-β
signaling and revealed that exogenous or upregulated TGF-β1
significantly increases BMP signaling (22). However, it is important to note
that this negative regulation does not fully agree with previously
published data and must therefore be further validated (23).
SMAD2 and 3 respond to TGF-β receptors. In response
to activation of TGF-β, the SMAD and p38 mitogen activated protein
kinase (MAPK) pathways, together with the RUNX2 gene,
control mesenchymal precursor cell differentiation. Induction of
TGF-β/BMP-2 signaling, MAPK-dependent phosphorylation and
RUNX2 results in osteoblast differentiation (24). The TGF-β isoforms and
TGF-β receptors are expressed in cartilage, bone and
synovial tissues. In osteoarthritis, there is an interaction
between TGF-β signaling (in particular between RUNX2 and
MMP-13) and WNT/β-catenin and Notch, as well as IHH
(25). TGF-β family signaling
occurs not only in differentiation but also in maintenance of
self-renewal and pluripotency of hESCs, due to the interplay
between TGF-β, activin, and Nodal signaling, whose activity
significantly decreases during early differentiation (26).
Cells differentiated in TGF-β3 and HC-402-05a
conditioned mediums expressed TGF-β receptors I, II and
III. Nevertheless, the profiles of expression varied
according to the culture medium: Cells differentiated in the
conditioned medium presented with a greater expression of
TβRII whereas cells cultured in the TGF-β3-medium
demonstrated higher expression of TβRI (Fig. 1). In this case the usage of
sequential administration of growth factors is likely to be an
interesting approach in effective chondrogenesis of stem cells
in vitro. The available evidence indicates that TGF-β
receptors are not specific markers of in vitro
chondrogenesis because these receptors are engaged in multiple
processes during several stages of development, including
maintenance of pluripotency, chondrogenesis and ossification, and
also in disease conditions including osteoarthritis (27). The results of the present study
confirm this characteristic: The TGF-β receptors were
expressed by numerous cell types, including all differentiated
cells, PHDFs, HC-402-05a, and hiPSCs (Fig. 1). This lack of specificity makes
them unsuitable as markers for chondrogenic differentiation.
The TGF-β superfamily is involved in regulating
chondrogenesis and is composed of two subfamilies: The TGF-β
subfamily (TGF-β1, TGF-β2, TGF-β3, activin, nodals, myostatin, and
Mullerian inhibiting substance) and the BMP subfamily (BMP-2,
BMP-4, BMP-10, GDFs) (28).
TGF-β1 and TGF-β3 are highly expressed in the
proliferative and hypertrophic zones. TGF-β2 expression is
observed in all zones, particularly the hypertrophic zone. Although
TGF-β1 induces chondrogenesis, TGF-β2 and TGF-β3 are even more
chondrogenic, resulting in a two-fold greater production of
glycosaminoglycan (28). Tan et
al (29) reported that
inhibition of TGF-β signaling through addition of the SB431542
molecule may substitute for octamer-binding transcription factor
3/4 in generating PSCs. Those authors also demonstrated that mPSCs
with an inhibited TGF-β signaling pathway had greater pluripotent
properties due to a decrease in extracellular signal-related kinase
(ERK) phosphorylation and consequent modulation of FGF/
mitogen-activated protein kinase kinase/ERK signaling (29). Overexpression of TGF-β
during enhanced cartilage repair, apart from the accumulation of
proteoglycans, may lead to synovial fibrosis. The intercellular
signaling molecule SMAD7 inhibits SMAD2 and SMAD3 phosphorylation,
thereby further blocking the TGF-β signaling pathway. The
simultaneous overexpression of TGF-β and SMAD7
prevents TGF-β-induced fibrosis by maintaining the
repair-stimulating effect of TGF-β on cartilage (30). Tekari et al (31) expanded chondrocytes in a monolayer
culture and reported that the chondrocytes maintained their
potential for cartilage-like tissue formation for up to three
passages. However, exogenous TGF-β1 had to be added following three
passages to induce the formation of cartilage-like tissue. The
authors hypothesized that this may be due to the lower expression
of TGF-β family members and TGF-β receptors during
prolonged culture (31). The
members of the TGF-β subfamily additionally possess osteogenic
potential, as reported by Li et al (32), who demonstrated that miPSC-derived
mesenchymal precursors differentiated into functional osteoblasts
following stimulation with TGF-β1 or -β3 in the presence of
retinoic acid.
In the present study, cells differentiated in the
conditioned medium expressed TGF-β3 at high levels but
TGF-β2 at lower levels. This result may indicate that these
cells possess desirable chondrogenic features at the mRNA level.
Cells differentiated in the presence of TGF-β3 also expressed of
TGF-β2 and TGF-β3 (Fig.
2). According to previously published data (32), the expression profile of these
cells is characteristic of chondrocytes in the hypertrophic zone.
