Introduction
Hepatitis B virus (HBV) infection is one of the
major etiological factors of HCC in China (1). Almost 360,000,000 individuals are
chronic HBV carriers worldwide (2). HBV infection can lead to liver
diseases, including hepatitis, liver cirrhosis and hepatocellular
carcinoma (HCC) (3). The HBV
genome has a partially double stranded circular structure,
consisting of four overlapping open reading frames (ORFs) (4) encoding the virus polymerase, S
protein, X protein and core protein, respectively. The ORF of
polymerase contains four regions: A carboxy terminal region
(nt2307-2840), spacer region (nt2841-129), reverse transcriptase
(RT) region (nt130-1161) and an RNase H region (nt1129-1621)
(5), of which the RT region is
crucial for HBV replication. Several mutations occur in the HBV
genome due to deficiency in the proofreading function in RT and HBV
replication through RNA-intermediated reverse transcription.
Therefore, in the course of HBV infection, mutations continuously
accumulate, and a number of these mutations may be used as viral
markers for evaluating the development and prognosis of
HBV-associated HCC.
Numerous studies in previous decades have focused on
the association between HBV mutations, including point mutations,
deletions and structure variation, and the risk of HCC. It has been
shown that nucleotide mutations in the S gene and pre C/C gene are
closely associated with increasing risks of HCC (6–9). In
the basal core promoter/enhancer II region, A1762T/G1764A has been
found to be significantly associated with HCC (10–12).
Increasing studies are focusing on the association between HBV gene
variation and the prognosis with HCC. Yeh et al (13) reported that the presence of the
A1762T/G1764A mutation in liver tissue within the BCP was an
independent predictor for disease-free survival (DFS) and overall
survival (OS) rates in HCC. A pre-S deletion located between codons
107 and 141 was found to be associated with poorer postoperative
prognosis, and Su et al (14) showed that the pre-S deletion was
crucial for post-operative tumor recurrence. However, studies
investigating mutations in RT associated with the HCC prognosis are
limited.
In the present study, HBV DNA was extracted from
liver tissues of patients with HCC, and viral quasispecies within
the RT/S region (RT overlapped with the S gene) were analyzed using
Sanger sequencing. Cox proportional hazard model analysis was used
to investigate the association between variations in the HBV RT/S
region and the prognosis of HCC.
Materials and methods
Patients and samples
A total of 84 patients with HCC were recruited
between March 2007 and May 2009, who received complete surgical
resection at the Eastern Hepatobiliary Surgery Hospital (Shanghai,
China). Serum samples, tumor tissue (TT) and paired adjacent
non-tumor tissue (ANTT) samples were collected. Written informed
consent was obtained from all patients. The present study was
approved by the Ethics Committee of Human Resources at the Second
Military Medical University (Shanghai, China).
Patients were included in the cohort for examination
if they fulfilled following inclusion criteria: i) serum hepatitis
B surface antigen (HBsAg)-positive for at least 6 months; ii) HBV
DNA levels >1,000 IU/ml; iii) nucleos(t)ide analogue (NA) naïve
prior to surgical resection. The exclusion criteria included
hepatitis C virus or human immunodeficiency virus co-infection, or
a history of liver transplantation, autoimmune liver diseases,
metastatic liver cancer, other malignancies, drug-associated liver
diseases, alcoholic hepatitis or other causes of chronic liver
disease diagnosed prior to enrollment.
HBV nucleic acid extraction and
polymerase chain reaction (PCR) amplification
HBV genomes were extracted from frozen TTs and ANTTs
with a QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. Due to the limitation of
sequencing length, the RT region was amplified as two overlapping
segments, respectively. The primers for the first segment were as
follows: Forward 5′-CTGCTGGTGGCTCCAGTTC-3′ (nucleotides 57–75) and
reverse 5′-TGGCTCAGTTTACTAGTGCCA-3′ (nucleotides 668–688). The
primers for the second segment were as follows: Forward
5′-TCAGTCCGTTTCTCCTGGCTCAG-3′ (nucleotides 653–675) and reverse
5′-GAGTTCCGCAGTATGGATCG-3′ (nucleotides 1,281–1,262). The RT
segments were amplified with Phusion High-Fidelity DNA polymerase
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). The final
composition of 20 µl PCR mixtures contained 5 µl DNA template, 0.5
µl Phusion High-Fidelity DNA polymerase, 4 µl 5X Phusion HF buffer,
0.5 µl 10 mM dNTPs, 1 µl forward primer (1 µM), 1 µl reverse primer
(1 µM) and 8 µl DNase-Free water. The RT segments were amplified
with the following thermocycling conditions: 95°C for 5 min
followed by 35 cycles of 98°C for 30 sec, 57°C for 30 sec and
extension at 72°C for 1 min. The mixtures were subjected to further
extension at 72°C for 10 min. The TaqMan probes for covalently
closed circular (ccc) DNA and intrahepatic HBV total DNA (tDNA)
quantification were FAM-ATC TGC CGG ACC GTG TGC -TAMARA and FAM-CTC
ACC AAC CTC CTG TCC TCC A-TAMARA, respectively.
Sanger sequencing and sequence
alignment
The products of the RT region segmented PCR
amplification were gel purified and sequenced using an ABI PRISM
BigDye sequencing kit on an ABI 3500 genetic analyzer (Applied
Biosystens; Thermo Fisher Scientific, Inc.). HBV genotypes were
identified using an online genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi).
The sequences from each of the samples were compared with the RT
sequence in the NCBI database (HBVgp1; NC_003977.2), which was
performed using DNAMAN software (version 4.0; Lynnon Corporation,
Pointe-Claire, QC, Canada).
