Introduction
Bone homeostasis is maintained by the correct
balance between bone formation, which is mediated by osteoblasts,
and bone resorption mediated by osteoclasts, to regulate bone
volume (1). Bone marrow-derived
mesenchymal stem cells (BMSCs) are a population of self-renewing
multipotent cells derived from bone marrow (2). Numerous studies have demonstrated
that BMSCs may differentiate into osteoblasts, which makes them a
suitable cell type for therapeutic use in bone repair and tissue
engineering (3,4). However, the regulation of these
cellular pathways remains to be fully elucidated. Therefore,
investigation of the regulatory mechanism of osteoblast
differentiation is essential for the development of therapeutic
strategies to treat bone loss (5).
MicroRNAs (miRNAs) are a class of endogenous
non-coding single-stranded RNA molecules of ~22 nucleotides in
length, which function at the post-transcriptional level by
negatively regulating gene expression via base pairing to
complementary sites in the target mRNA 3′ untranslated regions
(UTR) (6). miRNAs have emerged as
key post-transcriptional regulators of gene expression and are
associated with a range of various physiological and pathological
processes (7). It has previously
been demonstrated that osteogenic induction and differentiation are
regulated by post-transcriptional mechanisms, predominantly by
temporally expressed miRNAs. Various miRNAs (miRs), including
miR-30, miR-182 and miR-346 have been demonstrated to regulate the
osteogenic differentiation of BMSCs (8–10).
miR-30 inhibits osteoblast differentiation by targeting mothers
against decapentaplegic homolog 1 and runt-related transcription
factor 2 (Runx2) (8), miR-182
represses the osteogenesis of mesenchymal stem cells by targeting
forkhead box O1 (9) and miR-346
regulates human osteogenic differentiation by targeting the
Wnt/β-catenin pathway (10).
However, the specific role and function of miRNAs in osteogenic
differentiation remains to be elucidated.
miR-217 was previously demonstrated to act as a
tumor suppressor and is important in tumor cell proliferation and
migration. The dysregulation of miR-217 has been reported in
various tumor types including osteosarcoma (11), gastric cancer (12), pancreatic cancer (13) and clear cell renal cell carcinoma
(14). However, studies remain to
be conducted to investigate the role of miR-217 in the osteogenic
differentiation of BMSCs.
In the present study, the expression of miR-217
during the osteogenic differentiation of BMSCs was first detected
and it was observed that the expression of miR-217 was negatively
associated with bone formation and osteogenic differentiation.
Gain- and loss-of-function experiments subsequently demonstrated
that miR-217 is a negative regulator of osteogenic differentiation
in BMSCs. Luciferase assay and western blotting then verified that
Runx2 is a target of miR-217 and the regulatory mechanisms may be
mediated via extracellular signal-regulated kinase (ERK) and p38
mitogen-activated protein kinase (p38 MAPK) pathways.
Materials and methods
Isolation and culture of rat
BMSCs
The Animal Research Ethics Committee of Sheyang
County People's Hospital (Jiangsu, China) approved all animal
experiments. Sprague-Dawley male rats (6 weeks old; n=30) weighing
200–250 g were provided by the Experimental Animal Center of
Nanjing Medical University (Nanjing, China). They were housed under
controlled conditions with a 12-h light/dark cycle at a temperature
of 24±1°C and humidity of 50±5%. The rats were allowed free access
to standard rat chow and water. Rats were anesthetized via
intraperitoneal injection of urethane (20% in saline, 5 ml/kg;
Shanghai Qingxi Chemical Technology Co., Ltd., Shanghai, China).
Bone marrow was flushed out from the tibiae and femurs of rats with
sterilized PBS, and subsequently filtered through a strainer and
suspended in Dulbecco's modified Eagle's medium (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany), supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 10
U/ml penicillin and 100 µg/ml streptomycin (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA). Cells were subsequently cultured at
37°C in an atmosphere containing 5% CO2 for 24 h. When
cells reached 90% confluence, they were trypsinized and re-plated
in 25 cm2 culture flasks. The resulting cultures were
referred to as passage 0 (15).
The passage 3 of BMSCs was used in the present study.
Osteogenesis-induced
differentiation
The BMSCs were trypsinized and re-plated in 6-well
plates at a concentration of 1×105 cells/well. When
cells were 80–90% confluent, the medium was replaced by osteogenic
induction medium containing 100 nM dexamethasone, 50 mg/ml
ascorbate, 10 mM β-glycerophosphate and 25 ng/ml recombinant human
bone morphogenetic protein 2 precursor (Bmp2; Sigma-Aldrich; Merck
KGaA). The medium was replaced every 3 days for 3 weeks.
Alizarin red staining
Alizarin red staining was used to detect the calcium
deposits. A total of 1×105 cells were washed twice with
PBS after 21 days, following fixation with paraformaldehyde for 30
min at 4°C and then washed with double distilled water
(ddH2O) three times. The fixed cells were subsequently
stained with 0.1% alizarin red (Sigma-Aldrich; Merck KGaA) solution
for 30 min at room temperature. The cells were then washed twice
with PBS and observed under a microscope (Olympus Corporation,
Tokyo, Japan; ×50 magnification).