For this reason, it is important to administer TGF-β3 at an
optimized concentration and culture period to obtain
chondrocyte-like cells from early or advanced stages of
chondrogenesis, thus avoiding hypertrophy during in vitro
chondrogenesis. These findings revealed that expression of
TGF-β family members provides a good, specific marker of
chondrogenic progression. However, because TGF-βs are active during
the entire chondrogenic and osteogenic processes, it is very
difficult to classify the differentiating cells into the precise
stage. Notably, the presence of TGF-βs is observed in human
chondrocytes and chondrocyte-like cells but not in parental stem
cells and PHDFs, a finding that further underscores the selective
nature and value of these as markers.
BMP-2 is one of the predominant growth factors with
beneficial potential in cartilage repair and cartilage tissue
engineering. It has the capacity to stimulate proteoglycan
synthesis and enhance production of type II collagen. BMP-2
expression is elevated in areas surrounding cartilage injury and in
osteoarthritis (33). BMP-2
and BMP-4 induce the progression of chondrocyte hypertrophy.
Thus, increased expression of BMP-2 and/or BMP-4 is
detectable during chondrocyte proliferation and maturation to
endochondral bone development. BMP-2 stimulates the expression of
other hypertrophic markers including type X collagen through
SMAD1-RUNX2 interaction at the 5′ promoter region in addition to
ALPL. Shu et al (34) demonstrated that BMP-2 is
involved in endochondral bone development, while BMP-4 is
involved to a lesser extent. BMP-2 has osteogenic properties and
upregulates the transcription of osteogenic genes, including type I
collagen, ALPL, osteocalcin and bone sialoprotein. However,
the SMAD signaling pathway is required for activation of osterix,
another factor involved in osteoblastic differentiation. BMP-2
modulates the expression of osterix through dependent and
independent RUNX2 (via msh homeobox 2) mechanisms (35).
Cells differentiated in the two study mediums
expressed BMP-2 and 4 (Fig. 2), the expression of which
indicating efficient chondrogenic differentiation and also
hypertrophy. Unfortunately, this marker alone is not able to
indicate which of these processes is more prevalent. Nevertheless,
cells cultured in the TGF-β3 medium are assumed to possess
hypertrophic properties, and they express BMP-2 and
4, RUNX2, and ALPL at significantly higher
levels than in cells differentiated in the conditioned medium and
positive controls. In addition, these markers are also expressed in
PHDFs (BMP-2 and −4) and hiPSCs (BMP-2; Fig. 2). For this reason it is difficult
to use these markers to assess the chondrogenic process in
vitro.
GDF-5 belongs to the TGF-β superfamily and is
involved in cartilage development and differentiation. Mutations in
the GDF-5 gene often result in defects in the appendicular skeleton
during development (36). The
expression profile of GDF-5 confirms its involvement in
endochondral ossification. Exposure to GDF-5 increases the
expression of chondrogenic markers including type I collagen
and aggrecan. However, prolonged stimulation by
administration of GDF-5 results in increased transcription of
type X collagen and ALPL, which are common indicators
of chondrocyte hypertrophy and initial endochondral ossification.
GDF-5 may, therefore, provide efficient regeneration of damaged
bone together with other pro-osteogenic and angiogenic factors
(37). In osteoblast-like cells,
GDF-5 stimulates the activity of gelatinases and matrix
metalloproteinases (MMP-2, MMP-9 and MMP-13) at matrix formation
sites. The metalloproteinases degrade denatured and native
collagens (gelatin and type I collagen, respectively) and
proteoglycan core proteins. This phenomenon involves the activation
of the p38 MAPK signaling pathway. Expression of metalloproteinases
is also under control of other members of the TGF-β superfamily,
the BMPs (38).
GDF-5 expression in the differentiated cells
was similar to that observed in adult articular chondrocytes. As
with BMP-2 and BMP-4 expression, GDF-5
expression may indicate successful chondrogenesis and initiation of
hypertrophy. Furthermore, GDF-5 is also present in PHDFs
(Fig. 3). Consequently, to
identify the chondrogenic stage of the cultured cells or to rule
out dedifferentiation, other markers are needed. The majority of
the markers evaluated in the present study indicated the presence
of chondrocyte-like cells (differentiation via the conditioned
medium) and chondrocyte-like hypertrophic cells (differentiation
via TGF-β3).
SMAD3 is another relevant member of the TGF-β
signaling pathway. The TGF-βRI phosphorylates SMAD2 and SMAD3,
together with SMAD4, form a heteromeric complex. SMAD3 associates
with multiple transcription factors, including RUNX2. Furthermore,
the MH2 domain of SMAD2 and SMAD3 interacts with the co-activator
cAMP-response-element-binding-protein-binding protein, in addition
to its paralog p300, with acetyltransferase activity. SMAD2/3 is
one of the TGF-β signaling pathways, and is involved in maintaining
and developing the chondrocyte phenotype. SMAD3 enhances the
transcriptional activity of SOX9 and increases expression of the
type II collagen gene (39).