Statistical analysis
Mutations between TT and ANTT were analyzed using
the χ2 test. Forward stepwise multivariate regression
analysis was performed to obtain the hazard ratios of potential
risk factors for HCC prognosis. Matched clinicopathological
characteristics were subjected to stratified analysis. P<0.05
was considered to indicate a statistically significant difference.
All statistical analyses were performed on SPSS software (version
18.0; SPSS, Inc., Chicago, IL, USA).
Results
Patient characteristics
The baseline characteristics of the 84 patients
included in the present study are presented in Table I. The cohort, comprised of 72 (86%)
male subjects, had a median age of 50 years, and 74% were infected
with HBV genotype C. Over half of the patients (58%) were
histologically diagnosed with early stage tumors
(tumor-node-metastasis stages I–II). In addition, the median
duration of HCC recurrence was 10.0 months (range 0.43–50.9
months).
 | Table I.Clinical characteristics of patients
with HBV-associated hepatocellular carcinoma. |
Table I.
Clinical characteristics of patients
with HBV-associated hepatocellular carcinoma.
| Characteristic | Cohort (n=84) | P-value |
|---|
| Gender |
|
|
| Male | 72 (86%) |
|
|
Female | 12 (14%) |
|
| Age (years) | 50 (28–70) |
|
| AFP (ng/ml) | 1,067
(1.5->1,210) |
|
| TBIL (µmol/l) | 14.7 (7.1–50.5) |
|
| DBIL (µmol/l) | 5.4 (1.7–18.4) |
|
| ALT (U/l) | 49 (21–1,067) |
|
| AST (U/l) | 51 (22–1041) |
|
| Serum HBV
DNA(log10IU/ml) | 5.2 (3.0–7.6) |
|
| HBV tDNA
(log10copies/106cells) |
| 0.029 |
| TT | 6.6±1.2 |
|
|
ANTT | 7.0±0.9 |
|
| cccDNA
(log10copies/106cells) |
| 0.544 |
| TT | 4.9±1.4 |
|
|
ANTT | 5.0±1.0 |
|
| HBsAg
(log10IU/ml) | 3.1
(0.32–4.11) |
|
| Tumor size
(cm) | 8.0 (1.1–25.0) |
|
| HBeAg |
|
|
|
Positive | 40 (48%) |
|
|
Negative | 44 (52%) |
|
| HBV genotype |
|
|
| B | 22 (26%) |
|
| C | 62 (74%) |
|
| Ascites |
|
|
|
Yes | 13 (15%) |
|
| No | 67 (80%) |
|
| Tumor number |
|
|
|
Single | 59 (70%) |
|
|
Multiple | 22 (26%) |
|
| Cirrhosis |
|
|
|
Present | 72 (86%) |
|
|
Absent | 12 (14%) |
|
| Capsule |
|
|
|
Complete | 23 (27%) |
|
|
Incomplete | 47 (56%) |
|
| TNM stage |
|
|
|
I–II | 49 (58%) |
|
|
III–IV | 23 (27%) |
|
Association between intrahepatic HBV
DNA and serum HBV DNA