Reverse transcription-quantitative
polymerase chain reaction (qPCR)
Total RNA from the BMSCs was extracted using an
RNeasy Mini kit (Qiagen China Co., Ltd., Shanghai, China),
according to the manufacturer's protocol. The mRNA was reverse
transcribed to cDNA using the PrimeScript 1st strand cDNA synthesis
kit (Takara Biotechnology Co., Ltd., Dalian, China). The PCR
amplification was conducted in a total volume of 20 µl, containing
0.4 µl each primer, 10 ml SYBR® Green qPCR Master Mix
(Takara Biotechnology Co., Ltd.), 2 µl cDNA and 7.2 µl
H2O. Primer sequences are presented in Table I. Generation of standard curves and
qPCR were carried out using an ABI7500 Real-Time PCR instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were 95°C for 10 min followed by 40 cycles
of 95°C for 5 sec and 60°C for 30 sec. The expression of the target
gene was normalized to the geometric mean of the GAPDH housekeeping
gene.
 | Table I.Oligonucleotide primer sequences and
product sizes for reverse transcription-quantitative polymerase
chain reaction. |
Table I.
Oligonucleotide primer sequences and
product sizes for reverse transcription-quantitative polymerase
chain reaction.
| Gene | Forward primer
sequence | Reverse primer
sequence | Size (bp) |
|---|
| Runx2 |
GCCACTTACCACAGAGCTATTA |
GGCGGTCAGAGAACAAACTA | 106 |
| Alp |
CATGTTCCTGGGAGATGGTAT |
GTGTTGTACGTCTTGGAGAGA | 144 |
| Bmp2 |
TGTGAGGATTAGCAGGTCTTT |
TTGTGGAGTGGATGTCCTTTAC | 105 |
| OPN |
CCCATCTCAGAAGCAGAATCTT |
GTCATGGCTTTCATTGGAGTT | 109 |
| OC |
TGACTGCATTCTGCCTCTC |
CGGAGTCTATTCACCACCTTA | 109 |
| GAPDH |
ACTCCCATTCTTCCACCTTT |
CCCTGTTGCTGTAGCCATATT | 105 |
Total miRNA was extracted using the miRNeasy Mini
Kit (Qiagen China Co., Ltd.) and reverse transcribed to form cDNA
by TaqMan miRNA reverse transcription kit (Applied Biosystems,
Thermo Fisher Scientific Inc.). Their primer sequences are
presented in Table I. The
expression levels of miRNA were measured using the TaqMan miRNA
Assay kit (Applied Biosystems, Thermo Fisher Scientific Inc.) and
normalized to U6 snRNA transcript levels. All relative gene
expression levels were analyzed using the 2−ΔΔCq formula
(16). Each sample was examined in
triplicate and a no-template control was included for each
amplification.
Transfection of miRNA mimics and
inhibitor
miR-217 mimics, anti-miR-217 oligos and a scramble
negative oligo control were purchased from Applied Biosystems
(Rockville, MD, USA). Diethylpyrocarbonate-treated water was used
to dissolve the oligonucleotides. A total of 50 nM miRNA precursor
or 10 nM antisense oligos were transfected into cells using
Lipofectamine® 2000 (Invitrogen, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cells were harvested after 48 h. For long-term detection, mimics,
inhibitors and negative control were repeatedly transfected every 3
days. miR-217 mimic sequences were 5′-GUCAGUCAAGGACUACGTCAU-3′;
miR-217 inhibitor sequence was 5′-AUGACGUAGUCCUUGACUGAC-3′ and the
negative control sequence was 5′-CAGUACUUUUGUGTAGUACAA-3′.
Western blotting
Total protein was extracted from BMSCs using lysis
buffer (50 mmol/l Tris, 150 mmol/l NaCl, 1% Triton X-100, 1%
deoxycholic phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, 5.0
mmol sodium pyrophosphate, 1.0 g/ml leupeptin, 0.1 mmol
phenylmethylsulfonyl fluoride and 1 mmol/l dithiothreitol) on ice
for 30 min, and protein concentrations were quantified using the
Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.).
Proteins (25 µg) were subsequently separated using SDS-PAGE on a
12% gel and electrophoretically transferred onto polyvinylidene
difluoride membranes. The membranes were blocked with 5% fat-free
milk in TBS Tween-20 for 1 h at room temperature and then incubated
with primary antibodies at 4°C overnight. Blots were incubated with
horseradish peroxidase-conjugated rabbit anti-rat secondary
antibody (catalog no. ab6734; 1:5,000; Abcam, Cambridge, UK) for 1
h at room temperature. The immunoreactive bands were detected by
enhanced chemiluminescence detection reagents (GE Healthcare Life
Sciences, Chalfont, UK).
Antibodies
Anti-Runx2 (catalog no. ab23981; 1:1,000) and
anti-Bmp2 (catalog no. ab14933; 1:1,000) were obtained from Abcam.
Anti-alkaline phosphatase (ALP; catalog no. sc271431; 1:1,000),
anti-osteocalcin (OC; catalog no. sc18319; 1:1,000),
anti-osteopontin (OPN; catalog no. sc20788; 1:1,000),
anti-phosphorylated (p)-ERK (catalog no. sc7383; 1:500), anti-ERK
(catalog no. sc292838; 1:500), anti-p-p38 MAPK (catalog no.
sc101758; 1:500) and anti-p38 MAPK (catalog no. sc7972; 1:500) were
obtained from Santa Cruz Biotechnology Inc., Dallas, TX, USA.