C-terminal SMAD3 has an effect on β-catenin protein in a
TGF-β-dependent manner. It protects β-catenin from
ubiquitin-proteasome-dependent degradation and mediates its nuclear
transition. This SMAD3-mediated mechanism of β-catenin protein
stability promotes the activity of β-catenin, in turn affecting its
downstream target chondrogenesis-associated genes (40). SMAD3 is also responsible for the
balance between cartilage matrix synthesis and degradation through
increasing type II collagen expression with simultaneous
inhibition of RUNX2-induced MMP-13 expression. SMAD3
maintains chondrocyte homeostasis and prevents cells from
hypertrophy and osteoarthritis (41).
The expression of SMAD3 was detectable in all
cells, with the highest expression in cells differentiated in the
TGF-β3-medium (Fig. 3). Given the
elevated level of RUNX2, this may indicate that these cells
started to activate their SMAD3-mediated defence mechanisms to
prevent hypertrophy. As a marker of chondrogenesis, the specificity
of SMAD3 is questionable because its expression is observed
in chondrocytes, chondrocyte-like cells, and in the negative
controls (PHDF and hiPSCs).
Collagens represent a highly diverse group of ECM
proteins. The type II procollagen gene COL2A1 is widely
expressed in non-chondrogenic and chondrogenic tissues. As a result
of alternate splicing, type II collagen is synthesized and secreted
into ECM as two isoforms: II1 and IIB. The II1 isoform is expressed
in chondroprogenitor cells while the IIB isoform occurs in
differentiated chondrocytes (42).
Krug et al (43) examined
the change in patterns of gene expression (type II, IX and XI
collagen and aggrecan) in ESC-derived and primary chondrocytes. The
chondrocytes lost their characteristic phenotype (including type II
collagen and aggrecan) during monolayer culture (43). Type I collagen is a
dedifferentiation marker of chondrocyte-like cells because it is
widely expressed in primary human fibroblasts. It is characteristic
in bone, tendon, cornea and skin. During cartilage injury or
osteochondral defects, differentiation towards fibroblasts and
osteoblasts producing type I collagen and fibronectin is favoured
(44). Aggrecan, type X and type
II collagen are markers of late stage chondrocyte hypertrophy
associated with endochondral ossification. However, Mwale et
al (45) advise against
straightforward classification of these markers, as in certain
cases type X collagen may appear earlier than type II. Therefore,
caution must be exercised in using type X collagen as a marker of
chondrogenesis or chondrocyte hypertrophy (45). Type II collagen is synthesized as a
homotrimeric procollagen molecule [α1(II)]3 in cartilage
and is the most abundant fibril-forming collagen within joints.
Type XI collagen, a heterotrimeric collagen molecule [α1(XI)α2(XI)
α3(XI)], forms the core of the type II collagen fibril for type II
collagen fibrillogenesis and is responsible for fibril diameters in
cartilage. Collagen type II and XI collagens co-polymerize with
type IX collagen to form a heteropolymeric fibrillary framework
that corresponds with the tensile strength of cartilage (46).
Although chondrogenesis is usually assessed with
markers of the chondrocyte phenotype (including type II, X and XI
collagen), the usefulness of these markers may need to be
reconsidered given that expression of collagen (all types) may vary
as a function of the duration of the culture period and the number
of passages. In the present study, the cells cultured in the
conditioned medium primarily expressed type I and type X
collagen, which may indicate dedifferentiation or hypertrophic
processes. The human primary fibroblasts also expressed these
markers at high levels. In contrast, cells differentiated in the
HC-402-05a-condition medium did not present other hypertrophic
markers; furthermore, they had significantly higher levels of
desired chondrogenic markers (14). Cells cultured in the presence of
TGF-β3 also expressed high levels of type XI collagen in
addition to type I and X collagens (Fig. 4), which is a desirable marker
because it suggests that these cells present characteristics of
mature and hypertrophic chondrocytes. Although the most accurate
marker is type II collagen, this marker was not expressed in
any of the cells. This may be because all cultured cells were
obtained following the third passage or, alternatively, due to lack
of sex determining region Y-box 5 and SOX6
expression, which are required for the production of type II
collagen.