The levels of HBV tDNA in the ANTTs were higher,
compared with those in the TTs (7.0±0.9, vs. 6.6±1.2
log10copies/106cells; P=0.029; Table I). Serum HBV DNA was moderately
correlated with ANTT tDNA (r=0.419; P<0.001) and cccDNA
(r=0.370; P<0.001), but not with TT tDNA (r=0.154; P=0.166) or
cccDNA (r=0.123; P=0.281). These results suggested that serum HBV
DNA may derive from ANTTs, rather than TTs, therefore, serum HBV
DNA levels may not reflect the real level of HBV DNA in TTs.
Characteristics of amino acid
mutations in RT and HBsAg
According to the criteria of 5% frequency, a total
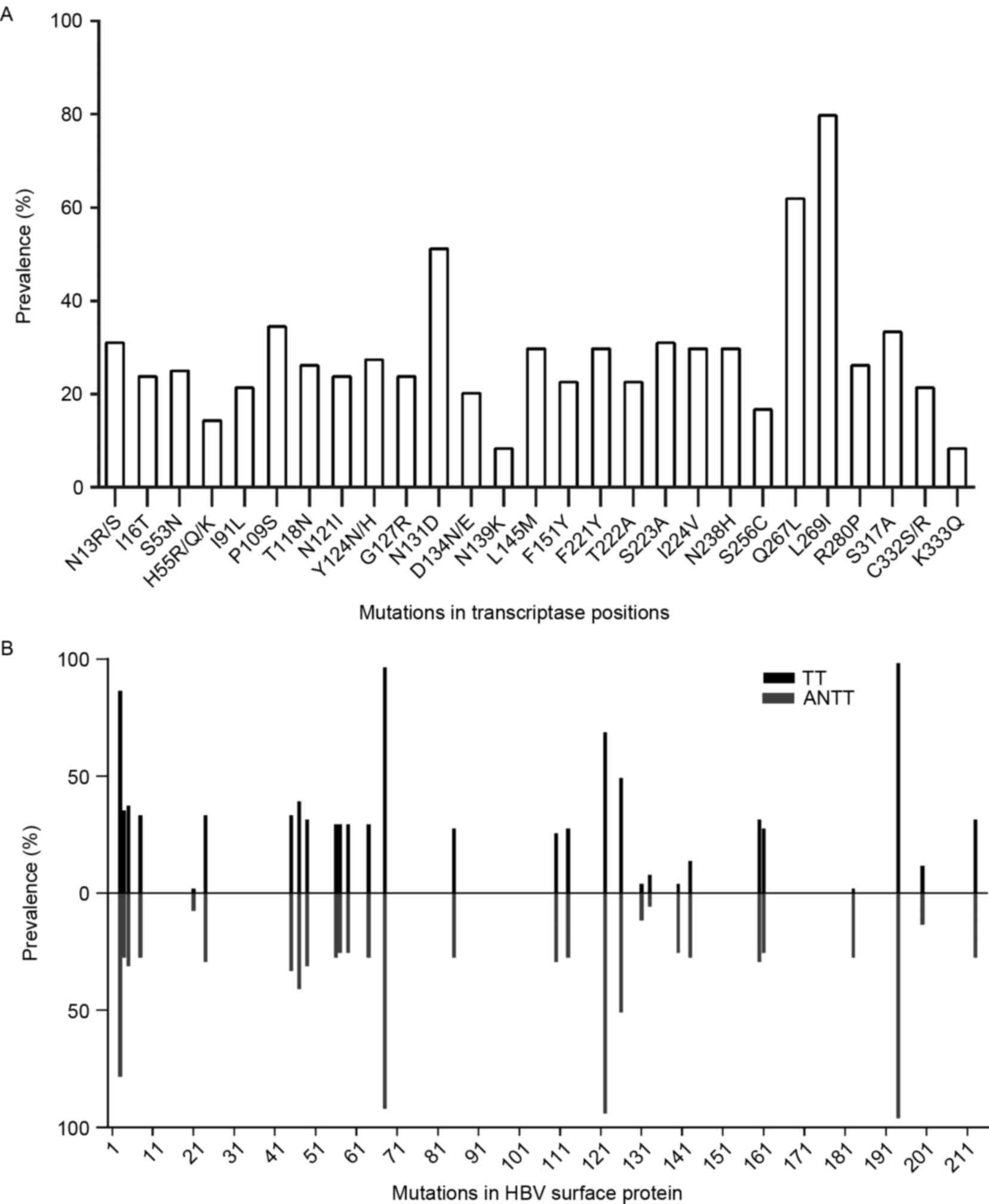
of 27 amino acid mutations in the RT domain were included (Fig. 1A). The majority of these mutations
occurred with a frequency of >20%, particularly rtQ267 L and
rtL269I, which had frequencies >50%. Common NA resistance
mutations, including rtI169T, rtA181T/V, rtT184A/C/F/G/I/L/M/S,
rtA194T, rtS202C/G/I, rtM204I/V/S, rtN236T, rtM250I/L/V, rtV173 L
and rtL80I/V were not detected during sequence alignment. The
substitutions of rtS53N, rtI91L, rtF221Y and rtN238H detected were
putative NA-resistance mutations. The pretreatment mutations,
rtY124 N/H and rtN139K, were also found in the cohort. As the HBV S
gene overlaps with the RT gene, the S gene originating from 51
pairs of TTs and ANTTs were analyzed, and 29 amino acid mutations
within HBsAg met the 5% frequency inclusion criteria (Fig. 1B). The frequencies of sA194V,
sT68I, sR122K, sS3N and sI126T were >50%, and 10 amino acid
mutations were located in the major hydrophilic region (MHR;
aa99-169) of HBsAg. No significant differences were found in HBsAg
mutations between TTs and ANTTs, with the exception of three
mutations, sR122 K (P=0.004), sT140S (P=0.001) and sF183V
(P=0.001), which occurred more frequently in ANTTs.
Mutations associated with the
prognosis of HCC
The Cox proportional hazard model was used to
analyze the association between clinicopathological and virological
factors associated with DFS and OS following surgical resection of
HBV-associated HCC (Table II).
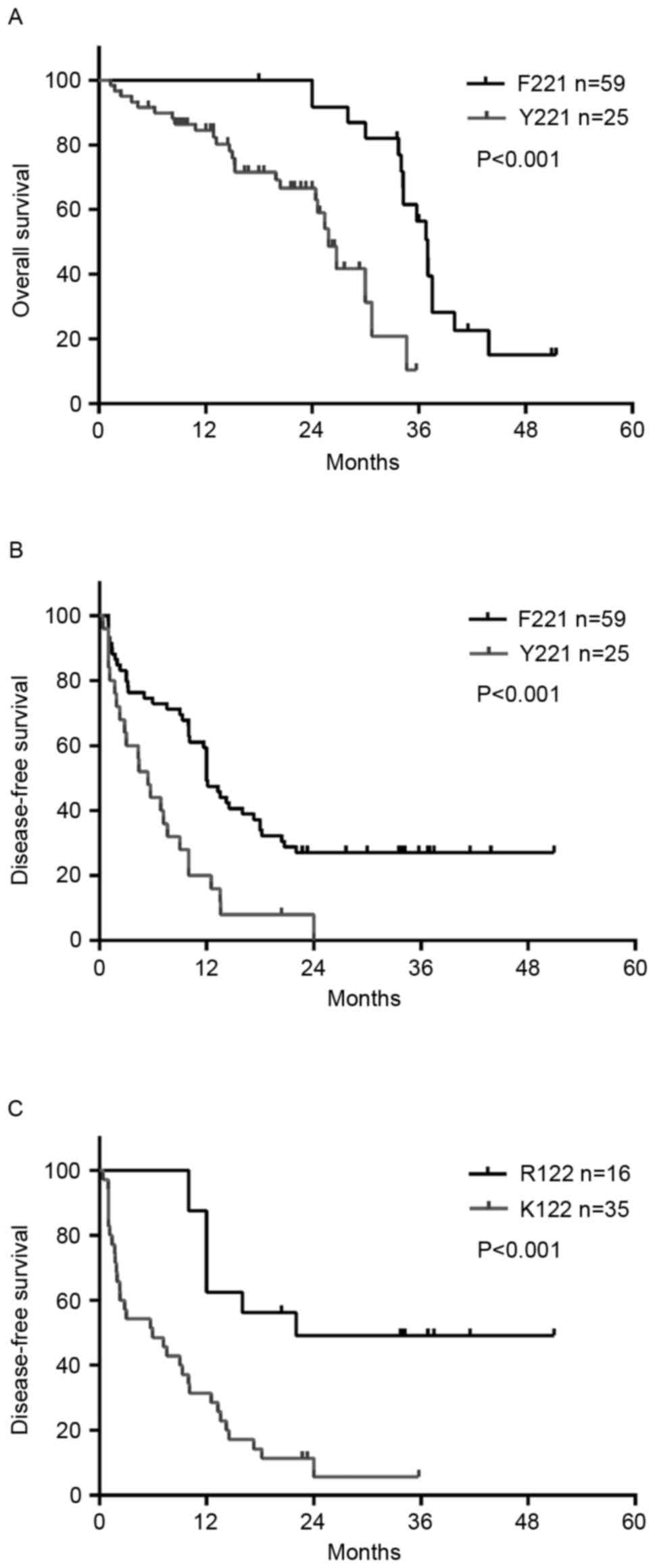
Tumor sizes >8 cm (HR=2.345; P=0.001) and rtF221Y (HR=1.838;
P=0.028) were associated with shorter DFS. rtF221Y (HR=2.557;
P=0.004) was also found to be an independent risk factor for OS.