Anti-GAPDH (catalog no. TA309157; 1:1,000) was purchased from
ZSGB-Bio (Beijing, China).
Luciferase assay
The full length Runx2 3′-UTR (Shanghai GenePharma
Co., Ltd., Shanghai, China) and miR-217 binding sites were cloned
into pGL3 vectors (Huada Genomics, Shenzheng, China). HEK293T cells
were purchased from American Type Culture Collection (Teddington,
UK) and cultured with high-glucose Dulbecco's modified Eagle's
medium (DMEM; HyClone, GE Healthcare Life Sciences) supplemented
with penicillin-streptomycin, fetal bovine serum and 0.25%
trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.). HEK293T cells
were transfected with miR-217 mimic, or control at a final
concentration of 50 nM using Lipofectamine® 2000. The
following day, cells were transfected with 200 ng 3′-UTR plasmids
along with Renilla luciferase plasmid (Promega Corporation,
Madison, WI, USA) using XtremeGENE 9 (Roche Diagnostics, Basel,
Switzerland), according to the manufacturer's protocol. Cell were
harvested and lysed after 48 h. Luciferase activity was measured
using dual-luciferase assay kit (Promega Corporation) at a
wavelength of 480 nm. Renilla luciferase activity at 560 nm was
used for normalization. The experiments were performed
independently three times.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Univariate comparison of means was evaluated using an unpaired
Student's t-test. A value of P<0.05 was considered to indicate a
statistically significant difference. All statistical analysis was
performed using SPSS 17.0 software (SPSS Inc., Chicago, IL,