Indian hedgehog belongs to the hedgehog family and
is involved in the regulation of chondrocyte proliferation and
differentiation through a negative feedback loop with parathyroid
hormone related protein (PTHrP), a hypertrophy regulator that acts
in an IHH-dependent manner during chondrocyte proliferation and
hypertrophy (47,48). IHH activates expression of
PTHrP and the resultant PTHrP protein signals through its
receptor PTHR1 and inhibits excessive expression of IHH to
prevent hypertrophy and maintain the proliferating state in
chondrocytes. Mak et al (49) demonstrated that IHH promotes
chondrocyte hypertrophy independently of PTHrP through BMP and
WNT/β-catenin signaling. The canonical WNT/β-catenin pathway
controls the function of IHH, via WNT family member 9a (WNT9a),
which is involved in skeletogenesis. WNT9a signaling may inhibit
the expression of WNT family member 4, which is
characteristic for prehypertrophic chondrocytes (50). In mature proliferating
chondrocytes, the production of IHH is increased, which has a
visible effect on the neighbouring cells and results in activation
of their PTCH1 receptor. This results in elevated production of
PTHrP and slower cell differentiation. Insulin-like growth factor 1
is a major growth-promoting signal for skeletogenesis: It
suppresses PTHrP, thus inducing VEGF expression and
controlling the production of hypoxia-inducible factor 1-induced
angiogenesis (51). The IHH
signaling pathway also interacts with RUNX2 and RUNX3 during
chondrocyte proliferation and differentiation. Induction of
osteoblast differentiation by IHH requires other effectors, that
remain to be identified, besides RUNX2. Inhibition of IHH activity
regulating RUNX2 and RUNX3 in the perichondrium during
chondrogenesis triggers limb shortening (52,53).
IHH expression was observed in all the
cultured cells, with the highest levels of expression in cells
differentiated in the conditioned medium. These cells may be from
the pre-hypertrophic stage that is enriched in proliferative cells
(Fig. 5). These cells did not
present the parathyroid hormone at the mRNA level, a finding that
indicated the presence of a negative feedback loop between IHH and
PTHrP. The selectivity of this marker is only moderate because it
was expressed by negative controls, in particular PHDFs, but there
was a difference between the cells differentiated in the TGF- β3
medium and the conditioned medium. In addition, the chondrocytes
obtained from TGF-β3-mediated chondrogenesis in vitro
demontrated decreased levels of IHH with elevated expression
of PTHrP (Fig. 5). This
result also confirmed the existence of a negative feedback loop
between IHH and PTHrP and consequently the hypertrophic features of
these cells. The value of PTHrP as a specific marker of
hypertrophy is significantly diminished by the fact that
PTHrP is also highly expressed in PHDFs.
PTCH1 is a transmembrane receptor expressed in the
cartilage-bone interface, the perichondrium, and in proliferating
chondrocytes. It negatively regulates and is transcriptionally
activated by the Hedgehog signaling pathway, thus creating a
negative autoregulatory feedback loop (54,55).
Sonic hedgehog is the most prominent member of Hedgehog family. It
induces the activation of GLI family transcription factors
regulating the transcription of target genes, including GLI1
and PTCH1 during the generation of induced pluripotent cells
from fibroblasts (56).
Cells differentiated in the presence of TGF-β3
possessed features characteristic for hypertrophic chondrocytes at
the genetic level. They expressed PTCH1, which is negatively
regulated by IHH, a hypertrophic marker. The negative feedback loop
between PTCH1 and IHH was also demonstrated. Compared with cells
cultured in the conditioned medium, IHH expression was
downregulated in cells differentiated with TGF-β3, potentially due
to activated PTCH1. However, it should be noted that high levels of
PTCH1 expression were observed in hiPSCs (Fig. 5). It is possible to ascribe this
phenomenon to a reprogramming process rather than to the
spontaneous chondrogenic differentiation of hiPSCs. For this
reason, the selectivity of this marker for chondrogenesis in
vitro is less valuable than would otherwise be expected.
The RUNX family of DNA-binding transcription
factors control cell fate determination in a variety of tissues.
They are essential for hematopoiesis, skeletal development, and
development of the digestive and nervous systems. RUNX belongs to
the small transcription factor family and is expressed in
pre-hypertrophic and hypertrophic chondrocytes, indicating its
involvement in cartilage development. Furthermore, there is
evidence that RUNX2, a key regulator of osteoblast differentiation,
is also involved in regulating chondrocyte and osteoclast
differentiation, vascular invasion and periosteal bone formation.
During cartilaginous condensation, RUNX2 forms a complex with RUNX3
to control early chondrocyte differentiation (57,58).
The expression and activity of RUNX2 is regulated by a
multiple growth factors, including fibroblast-like growth factor 2,
TGF-β/BMP-2 and parathyroid hormone (59). RUNX2 is highly expressed in
immature osteoblasts but its expression is abruptly downregulated
during osteoblast maturation, when mature bone is formed. Hence,
RUNX2 determines the stage of osteoblast maturation, bone maturity
and the bone turnover rate, and is generally considered a major
driver for the later stages of endochondral ossification (60,61).