Kaplan-Meier survival analysis indicated that rtF221Y was
significantly associated with poorer DFS (P=0.0027) and OS
(P<0.001; Fig. 2A and B). The
DFS and OS rates were significantly shorter in those with the
rtF221Y mutation, compared with those with the wild-type amino
acid. In addition, when Kaplan-Meier survival analysis and log-rank
tests were used to compare survival probability, it was found that
the HBsAg R122 K mutation in the TT was closely associated with
tumor recurrence (P<0.001; Fig.
2C) and patients with the sR122 K mutation had a higher rate of
recurrence (P=0.002), which was determined using the χ2
test.
 | Table II.Univariate and multivariate analyses
of clinicopathologial and virological characteristic for DFS and OS
in patients with HBV-associated hepatocellular carcinoma. |
Table II.
Univariate and multivariate analyses
of clinicopathologial and virological characteristic for DFS and OS
in patients with HBV-associated hepatocellular carcinoma.
|
| DFS | OS |
|---|
|
|
|
|
|---|
|
| Univariate | Multivariate | Univariate | Multivariate |
|---|
|
|
|
|
|
|
|---|
| Characteristic | P-value | HR | (95% CI) | P-value | P-value | HR | (95% CI) | P-value |
|---|
| Age >50
years | 0.717 |
|
|
| 0.985 |
|
|
|
| Gender (male) | 0.680 |
|
|
| 0.952 |
|
|
|
| AFP >400
ng/ml | 0.736 |
|
|
| 0.735 |
|
|
|
| ALT >40 U/l | 0.274 |
|
|
| 0.475 |
|
|
|
| Tumor size >8
cm | <0.001 | 2.345 | 1.391–3.953 | 0.001 | 0.688 |
|
|
|
| HBV DNA
(log10 IU/ml) | 0.497 |
|
|
| 0.375 |
|
|
|
| ANTT HBV tDNA
(log10copies/106cells) | 0.673 |
|
|
| 0.452 |
|
|
|
| ANTT HBV cccDNA
(log10copies/106cells) | 0.892 |
|
|
| 0.695 |
|
|
|
| N13R/S | 0.024 |
|
|
| 0.130 |
|
|
|
| I16T | 0.002 |
|
|
| 0.015 |
|
|
|
| S53N | 0.002 |
|
|
| 0.020 |
|
|
|
| H55R/Q/K | 0.509 |
|
|
| 0.056 |
|
|
|
| I91L | <0.001 |
|
|
| 0.017 |
|
|
|
| P109S | 0.419 |
|
|
| 0.612 |
|
|
|
| T118N | 0.118 |
|
|
| 0.321 |
|
|
|
| N121I | 0.023 |
|
|
| 0.084 |
|
|
|
| Y124N/H | 0.004 |
|
|
| 0.160 |
|
|
|
| G127R | 0.005 |
|
|
| 0.084 |
|
|
|
| N131D | 0.001 |
|
|
| 0.058 |
|
|
|
| D134N/E | 0.994 |
|
|
| 0.405 |
|
|
|
| N139K | 0.211 |
|
|
| 0.197 |
|
|
|
| L145M | 0.190 |
|
|
| 0.119 |
|
|
|
| F151Y | 0.003 |
|
|
| 0.033 |
|
|
|
| F221Y | <0.001 | 1.838 | 1.069–3.161 | 0.028 | 0.003 | 2.557 | 1.344–4.866 | 0.004 |
| T222A | 0.003 |
|
|
| 0.033 |
|
|
|
| S223A | 0.210 |
|
|
| 0.311 |
|
|
|
| I224V | 0.784 |
|
|
| 0.638 |
|
|
|
| N238H | 0.272 |
|
|
| 0.543 |
|
|
|
| S256C | 0.959 |
|
|
| 0.092 |
|
|
|
| Q267L | 0.146 |
|
|
| 0.679 |
|
|
|
| L269I | 0.012 |
|
|
| 0.272 |
|
|
|
| R280P | 0.077 |
|
|
| 0.097 |
|
|
|
| S317A | 0.027 |
|
|
| 0.056 |
|
|
|
| C332S/R | 0.007 |
|
|
| 0.085 |
|
|
|
| K333Q | 0.002 |
|
|
| 0.154 |
|
|
|
Association between HBV mutations and
the clinical characteristics of patients with HCC
The levels of α-fetoprotein (AFP) in patients with
the F221Y mutation were higher, compared with those without the
mutation (P=0.048). In addition, patients harboring the F221Y
mutation had larger tumor sizes, compared with patients without the
F221Y mutation (P=0.001; Table
III). No significant differences in age, alanine
aminotransferase (ALT), aspartate aminotransferase,
γ-glutamyltransferase, serum HBV DNA levels or tumor numbers were
found between the patients with or without the F221Y mutation.
 | Table III.Comparison of clinical
characteristics of patients with and without the rtF221Y
mutation. |
Table III.