USA).
Bioinformatics predictions
To predict the target genes of miR-217 during the
osteogenic differentiation, the present study selected two miRNA
target prediction databases: TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org).
Results
Osteogenic differentiation of rat
BMSCs
BMSCs were treated with standard
osteogenic-induction medium in order to induce osteogenic
differentiation. This was detected by increased mRNA (data not
shown) and protein expression levels of osteogenic-associated
genes, including Runx2, ALP, OC, OPN, and Bmp2, at days 14 and 21
(Fig. 1A). Alizarin red staining
for matrix mineralization confirmed the osteoblast phenotype
(Fig. 1B). Increased expression of
osteogenic-associated genes and the observed osteoblast phenotype
were in accordance with previous reports describing rat BMSCs
differentiation (17,18).
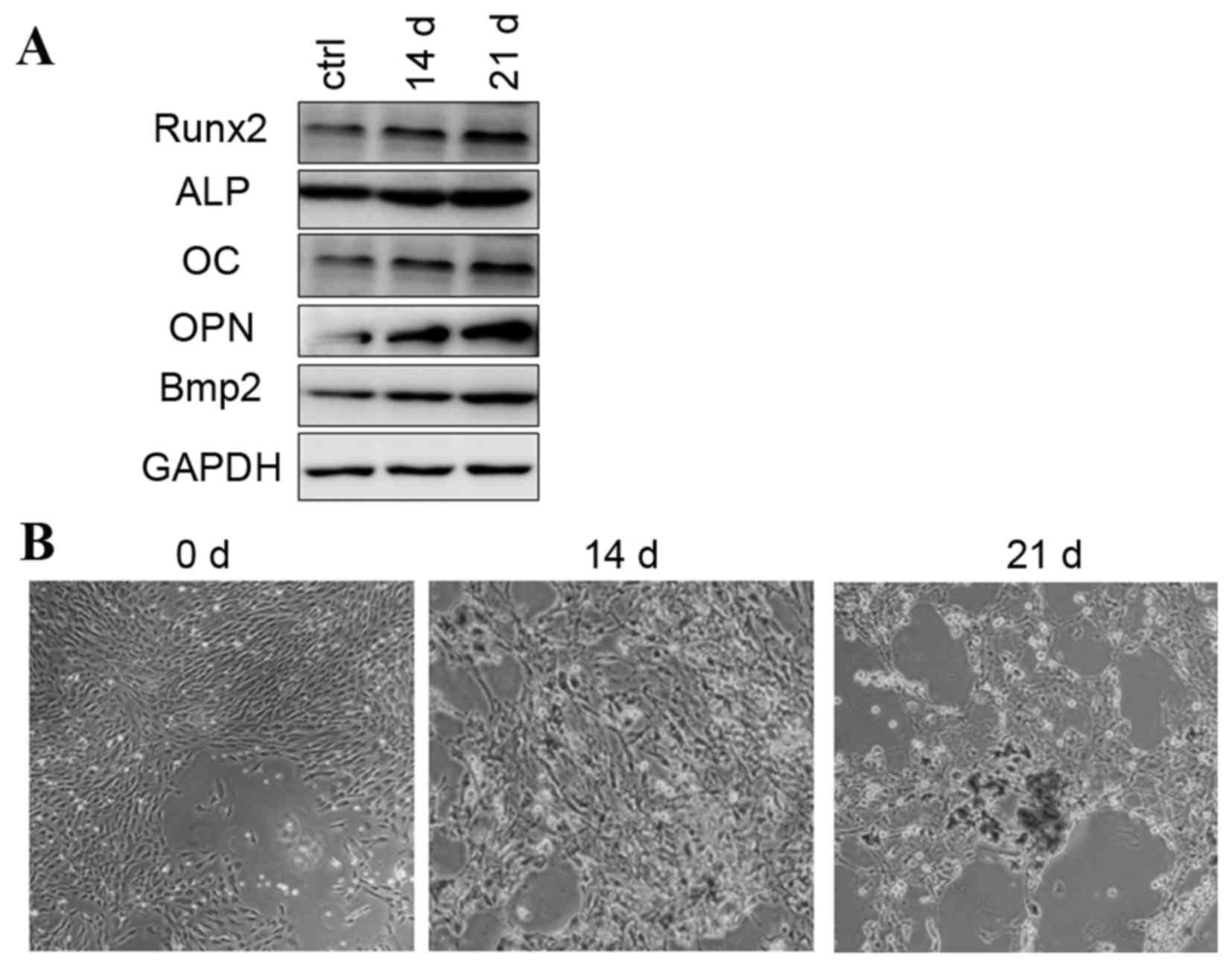 | Figure 1.Osteogenic differentiation was induced
by osteogenic-induction medium. (A) Protein expression of Runx2,
ALP, OC, OPN and Bmp2 during osteogenic differentiation. (B)
Alizarin red staining images are presented at 0, 14 and 21 days.
ALP, alkaline phosphatase; OC, osteocalcin; OPN, osteopontin; Bmp2,
bone morphogenetic protein 2 precursor; ctrl, control; Runx2, runt
related transcription factor 2. |
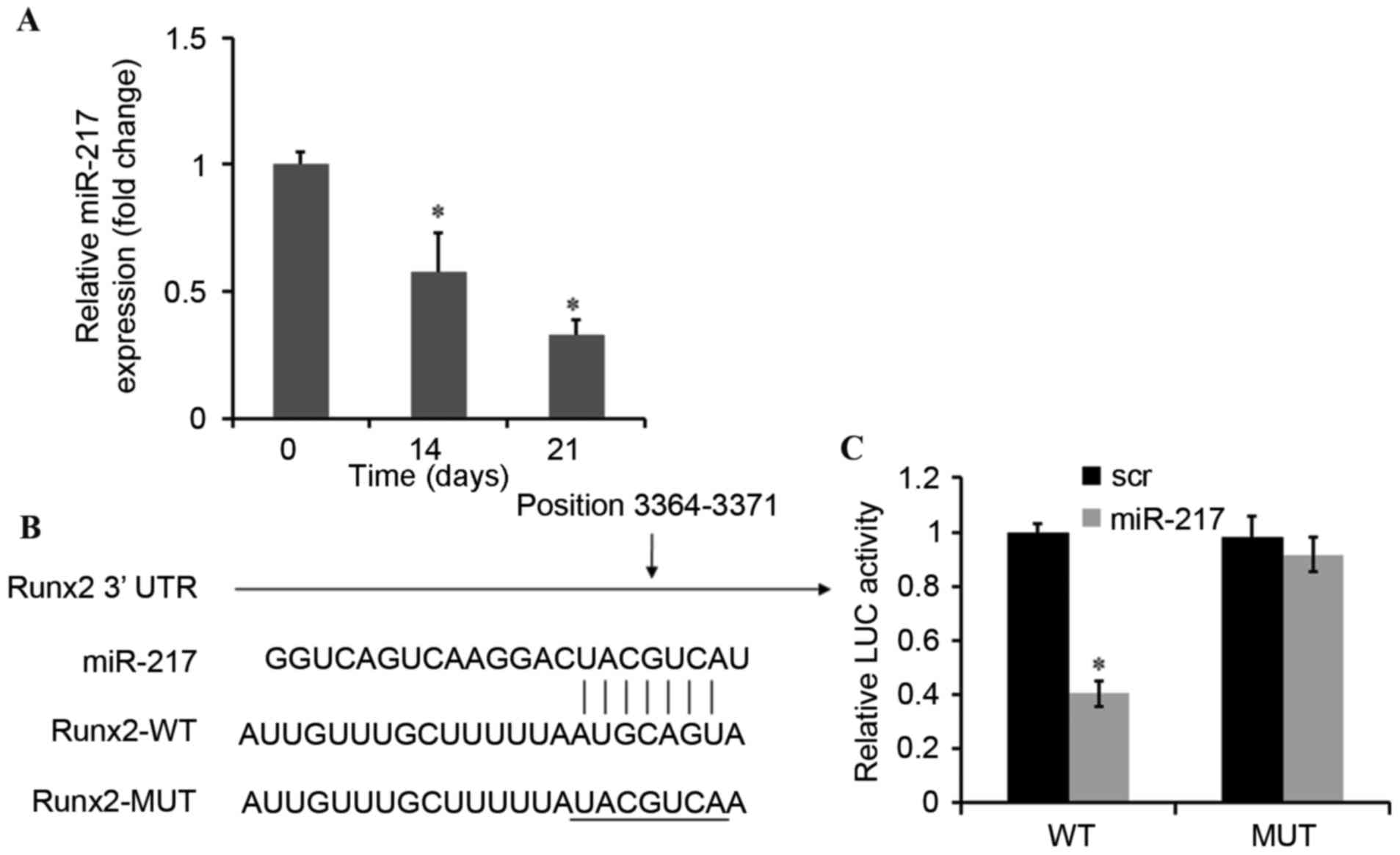
miR-217 directly represses Runx2
expression in BMSCs
The expression of miR-217 was significantly
decreased in a time-dependent manner during osteogenic
differentiation of BMSCs (Fig.