RUNX2 expression was particularly detectable
in the chondrocyte-like cells differentiated in the presence of
TGF-β3 (Fig. 5). This finding
suggests that differentiation in the presence of these growth
factors, in contrast to culture in the conditioned medium, favours
the creation of hypertrophic chondrocytes. Other results obtained
in the present study also confirm this, particularly the strong
association between RUNX, FGF-2,
TGF-β/BMP-2, and PTHrP.
Finally, in terms of markers characteristic for
chondrocytes with osteoarthritic properties and cells undergoing
ossification, hiPSCs differentiated in TGF-β3 medium expressed mRNA
indicative of cartilage degradation (Fig. 6).
In terms of study limitations, this was a two-part
study that demonstrated that hiPSCs differentiated in vitro
acquire a characteristic gene expression profile. Although this
gene expression profile is informative, it does not provide
unambiguous data to ascertain the precise chondrogenic stage of the
differentiated cells. In addition, due to the close interrelation
among the pathways activated during chondrogenesis, it is difficult
to identify the specific contribution of each pathway to the
chondrogenic process. More research is required to elucidate this
poorly understood area of chondrogenic differentiation. Thus, it is
important to emphasize that the classification of markers in the
present study is subjective and is based on the authors' knowledge
and understanding of the available literature.
To conclude, in the present study, chondrocytes
obtained by differentiating hiPSCs via EB formation in different
mediums resulted in divergent gene profiles. Cells differentiated
in the presence of TGF-β3 expressed genes associated with
hypertrophy, whereas hiPSCs differentiated in a medium conditioned
with HC-402-05a cells expressed genes indicative of early and
advanced chondrogenesis, thus indicating the superiority of this
medium. The most promising gene markers of hiPSC differentiation
were as follows: RUNX2, TGF-β2, TGF-β3,
type XI collagen, type X collagen, PTHLH,
MMP-13 and VEGF. Useful but less valuable markers
included TGF-β receptors and the remaining members of the TGF-β
superfamily.
The present study contributes to the development of
a novel, cost-effective protocol based on the use of endogenous
growth factors and metabolites in HC-402-05a-conditioned medium.
Notably, this approach does not require the use of expensive
exogenous growth factors. These preliminary results suggest that
the conditioned medium is likely to be more efficient than the
standard TGF-β3-based protocol, which may result in hypertrophy.
However, the chondrocyte-like cells obtained in these processes
consist of a mixed population that includes partially and fully
differentiated cells as well as undifferentiated cells. As a
result, evaluation of the chondrogenic process is challenging.
Furthermore, the origin of hiPSCs has a substantial impact on their
further differentiation toward chondrocyte-like cells deriving from
the same germ layer as the parental reprogrammed fibroblasts. The
present study contributes to an improved understanding of
chondrogenic differentiation and may help further the development
of regenerative medicine with hiPSCs.
Acknowledgements
The authors would like to thank Mr. Bradley Londres
for his assistance in editing the manuscript. The present study was
supported by the National Science Centre (grant no.
2012/07/E/NZ3/01819).
Glossary
Abbreviations
Abbreviations:
|
ALPL
|
alkaline phosphatase
|
|
BMP-2
|
bone morphogenetic protein-2
|
|
BMP-4
|
bone morphogenetic protein-4
|
|
CH13L1
|
chitanise-3-like protein
|
|
COL2A1
|
type II collagen
|
|
EBs
|
embryoid bodies
|
|
ECM
|
extracellular matrix
|
|
FGF-2
|
fibroblast growth factor 2
|
|
GDF-5
|
growth differentiation factor 5
|
|
HC-402-05a
|
human primary chondrocyte cell
line
|
|
hESCs
|
human embryonic stem cells
|
|
hiPSCs
|
human induced pluripotent stem
cells
|
|
IHH
|
indian hedgehog
|
|
MMP
|
matrix metalloproteinase
|
|
PHDFs
|
primary human dermal fibroblasts
|
|
PTHLH
|
parathyroid hormone related
protein
|
|
PTHrP
|
parathyroid hormone-like hormone
|
|
RUNX2
|
runt-related transcription factor
2
|
|
SOX5
|
6, 9, sex determining region Y-box 5,
6, 9
|
|
TGF-β2
|
transforming growth factor β2
|
|
TGF-β3
|
transforming growth factor β3
|
|
VEGF
|
vascular endothelial growth
factor
|
References
|
1
|
Xie A, Nie L, Shen G, Cui Z, Xu P, Ge H
and Tan Q: The application of autologous platelet-rich plasma gel
in cartilage regeneration. Mol Med Rep. 10:1642–1648.