Comparison of clinical
characteristics of patients with and without the rtF221Y
mutation.
| Characteristic | MT (n=25) | WT (n=59) | P-value |
|---|
| Age (years) | 49.2±8.5 | 49.2±9.8 | 0.748 |
| ALT (U/l) | 50
(21.2–360.5) | 49
(21.4–1,067.5) | 0.773 |
| DBIL (µmol/l) | 4.9±1.8 | 6.4±3.0 | 0.021 |
| AFP (ng/ml) | 1,210
(39.7–1,210) | 496 (10–1,210) | 0.048 |
| CREA (µmol/l) | 71.5±11.1 | 65.6±9.5 | 0.018 |
| Serum HBV DNA
(log10IU/ml) | 5.1±1.3 | 5.3±1.0 | 0.525 |
| ANTT tDNA
(log10copies/106cells) | 6.8±1.0 | 7.0±0.9 | 0.461 |
| ANTT
cccDNA(log10copies/106cells) | 5.3±0.9 | 4.8±1.1 | 0.090 |
| Tumor diameter
(cm) |
|
|
|
| ≤8 | 4 | 24 | 0.001 |
|
>8 | 18 | 35 |
|
| Ascites |
|
|
|
|
Yes | 3 | 10 | 0.487 |
| No | 22 | 45 |
|
| Capsule |
|
|
|
|
Complete | 15 | 32 | 0.994 |
|
Incomplete | 8 | 17 |
|
| Tumor number |
|
|
|
|
Single | 14 | 45 | 0.255 |
|
Multiple | 8 | 24 |
|
Due to the negative relevance between HBsAg
mutations in ANTTs with clinicopathological characteristics, amino
acid substitutions within HBsAg in TTs were subjected to stratified
analysis (Table IV). The results
revealed that the occurrence of the sS3 N mutation was associated
with higher TT cccDNA levels (3.96±0.7, vs. 5.09±1.0
log10copies/106cells; P=0.004). In addition,
patients with sI126T had higher AFP levels, compared with those
with wild-type HBsAg (P=0.007). Patients with sR122 K or sI126T had
larger tumor sizes (P=0.017 and P=0.049, respectively). However, no
associations were found between HBsAg mutations in the ANTTs and
the clinicopathological features.
 | Table IV.Association between TT HBsAg
mutations and patient characteristics. |
Table IV.
Association between TT HBsAg
mutations and patient characteristics.
|
| sS3N | sR122K | sI126T |
|---|
|
|
|
|
|
|---|
| Characteristic | WT n=7 | MT n=44 | P-value | WT n=16 | MT n=35 | P-value | WT n=26 | MT n=25 | P-value |
|---|
| Age (years) | 50.4 | 49 | 0.710 | 51.1 | 48.3 | 0.296 | 50.3 | 48.1 | 0.392 |
| Gender |
|
|
|
|
|
|
|
|
|
|
Male | 1 | 2 | 0.364 | 15 | 30 | 0.999 | 25 | 23 | 0.610 |
|
Female | 6 | 42 |
| 1 | 2 |
| 1 | 2 |
|
| ALT (U/l) | 148 | 65 | 0.925 | 97.1 | 70.2 | 0.948 | 95 | 58 | 0.105 |
| AFP (ng/ml) | 866 | 730 | 0.620 | 767 | 772 | 0.736 | 570 | 935 | 0.007 |
| HBsAg (S/CO) | 2,696 | 1,434 | 0.378 | 1,809 | 1,455 | 0.543 | 2,006 | 1,134 | 0.162 |
| Serum DNA | 4.86 | 5.46 | 0.201 | 5.07 | 5.51 | 0.166 | 5.53 | 5.24 | 0.354 |
|
(log10IU/ml) |
|
|
|
|
|
|
|
|
|
| HBV tDNA | 6.56 | 7.02 | 0.215 | 7.1 | 6.9 | 0.428 | 7.12 | 6.79 | 0.204 |
|
(log10copies/106cells) |
|
|
|
|
|
|
|
|
|
| cccDNA | 3.96 | 5.09 | 0.004 | 4.8 | 4.9 | 0.687 | 4.88 | 5 | 0.675 |
|
(log10copies/106cells) |
|
|
|
|
|
|
|
|
|
| Tumor size
(cm) | 9.7 | 8.4 | 0.565 | 5.9 | 9.7 | 0.017 | 7.12 | 10.04 | 0.049 |
| Recurrence |
|
|
|
|
|
|
|
|
|
|
Yes | 7 | 33 | 0.323 | 8 | 32 | 0.002 | 21 | 19 | 0.679 |
| No | 0 | 11 |
| 8 | 3 |
| 5 | 6 |
|
| Cirrhosis |
|
|
|
|
|
|
|
|
|
|
Present | 6 | 39 | 1.000 | 13 | 31 | 0.999 | 23 | 22 | 1.000 |
|
Absent | 0 | 5 |
| 1 | 4 |
| 2 | 3 |
|
| Tumor number |
|
|
|
|
|
|
|
|
|
|
Single | 4 | 33 | 0.643 | 10 | 27 | 0.493 | 18 | 19 | 0.747 |
|
Multiple | 2 | 11 |
| 5 | 8 |
| 7 | 6 |
|
| Capsule |
|
|
|
|
|
|
|
|
|
|
Complete | 3 | 26 | 1.000 | 4 | 25 | 0.019 | 14 | 15 | 0.936 |
|
Incomplete | 2 | 15 |
| 8 | 9 |
| 8 | 9 |
|
| TNM stage |
|
|
|
|
|
|
|
|
|
|
I–II | 5 | 29 | 0.573 | 9 | 25 | 0.411 | 16 | 18 | 0.734 |
|
III–IV | 0 | 10 |
| 1 | 9 |
| 4 | 6 |
|
Discussion
Although several studies have focused on the
association between HBV variation and HCC, the majority of these
have been performed using serum samples (11). Previous data have reported that
serum HBV DNA shows moderate correlation with ANTT viral DNA, but
no correlation with that of TT, which suggests that the presence of
variants in serum HBV DNA may not reveal the actual status in the
liver. Therefore, resected tissue specimens may have preponderance
when designing investigations. Furthermore, the quantitative
results of HBV tDNA and cccDNA from the liver tissues in the
present study suggested that the levels of tDNAs in ANTT were
significantly higher, compared with those in paired TT samples
(P=0.029). The microenvironment in ANTT may be more suitable for
HBV replication, and viruses in TT may have deficient replication
activity due to an unfavorable microenvironment. Therefore,
numerous missense mutations are likely to accumulate during the
replication of HBV in the ANTT. HBV DNA in TT is more likely to
integrate with the hepatocyte genome, which may also contribute to