2A). Bioinformatic analyses were then conducted to predict the
target genes of miR-217 using Targetscan (www.targetscan.org) and miRanda (www.microrna.org) (19). The analysis of the 3′-UTR of the
Runx2 mRNA revealed potential binding sites for miR-217, which
suggested the existence of a regulatory association between miR-217
and Runx2 (Fig. 2B). To assess
whether Runx2 is a direct target of miR-217, the dual-luciferase
reporter gene assay was performed. The results demonstrated that
upregulation of miR-217 may decrease the luciferase activity of the
WT-Runx2–3′-UTRs of Runx2 and the luciferase activity was observed
to be ~40% compared with negative control. Furthermore, no
significant effect was observed on the Mut-Runx2–3′-UTRs (Fig. 2C). These results suggested that
miR-217 mimics may inhibit Runx2 expression via a pairing with the
Runx2 3′-UTR binding site.
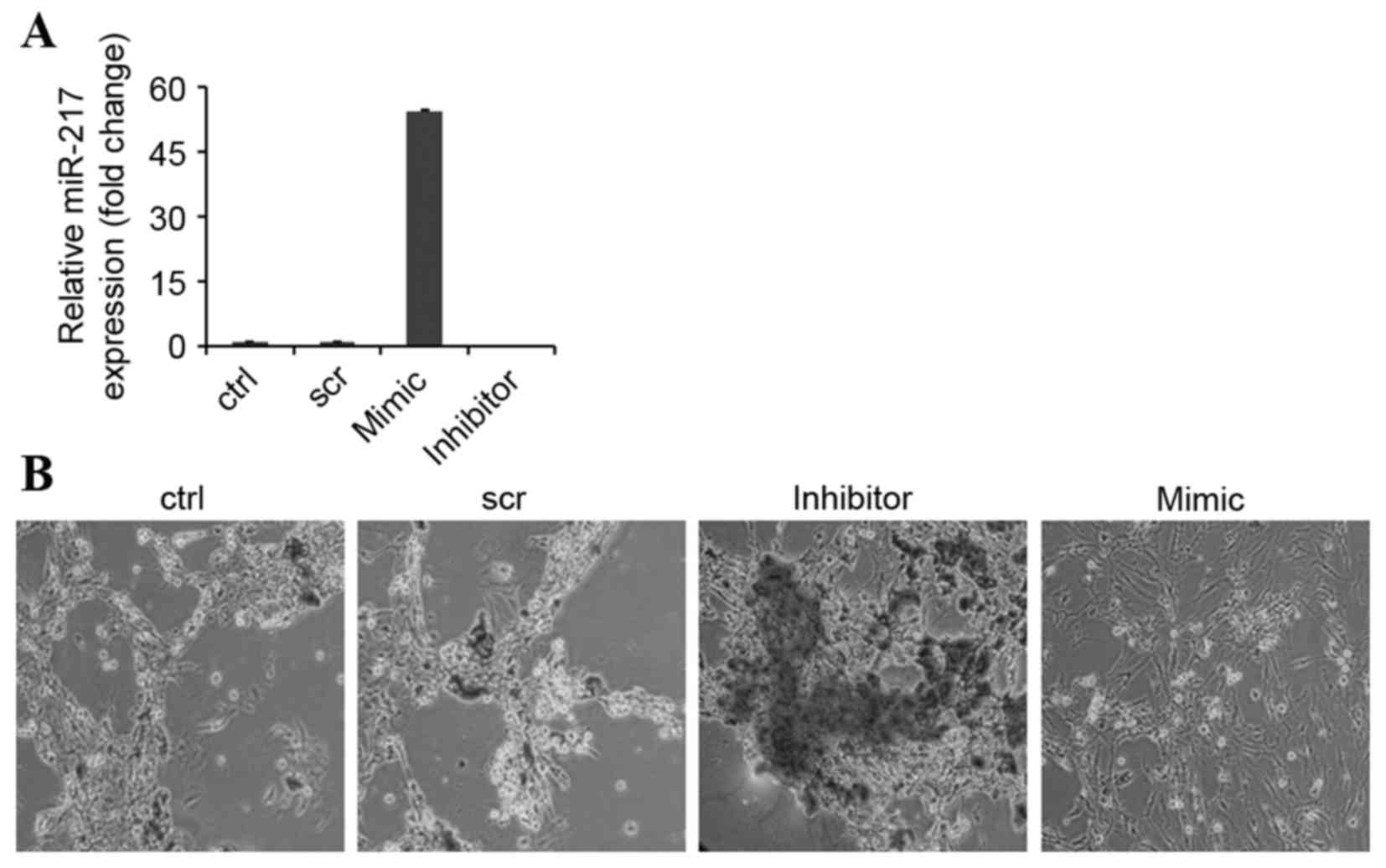
miR-217 inhibits osteogenic
differentiation
To investigate the role of miR-217 in osteogenic
differentiation, BMSCs were induced to differentiate into
osteoblasts following transfection with miR-217 mimics or
anti-miR-217 (Fig. 3A). Inhibiting
miR-217 dramatically enhanced osteoblastic differentiation, which
was indicated by enhanced in vitro matrix mineralization
visualized by alizarin red staining (Fig. 3B). By contrast, matrix
mineralization was observed to be reduced in miR-217
mimics-transfected BMSCs and cells transfected with negative
control (Fig. 3B). In addition,
the expression levels of the osteoblast-specific genes Runx2, ALP,
OC, OPN and Bmp2 appeared to be increased by inhibition of miR-217,
and decreased in the mimic and control groups (Fig. 4A).