2014.PubMed/NCBI
|
|
2
|
Saito T, Yano F, Mori D, Kawata M, Hoshi
K, Takato T, Masaki H, Otsu M, Eto K, Nakauchi H, et al: Hyaline
cartilage formation and tumorigenesis of implanted tissues derived
from human induced pluripotent stem cells. Biomed Res. 36:179–186.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang J, Cui W, Song F, Zhai C, Hu H, Zuo Q
and Fan W: Effects of mesenchymal stem cell on
interleukin-1β-treated chondrocytes and cartilage in a rat
osteoarthritic model. Mol Med Rep. 12:1753–1760. 2015.PubMed/NCBI
|
|
4
|
Suchorska WM, Augustyniak E and Łukjanow
M: Genetic stability of pluripotent stem cells during anti-cancer
therapies. Exp Ther Med. 11:695–702. 2016.PubMed/NCBI
|
|
5
|
Sommer AG, Rozelle SS, Sullivan S, Mills
JA, Park SM, Smith BW, Iyer AM, French DL, Kotton DN, Gadue P, et
al: Generation of human induced pluripotent stem cells from
peripheral blood using the STEMCCA lentiviral vector. J Vis Exp.
2012:43272012.
|
|
6
|
Barczak W, Suchorska W, Rubiś B and
Kulcenty K: Universal real-time PCR-based assay for lentiviral
titration. Mol Biotechnol. 57:195–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye J, Hong J and Ye F: Reprogramming rat
embryonic fibroblasts into induced pluripotent stem cells using
transposon vectors and their chondrogenic differentiation in vitro.
Mol Med Rep. 11:989–994. 2015.PubMed/NCBI
|
|
8
|
Oldershaw RA, Baxter MA, Lowe ET, Bates N,
Grady LM, Soncin F, Brison DR, Hardingham TE and Kimber SJ:
Directed differentiation of human embryonic stem cells toward
chondrocytes. Nat Biotechnol. 28:1187–1194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Phillips MD, Kuznetsov SA, Cherman N, Park
K, Chen KG, McClendon BN, Hamilton RS, McKay RD, Chenoweth JG,
Mallon BS and Robey PG: Directed differentiation of human induced
pluripotent stem cells toward bone and cartilage: In vitro versus
in vivo assays. Stem Cell Transl Med. 3:867–878. 2014. View Article : Google Scholar
|
|
10
|
Toh WS and Cao T: Derivation of
chondrogenic cells from human embryonic stem cells for cartilage
tissue engineering. Methods Mol Biol. Jul 12–2014.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mardani M, Hashemibeni B, Ansar MM,
Esfahani Zarkesh SH, Kazemi M, Goharian V, Esmaeili N and
Esfandiary E: Comparison between chondrogenic markers of
differentiated chondrocytes from adipose deived stem cells and
articular chondrocytes in vitro. Iran J Basic Med Sci. 16:763–773.
2013.PubMed/NCBI
|
|
12
|
Lee HJ, Choi BH, Min BH and Park SR:
Changes in Surface markers of human mesenchymal stem cells during
the chondrogenic differentiation and dedifferentiation processes in
vitro. Arthritis Rheum. 60:2325–2332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trzeciak T, Augustyniak E, Richter M,
Kaczmarczyk J and Suchorska W: Induced pluripotent and mesenchymal
stem cells as a promising tool for articular cartilage
regeneration. J Cell Sci Ther. 5:42014. View Article : Google Scholar
|
|
14
|
Suchorska WM, Augustyniak E, Richter M and
Trzeciak T: Gene expression profile in human induced pluripotent
stem cells: Chondrogenic differentiation in vitro-part A.
Mol Med Rep. 15:2387–2401. 2017.
|
|
15
|
Wróblewska J: A new method to generate
human induced pluripotent stem cells (iPS) and the role of the
protein KAP1 in epigenetic regulation of self-renewal. PhD
dissertation. Poznan University of Medical Sciences. http://www.wbc.poznan.pl/Content/373798/index.pdf2015.
|
|
16
|
Suchorska WM, Lach MS, Richter M,
Kaczmarczyk J and Trzeciak T: Bioimaging: An useful tool to monitor
differentiation of human embryonic stem cells into chondrocytes.
Ann Biomed Eng. 44:1845–1859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nejadnik H, Diecke S, Lenkov OD, Chapelin
F, Doing J, Tong X, Derugin N, Chan RC, Gaur A, Yang F, et al:
Improved approach for chondrogenic differentiation of human induced
pluripotent stem cells. Stem Cell Rev. 11:242–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Polacek M, Bruun JA, Johansen O and
Martinez I: Comparative analyses of the secretome from
dedifferentiated and redifferentiated adult articular chondrocytes.