the lower level of replication of viral DNA (15). HBV integration can be detected at
any stage of HBV infection and it is reasonable to suggest that
fewer mutations in the integrated form of the HBV genome occur in
TT, compared with the replicative form of the HBV genome in ANTT
(16,17). When the distribution of HBV
mutations between the TT and ANTT samples were compared in the
present study, there were only nuance were detected. As a result,
gene variations in the HBV genome derived from ANTT may more
suitable for investigations on the effect of HBV mutations on
HCC.
Several studies have indicated that mutations in the
S, C and X genes of the HBV genome are associated with a high risk
of HCC (18,19), however, there are few reports on
mutations in the RT region of the HBV genome, particularly in TTs
and ANTTs, and its effect on development and prognosis of HCC. The
present study focused on the genetic diversity in RT sequences
isolated from liver tissues. When the alignment was completed, 27
mutations were obtained when the occurrence rate threshold was set
as 5%. In addition, there was minimal observation of common NAs
resistance mutations in sequence alignment, including L80I/V,
I169T, V173L, A181T/V, T184A/C/F/G/I/L/M/S, A194T, S202C/G/I,
M204I/V/S, N236T and M250I/L/V (20,21),
which indicated that these variants were absent or were present at
a low frequency due to the limitations of Sanger sequencing. This
can also be attributed to the lack of regular NAs therapy. In the
present study, S53N, I91L, N139K, F221Y and N238H were identified
as putative drug resistance mutations; however, their functions
remain to be fully elucidated.
The present study demonstrated that rtF221Y was an
independent risk factor for DFS and OS in HCC. rtF221Y was also
linked to a higher level of AFP and larger tumor size. To the best
of our knowledge, the present study is the first to show that
rtF221Y was associated with HCC in a relatively large cohort of
patients. Therefore, rtF221Y may be used as a potential viral
marker for predicting HCC prognosis following surgery. However,
previous studies have suggested that rtF221Y may belong to putative
antiviral drug resistance mutations. Mirandola et al
(22) and Lee et al
(23) reported that rtF221Y was
associated with adefovir dipivoxil (ADV) or lamivudine+ADV
experience. Of note, data have shown that one point mutation,
L213I, observed in the surface protein, which leads to F221Y and
A222T dual mutations in the RT domain of polymerase, in combination
with the classical BCP mutations, A1762T/G1764A, are associated
with the development of HCC in HBeAg-positive patients (24). In the present study, stratified
analysis revealed that patients with the rtF221Y mutation had lower
intrahepatic cccDNA levels (P=0.090), suggesting viral replication
was less active. rtF221 is located at the nucleic acid binding
domain, based on three-dimensional modeling analysis, therefore the
rtF221Y mutation is likely to affect the enzyme structure and
impair its polymerase activity. Patients with rtF221Y may require
regular monitoring and caution when using ADV.
For HBV-associated HCC, a high copy number of HBV
DNA is one of the important factors promoting the development of
HCC (25,26). RT is important in the process of
HBV replication and key mutations may occur during the immune
response, affecting the activity of RT. When a guanine was
substituted by cytosine at nucleotide 162 in the RT region, which
induced the S3 N mutation in HBsAg synchronously, the levels of
cccDNA were found to be significantly elevated, compared with those
in the wild-type group (P=0.004). NAs treatment is considered to be
an effective method to suppress HBV replication, normalize liver
function, reduce HBV-associated HCC recurrence and improve
postoperative survival rates. The occurrence of mutations
association with the prognosis of HCC, including rtF221Y, may also
decrease when receiving NAs therapy. Several meta-analyses have
also revealed that regular antiviral treatment prior to or
following surgery can significantly prolong OS and decrease
recurrence rates (27–29). It is necessary for patients with
HBV-associated HCC to adhere to standard antiviral therapy.
The frequency of sR122K, sT140S and sF183 V within
the HBsAg between ANTTs and TTs were significantly different, and
these mutation sites were all located in the MHR, the primary
B-cell epitope of HBV. Of note, the amino acid at position 122
within an antigenic loop determinates the serological HBs subtype.