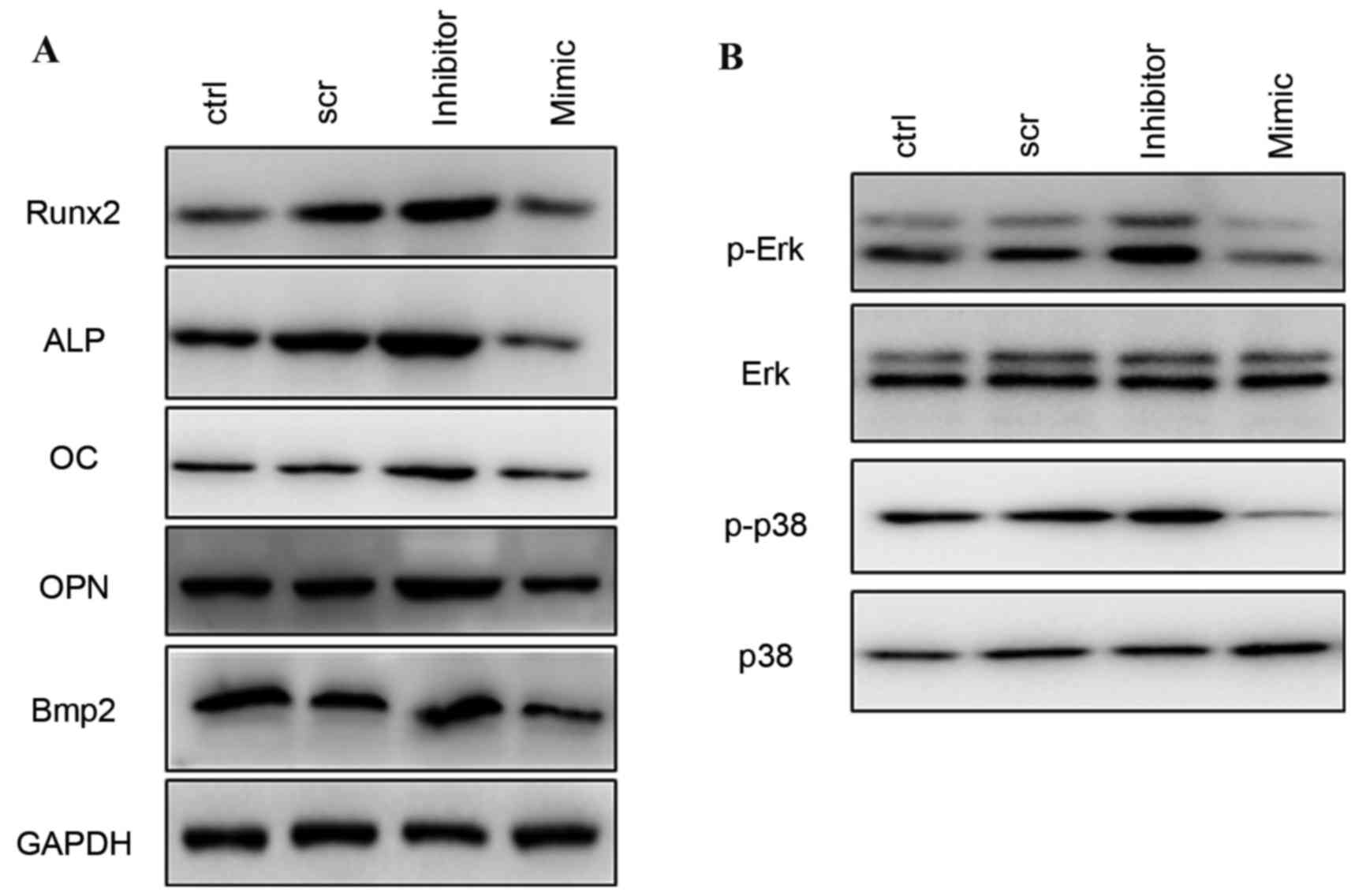 | Figure 4.Western blotting of osteogenic
differentiation-associated proteins. (A) Protein expression levels
of Runx2, ALP, OC, OPN, and Bmp2 following treatment with scr,
miR-217 inhibitor or miR-217 mimics. (B) Phosphorylation levels of
ERK and p38 MAPK following transfection with scr, miR-217 inhibitor
or mimics. ERK, extracellular signal-regulated kinase; MAPK,
mitogen-activated protein kinase; scr, scrambled; ctrl, control;
miRNA, microRNA; ALP, alkaline phosphatase; OC, osteocalcin; OPN,
osteopontin; Bmp2, bone morphogenetic protein 2 precursor, ctrl,
control; Runx2, runt related transcription factor 2. |
Silencing of miR-217 increases ERK and
p38 activation
To investigate the mechanism involved in the
regulation of osteogenic differentiation by miR-217, the present
study detected the phosphorylation level of ERK and p38 MAPK in
response to osteogenic stimulation. Following transfection with
negative control, miR-217 inhibitor or mimics for 48 h, it was
observed that inhibition of miR-217 markedly increased
phosphorylation of ERK1/2 and p38 MAPK, whereas overexpression of
miR-217 decreased phosphorylated ERK1/2 and p38 MAPK during
osteogenic differentiation (Fig.