Cartilage. 2:186–196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng W, Xia Q, Wu L, Chen S, He X, Zhang
L, Gao Q and Zhou H: Downregulation of TGF-beta receptors types II
and III in squamous cell carcinoma and oral carcinoma-associated
fibroblasts. BMC Cancer. 11:882011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signaling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keller B, Yang T, Chen Y, Munivez E,
Bertin T, Zabel B and Lee B: Interaction of TGFβ and BMP signaling
pathways during chondrogenesis. PLoS One. 6:e164212011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen B, Wei A, Tao H, Diwan AD and Ma DD:
BMP-2 enhances TGF-beta3-mediated chondrogenic differentiation of
human bone marrow multipotent mesenchymal stromal cells in alginate
bead culture. Tissue Eng Part A. 15:1311–1320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen G, Deng C and Li YP: TGF-β and BMP
signaling in osteoblast differentiation and bone formation. Int J
Biol Sci. 8:272–288. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang F and Chen YG: Regulation of TGF-β
receptor activity. Cell Biosci. 2:92012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen J, Li S and Chen D: TGF-β signaling
and the development of osteoarthritis. Bone Res. 2:140022014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Watabe T and Miyazono K: Roles of TGF-beta
family signaling in stem cell renewal and differentiation. Cell
Res. 19:103–115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang W, Rigueur D and Lyons KM: TGFβ
signaling in cartilage development and maintenance. Birth Defects
Res C Embryo Today. 102:37–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan F, Quian C, Tanq K, Abd-Allach SM and
Jing N: Inhibition of transforming growth factor β (TGF-β)
signaling can substitute for Oct4 protein in reprogramming and
maintain pluripotency. J Biol Chem. 290:4500–4511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davidson Blaney EN, Vitters EL, van den
Berg WB and van der Kraan PM: TGF beta-induced cartilage repair is
maintained but fibrosis is blocked in the presence of Smad7.
Arthritis Res Ther. 8:R652006. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tekari A, Luginbuehl R, Hofstetter W and
Egli RJ: Transforming growth factor beta signaling is essential for
the autonomous formation of cartilage-like tissue by expanded
chondrocytes. PLoS One. 10:e01208572015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li F and Niyibizi C: Cells derived from
murine induced pluripotent stem cells (iPSC) by treatment with
members of TGF-beta family give rise to osteoblasts differentiation
and form bone in vivo. BMC Cell Biol. 13:352012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun J, Li J, Li C and Yu Y: Role of bone
morphogenetic protein-2 in osteogenic differentiation of
mesenchymal stem cells. Mol Med Rep. 12:4230–4237. 2015.PubMed/NCBI
|
|
34
|
Shu B, Zhang M, Xie R, Wang M, Jin H, Hou
W, Tang D, Harris SE, Mishina Y, O'Keefe RJ, et al: BMP-2, but not
BMP-4, is crucial for chondrocyte proliferation and maturation
during endochondral bone development. J Cell Sci. 124:3428–3440.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matsubara T, Kida K, Yamaguchi A, Hata K,
Ichida F, Meguro H, Aburatani H, Nishimura R and Yoneda T: Bmp2
regulates Osterix through Msx2 and Runx2 during osteoblast
differentiation. J Biol Chem. 283:29119–29125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun Z, Zhang Y, Yang S, Jia J, Ye S, Chen
D and Mo F: Growth differentiation factor 5 modulation of
chondrogenesis of self-assembled constructs involves gap
junction-mediated intercellular communication. Dev Growth Differ.
54:809–817. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Coleman CM, Vaughan EE, Browe DC, Mooney
E, Howard L and Barry F: Growth differentiation factor-5 enhances
in vitro mesenchymal stromal cell chondrogenesis and hypertrophy.
Stem Cells Dev. 22:1968–1976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hatakeyama Y, Hatakeyama J, Maruya Y, Oka
K, Tsuruga E, Inai T and Sawa Y: Growth differentiation factor 5
(GDF-5) induces matrix metalloproteinase 2 (MMP-2) expression in
peridontal ligament cells and modulates MMP-2 and MMP-13 activity
in osteoblasts. Bone and Tissue Regeneration Insights. 3:1–10.
2010.
|
|
39
|
Furumatsu T, Tsuda M, Taniguchi N, Tajima
Y and Asahara H: Smad3 induces chondrogenesis through the
activation of SOX9 via CREB-binding protein/p300 recruitment. J
Biol Chem. 280:8343–8350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang M, Wang M, Tan X, Li TF, Zhang YE
and Chen D: Smad3 prevents beta-catenin degradation and facilitates
beta-catenin nuclear translocation in chondrocytes. J Biol Chem.