Subtype determinant d have a lysine (K) at this positions, whereas
an arginine (R) indicates subtype determinant y. The positive
charged amino acid R at position 122 primarily contributes to
electrostatic interaction with negatively charged heparan sulfate
proteoglycans at the plasma membrane of hepatocytes during
infection (30). However, sR122K
is closely associated with immune evasion (31). Patients who harbored R122 K in TTs
suffered from larger tumor size and had higher recurrence rates in
the present study. Further investigations are required to clarify
the association between R122K and HCC. There were certain
limitations in the present study; a lack of liver biopsies from
patients with chronic hepatitis prevented deeper probing into
sequence discrepancies in the HBV genome between patients suffering
from HCC and chronic hepatitis. Therefore, the exact role of
mutations, particularly those located in RT/S, during the
development of HCC requires further investigation. In addition, due
to the methodological limitations of Sanger sequencing, limited
sensitivity hinders the detection of low abundance mutations
(32). Therefore, next-generation
sequencing technology is required to dissect viral quasispecies at
the RT/S region, to understand the variation of the whole virus
population in patients, to analyze minor antiviral resistance
mutations and to provide a guide for HBV treatment.
In conclusion, the use of Sanger sequencing of virus
derived from TT and ANTT samples revealed two novel substitutions,
rtF221Y and sR122K, which were found to be associated with
HBV-associated HCC recurrence. In particular, rtF221Y may serve as
a viral marker for predicting HCC prognosis. In view of HBV RT
mutations being one of the important factors linked to recurrence,
it is necessary for NAs to be timely and selected deliberately for
limiting the copy numbers of HBV DNA and preventing drug
resistance. This can assist in decreasing recurrence rate and
improving postoperative survival rates of patients with
HBV-associated HCC.
Acknowledgements
This study was supported by the China National Key
Projects for Infectious Disease (grant no. 2012ZX10002-016), the
National Natural Science Foundation of China (grant nos. 81572,072
and 81171664) and the Key Projects of Science and Technology
Commission of Shanghai Municipality (grant no. 11JC1416400).
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HCC
|
hepatocellular carcinoma
|
|
RT
|
reverse transcriptase
|
|
tDNA
|
HBV total DNA
|
|
cccDNA
|
covalently closed circular DNA
|
|
TT
|
tumor tissue
|
|
ANTT
|
adjacent non-tumor tissue
|
|
NA
|
nucleos(t)ide analogue
|
|
ADV
|
adefovir dipivoxil
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
MHR
|
major hydrophilic region
|
|
WT
|
wild-type
|
|
MT
|
mutant type.
|
References
|
1
|
Ngamruengphong S and Patel T: Molecular
evolution of genetic susceptibility to hepatocellular carcinoma.
Dig Dis Sci. 59:986–991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu Y, Gan Y, Gao F, Zhao Z, Jin Y, Zhu Y,
Sun Z, Wu H, Chen T, Wang J, et al: Novel natural mutations in the
hepatitis B virus reverse transcriptase domain associated with
hepatocellular carcinoma. PLoS One. 9:e948642014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nischalke HD, Lutz P, Krämer B, Söhne J,
Müller T, Rosendahl J, Fischer J, Berg T, Hittatiya K, Fischer HP,
et al: A common polymorphism in the NCAN gene is associated with
hepatocellular carcinoma in alcoholic liver disease. J Hepatol.
61:1073–1079. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Bömmel F, Bartens A, Mysickova A,
Hofmann J, Krüger DH, Berg T and Edelmann A: Serum hepatitis B
virus RNA levels as an early predictor of hepatitis B envelope
antigen seroconversion during treatment with polymerase inhibitors.
Hepatology. 61:66–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guirgis BS, Abbas RO and Azzazy HM:
Hepatitis B virus genotyping: Current methods and clinical
implications. Int J Infect Dis. 14:e941–e953. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie JX, Zhao J, Yin JH, Zhang Q, Pu R, Lu
WY, Zhang HW, Wang HY and Cao GW: Association of novel mutations
and haplotypes in the preS region of hepatitis B virus with
hepatocellular carcinoma. Front Med China. 4:419–429. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park YM, Jang JW, Yoo SH, Kim SH, Oh IM,
Park SJ, Jang YS and Lee SJ: Combinations of eight key mutations in
the X/preC region and genomic activity of hepatitis B virus are
associated with hepatocellular carcinoma. J Viral Hepat.
21:171–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu S, Zhang H, Gu C, Yin J, He Y, Xie J
and Cao G: Associations between hepatitis B virus mutations and the
risk of hepatocellular carcinoma: A meta-analysis. J Natl Cancer
Inst. 101:1066–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen CH, Changchien CS, Lee CM, Tung WC,
Hung CH, Hu TH, Wang JH, Wang JC and Lu SN: A study on sequence
variations in pre-S/surface, X and enhancer II/core
promoter/precore regions of occult hepatitis B virus in non-B,
non-C hepatocellular carcinoma patients in Taiwan. Int J Cancer.
125:621–629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang D, Ma S, Zhang X, Zhao H, Ding H and
Zeng C: Prevalent HBV point mutations and mutation combinations at
BCP/preC region and their association with liver disease
progression. BMC Infect Dis. 10:2712010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin J, Xie J, Liu S, Zhang H, Han L, Lu W,
Shen Q, Xu G, Dong H and Shen J: Association between the various
mutations in viral core promoter region to different stages of
hepatitis B, ranging of asymptomatic carrier state to
hepatocellular carcinoma. Am J Gastroenterol. 106:81–92. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen QY, Harrison TJ, Sabin CA, Li GJ,
Huang GM, Yang JY, Wang XY, Li H, Liu MH and Fang ZL: The Effect of
HBV Genotype C on the Development of HCC Differs Between Wild-Type
Viruses and Those With BCP Double Mutations (T(1762)A(1764)). Hepat
Mon. 14:e162142014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai
MW, Chen TC, Lin CY, Yeh TS and Lee WC: Hepatitis B virus-DNA level
and basal core promoter A1762T/G1764A mutation in liver tissue
independently predict postoperative survival in hepatocellular
carcinoma. Hepatology. 52:1922–1933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su CW, Chiou YW, Tsai YH, Teng RD, Chau
GY, Lei HJ, Hung HH, Huo TI and Wu JC: The influence of hepatitis b
viral load and pre-s deletion mutations on post-operative
recurrence of hepatocellular carcinoma and the tertiary preventive
effects by anti-viral therapy. PLoS One. 8:e664572013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu Y, Jin Y, Cai X, Bai X, Chen M, Chen
T, Wang J, Qian G, Gu J, Li J and Tu H: Hepatitis B virus core
protein variations differ in tumor and adjacent nontumor tissues
from patients with hepatocellular carcinoma. Intervirology.