4B).
Discussion
BMSCs are multipotent cells that, when under the
appropriate conditions, differentiate into osteoblastic cells. The
present study identified miR-217 as a negative regulator of BMSC
osteogenic differentiation via inhibition of the expression of
Runx2, and the underlying molecular mechanism may be mediated in
part by the ERK and p38 MAPK signaling pathways.
Deregulation of miRNA mediated mechanisms is
pathologically associated with osteoporosis and other
bone-associated diseases (20,21).
Increasing evidence in recent years has demonstrated that miRNA are
important regulators in osteogenic differentiation of BMSCs
(22–25). The present study demonstrated that
miR-217 is downregulated during osteogenic differentiation.
Inhibition of miR-217 function promoted osteogenic differentiation
of BMSCs, whereas overexpression of miR-217 attenuated it. These
findings suggest that miR-217 is important in bone formation by
negatively regulating osteogenic differentiation.
To further investigate the underlying molecular
mechanism of miR-217 in regulating osteoblast differentiation of
BMSCs, the present study searched for potential target genes with
an established or potential function in osteogenesis. Notably, a
match between the miR-217 seed region and the 3′ UTR of Runx2 was
identified. It was subsequently demonstrated that miR-217
overexpression results in downregulation of Runx2, whereas
inhibition of miR-217 reverses this effect, suggesting that Runx2
is regulated by miR-217 during osteogenic differentiation. Runx2 is
a key transcription factor associated with osteogenic
differentiation. Targeted disruption of Runx2 in mice results in
the maturational arrest of osteoblasts and a complete lack of
mineralized bone (26,27). The epigenetic functions of Runx2
regulate expression of bone-associated genes (28), which accounts for the observed
miR-217-induced downregulation of ALP, OC, OPN, and Bmp2. miR-217
was a primary inhibitor of osteoblastic differentiation by directly
targeting Runx2.
Experimental evidence suggests that the MAPK
signaling pathway is essential during the initiation stage of
osteogenic differentiation (29).
Critical members of the MAPKs include ERK1/2, which is an essential
molecule for mechanotransduction (30). Runx2 is phosphorylated and
activated by the ERK1/2 signaling pathways (31). However, the association between
miR-217 and the ERK/p38 pathways remains to be elucidated.
Therefore, the present study further detected the phosphorylation
of ERK and p38 MAPK in BMSCs. Upregulation of miR-217 resulted in a
decrease in phosphorylation of ERK and p38 MAPK, suggesting that
miR-217 may negatively regulate osteogenic differentiation via the
MAPK/ERK signaling pathway. The present study therefore
hypothesized that miR-217 may negatively regulate osteogenic
differentiation in BMSCs, via alteration of the phosphorylation of
ERK and p38 MAPK.
In conclusion, the results demonstrated that the
expression of miR-217 was decreased during osteogenic
differentiation of rat BMSCs and miR-217 inhibited osteogenic
differentiation via directly suppressing Runx2 expression and
thereby inhibiting expression of osteoblast-associated genes.
Furthermore, the possible mechanisms may partly be mediated by the
ERK and p38 MAPK pathways. These observations are novel in
describing the role of miRNAs in osteoblast differentiation.
References
|
1
|
Corral DA, Amling M, Priemel M, Loyer E,
Fuchs S, Ducy P, Baron R and Karsenty G: Dissociation between bone
resorption and bone formation in osteopenic transgenic mice. Proc
Natl Acad Sci USA. 95:13835–13840. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Charbord P: Bone marrow mesenchymal stem
cells: Historical overview and concepts. Hum Gene Ther.
21:1045–1056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sacchetti B, Funari A, Michienzi S, Di
Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG,
Riminucci M and Bianco P: Self-renewing osteoprogenitors in bone
marrow sinusoids can organize a hematopoietic microenvironment.