285:8703–8710. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen CG, Thuillier D, Chin EN and Alliston
T: Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix
metalloproteinase 13 expression to maintain articular cartilage and
prevent osteoarthritis. Arthritis Rheum. 64:3278–3289. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Savontaus M, Ihanamäki T, Perälä M,
Metsäranta M, Sandberg-Lall M and Vuorio E: Expression of type II
and IX collagen isoforms during normal and pathological cartilage
and eye development. Histochem Cell Biol. 110:149–159. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krug D, Klinger M, Haller R, Hargus G,
Büning J, Rohwedel J and Kramer J: Minor cartilage collagens type
IX and XI are expressed during embryonic stem-cell derived in vitro
chondrogenesis. Ann Ant. 195:88–97. 2013. View Article : Google Scholar
|
|
44
|
Goessler UR, Bugert P, Bieback K, Baisch
A, Sadick H, Verse T, Klüter H, Hörmann K and Riedel F: Expression
of collagen fiber-associated proteins in human septal cartilage
during in vitro dedifferentiation. Int J Mol Med.
14:1015–1022. 2004.PubMed/NCBI
|
|
45
|
Mwale F, Stachura D, Roughley P and
Antoniou J: Limitations of using aggrecan and type X collagen as
markers of chondrogenesis in mesenchymal stem cell differentiation.
J Orthop Res. 24:1791–1798. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
McAlinden A, Traeger G, Hansen U, Weis MA,
Ravindran S, Wirthlin L, Eyre DR and Fernandes RJ: Molecular
properties and fibril ultrastructure of type II and XI collagens in
cartilage of mice expressing exclusively the α1 (IIA) collagen
isoform. Matrix Biol. 34:105–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma RS, Zhou ZL, Luo JW, Zhang H and Hou
JF: The Ihh signal is essential for regulating proliferation and
hypertrophy of cultured chicken chondrocytes. Comp Biochem Physiol
B Biochem Mol Biol. 166:117–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Katoh Y and Katoh M: Hedgehog signaling
pathway and gastrointestinal stem cell signaling network (Review).
Int J Mol Med. 18:1019–1023. 2006.PubMed/NCBI
|
|
49
|
Mak KK, Konenberg HM, Chuang PT, Mackem S
and Yang Y: Indian hedgehog signals independently of PTHrP to
promote chondrocyte hypertrophy. Development. 135:1947–1956. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Später D, Hill TP, O'sullivan RJ, Gruber
M, Conner DA and Hartmann C: Wnt9asignaling is required for joint
integrity and regulation of Ihh during chondrogenesis. Development.
133:3039–3049. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Y, Cheng Z, Elalieh HZ, Nakamura E,
Nguyen MT, Mackem S, Clemens TL, Bikle DD and Chang W: IGF-1R
signaling in chondrocytes modulates growth plate development by
interacting with the PTHrP/Ihh pathway. J Bone Miner Res.
26:1437–1446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kim EJ, Cho SW, Shin JO, Lee MJ, Kim KS
and Jung HS: Ihh and Runx2/Runx3 signaling interact to coordinate
early chondrogenesis: A mouse model. PLoS One. 8:e552962013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tu X, Joeng KS and Long F: Indian hedgehog
requires additional factors besides Runx2 to induces osteoblast
differentiation. Dev Biol. 362:76–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Z, Xu J, Colvin JS and Ornitz DM:
Coordination of chondrogenesis and osteogenesis by fibroblast
growth factor 18. Genes Dev. 16:859–869. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bruce SJ, Butterfield NC, Metzis V, Town
L, McGlinn E and Wicking C: Inactivation of Patched 1 in the mouse
limb has novel inhibitory effects on the chondrogenic program. J
Biol Chem. 285:27967–27981. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Moon JH, Heo JS, Kim JS, Jun EK, Lee JH,
Kim A, Kim J, Whang KY, Kang YK, Yeo S, et al: Reprogramming
fibroblasts into induced pluripotent stem cells with Bmi1. Cell
Res. 21:1305–1315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vimalraj S, Arumugam B, Miranda PJ and
Selvamurugan N: Runx2: Structure, function, and phosphorylation in
osteoblast differentiation. Int J Biol Macromol. 78:202–208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun J, Zhou H, Deng Y, Zhang Y, Gu P, Ge S
and Fan X: Conditioned medium from bone marrow mesenchymal stem
cells transiently retards osteoblast differentiation by
downregulating runx2. Cells Tissues Organs. 196:510–522. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ann EJ, Kim HY, Choi YH, Kim MY, Mo JS,
Jung J, Yoon JH, Kim SM, Moon JS, Seo MS, et al: Inhibition of
Notch1 signaling by Runx2 during osteoblast differentiation. J Bone
Miner Res. 26:317–330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Komori T: Regulation of bone development
and extracellular matrix protein genes by RUNX2. Cell Tissue Res.
339:189–195. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Du F, Wu H, Zhou Z and Liu YU:
microRNA-375 inhibits osteogenic differentiation by targeting
runt-related transcription factor 2. Exp Ther Med. 10:207–212.
2015.PubMed/NCBI
|