55:29–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan H, Yang Y, Zhang L, Tang G, Wang Y,
Xue G, Zhou W and Sun S: Characterization of the genotype and
integration patterns of hepatitis B virus in early- and late-onset
hepatocellular carcinoma. Hepatology. 61:1821–1831. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang XH and Wang PL: Relationship between
mutation on precore region of integrated HBV DNA and p53 gene
mutation in hepatocellular carcinoma. Ai Zheng. 22:715–718.
2003.(In Chinese). PubMed/NCBI
|
|
18
|
Chen CH, Changchien CS, Lee CM, Hung CH,
Hu TH, Wang JH, Wang JC and Lu SN: Combined mutations in
pre-s/surface and core promoter/precore regions of hepatitis B
virus increase the risk of hepatocellular carcinoma: A case-control
study. J Infect Dis. 198:1634–1642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qu LS, Liu TT, Jin F, Guo YM, Chen TY, Ni
ZP and Shen XZ: Combined pre-S deletion and core promoter mutations
related to hepatocellular carcinoma: A nested case-control study in
China. Hepatol Res. 41:54–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li MW, Hou W, Wo JE and Liu KZ: Character
of HBV (hepatitis B virus) polymerase gene rtM204V/I and rtL180M
mutation in patients with lamivudine resistance. J Zhejiang Univ
Sci B. 6:664–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu BM, Li T, Xu J, Li XG, Dong JP, Yan P,
Yang JX, Yan L, Gao ZY, Li WP, et al: Characterization of potential
antiviral resistance mutations in hepatitis B virus reverse
transcriptase sequences in treatment-naïve Chinese patients.
Antiviral Res. 85:512–519. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mirandola S, Sebastiani G, Rossi C, Velo
E, Erne EM, Vario A, Tempesta D, Romualdi C, Campagnolo D and
Alberti A: Genotype-specific mutations in the polymerase gene of
hepatitis B virus potentially associated with resistance to oral
antiviral therapy. Antiviral Res. 96:422–429. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee HW, Chang HY, Yang SY and Kim HJ:
Viral evolutionary changes during tenofovir treatment in a chronic
hepatitis B patient with sequential nucleos(t)ide therapy. J Clin
Virol. 60:313–316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Datta S, Ghosh A, Dasgupta D, Ghosh A,
Roychoudhury S, Roy G, Das S, Das K, Gupta S, Basu K, et al: Novel
point and combo-mutations in the genome of hepatitis B
virus-genotype D: Characterization and impact on liver disease
progression to hepatocellular carcinoma. PLoS One. 9:e1100122014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakamura M, Kanda T, Nakamoto S, Haga Y,
Sasaki R, Jiang X, Yasui S, Arai M and Yokosuka O: Reappearance of
serum hepatitis B viral DNA in patients with hepatitis B surface
antigen seroclearance. Hepatology. 62:13292015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC,
Chen CL, Kuo SF, Liu CH, Chen PJ, Chen DS and Kao JH: High levels
of hepatitis B surface antigen increase risk of hepatocellular
carcinoma in patients with low HBV load. Gastroenterology.
142:1140–1149, .e3; quiz e13-4. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu CY, Lin JT, Ho HJ, Su CW, Lee TY, Wang
SY, Wu C and Wu JC: Association of nucleos(t)ide analogue therapy
with reduced risk of hepatocellular carcinoma in patients with
chronic hepatitis B: A nationwide cohort study. Gastroenterology.
147:143–151.e5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong JS, Wong GL, Tsoi KK, Wong VW, Cheung
SY, Chong CN, Wong J, Lee KF, Lai PB and Chan HL: Meta-analysis:
The efficacy of anti-viral therapy in prevention of recurrence
after curative treatment of chronic hepatitis B-related
hepatocellular carcinoma. Aliment Pharmacol Ther. 33:1104–1112.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sinn DH, Lee J, Goo J, Kim K, Gwak GY,
Paik YH, Choi MS, Lee JH, Koh KC, Yoo BC, et al: Hepatocellular
carcinoma risk in chronic hepatitis B virus-infected compensated
cirrhosis patients with low viral load. Hepatology. 62:694–701.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Glebe D and Bremer CM: The molecular
virology of hepatitis B virus. Semin Liver Dis. 33:103–112. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Avellón A and Echevarria JM: Frequency of
hepatitis B virus ‘a’ determinant variants in unselected Spanish
chronic carriers. J Med Virol. 78:24–36. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gong L, Han Y, Chen L, Liu F, Hao P, Sheng
J, Li XH, Yu DM, Gong QM, Tian F, et al: Comparison of
next-generation sequencing and clone-based sequencing in analysis
of hepatitis B virus reverse transcriptase quasispecies
heterogeneity. J Clin Microbiol. 51:4087–4094. 2013. View Article : Google Scholar : PubMed/NCBI
|