Cell. 131:324–336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mathews S, Bhonde R, Gupta PK and Totey S:
Extracellular matrix protein mediated regulation of the osteoblast
differentiation of bone marrow derived human mesenchymal stem
cells. Differentiation. 84:185–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khosla S, Westendorf JJ and Oursler MJ:
Building bone to reverse osteoporosis and repair fractures. J Clin
Invest. 118:421–428. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghildiyal M and Zamore PD: Small silencing
RNAs: An expanding universe. Nat Rev Genet. 10:94–108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu T, Zhou H, Hong Y, Li J, Jiang X and
Huang H: miR-30 family members negatively regulate osteoblast
differentiation. J Biol Chem. 287:7503–7511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar
G, Ajita J, Rhee Y, Kim CH and Lim SK: miR-182 is a negative
regulator of osteoblast proliferation, differentiation, and
skeletogenesis through targeting FoxO1. J Bone Miner Res.
27:1669–1679. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Q, Cai J, Cai XH and Chen L: miR-346
regulates osteogenic differentiation of human bone marrow-derived
mesenchymal stem cells by targeting the Wnt/β-catenin pathway. PLoS
One. 8:e722662013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei R, Deng Z and Su J: miR-217 targeting
Wnt5a in osteosarcoma functions as a potential tumor suppressor.
Biomed Pharmacother. 72:158–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng
ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al:
microRNA-217 inhibits tumor progression and metastasis by
downregulating EZH2 and predicts favorable prognosis in gastric
cancer. Oncotarget. 6:10868–10879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM and
Chen J: The miR-217 microRNA functions as a potential tumor
suppressor in pancreatic ductal adenocarcinoma by targeting KRAS.
Carcinogenesis. 31:1726–1733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Zhao J, Zhang JW, Huang QY, Huang
JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ and Ma WM: MicroRNA-217,
down-regulated in clear cell renal cell carcinoma and associated
with lower survival, suppresses cell proliferation and migration.
Neoplasma. 60:511–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan GZ, Yang Y, Zhang J, Liu W, Wang GY,
Zhang YC, Yang Q, Zhai FX, Tai Y, Liu JR, et al: Bone marrow
mesenchymal stem cells ameliorate hepatic ischemia/reperfusion
injuries via inactivation of the MEK/ERK signaling pathway in rats.
J Surg Res. 178:935–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qu B, Xia X, Wu HH, Tu CQ and Pan XM:
PDGF-regulated miRNA-138 inhibits the osteogenic differentiation of
mesenchymal stem cells. Biochem Biophys Res Commun. 448:241–247.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu L, Liu Y, Hou Y, Wang K, Wong Y, Lin S
and Li G: U0126 promotes osteogenesis of rat bone-marrow-derived
mesenchymal stem cells by activating BMP/Smad signaling pathway.
Cell Tissue Res. 359:537–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krzeszinski JY, Wei W, Huynh H, Jin Z,
Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et
al: miR-34a blocks osteoporosis and bone metastasis by inhibiting
osteoclastogenesis and Tgif2. Nature. 512:431–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garnero P: New developments in biological
markers of bone metabolism in osteoporosis. Bone. 66:46–55. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qadir AS, Um S, Lee H, Baek K, Seo BM, Lee
G, Kim GS, Woo KM, Ryoo HM and Baek JH: miR-124 negatively
regulates osteogenic differentiation and in vivo bone formation of
mesenchymal stem cells. J Cell Biochem. 116:730–742. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huszar JM and Payne CJ: MIR146A inhibits
JMJD3 expression and osteogenic differentiation in human
mesenchymal stem cells. FEBS Lett. 588:1850–1856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang K, Fu J, Zhou W, Li W, Dong S, Yu S,
Hu Z, Wang H and Xie Z: MicroRNA-125b regulates osteogenic
differentiation of mesenchymal stem cells by targeting Cbfβ in
vitro. Biochimie. 102:47–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y,
Gu P and Fan X: Effects of a miR-31, Runx2, and Satb2 regulatory
loop on the osteogenic differentiation of bone mesenchymal stem
cells. Stem Cells Dev. 22:2278–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturational arrest of osteoblasts. Cell.
89:755–764. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshida CA, Furuichi T, Fujita T, Fukuyama
R, Kanatani N, Kobayashi S, Satake M, Takada K and Komori T:
Core-binding factor beta interacts with Runx2 and is required for
skeletal development. Nat Genet. 32:633–638. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lian JB and Stein GS: Runx2/Cbfa1: A
multifunctional regulator of bone formation. Curr Pharm Des.
9:2677–2685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lai CF, Chaudhary L, Fausto A, Halstead
LR, Ory DS, Avioli LV and Cheng SL: Erk is essential for growth,
differentiation, integrin expression, and cell function in human
osteoblastic cells. J Biol Chem. 276:14443–14450. 2001.PubMed/NCBI
|
|
30
|
Simmons CA, Matlis S, Thornton AJ, Chen S,
Wang CY and Mooney DJ: Cyclic strain enhances matrix mineralization
by adult human mesenchymal stem cells via the extracellular
signal-regulated kinase (ERK1/2) signaling pathway. J Biomech.
36:1087–1096. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao G, Jiang D, Thomas P, Benson MD, Guan
K, Karsenty G and Franceschi RT: MAPK pathways activate and
phosphorylate the osteoblast-specific transcription factor, Cbfa1.
J Biol Chem. 275:4453–4459. 2000. View Article : Google Scholar : PubMed/NCBI
|