Introduction
Cervical cancer is the fourth highest cause of death
from cancer in women worldwide and the third highest cause of death
in Thailand and other developing countries (1). High-risk human papillomavirus
(HR-HPV) leads to nearly all cases of cervical cancer, and among
HR-HPV genotypes, HPV16 and HPV18 are the most prevalent. HPV16
exhibits the highest frequency as the cause of cervical cancers in
women in Thailand and worldwide (2–4).
Many types of commercial HPV tests are available. Most commercial
tests are designed to detect the presence of DNA from high-risk HPV
in patient samples. For example, signal amplification methods,
including Hybrid Capture 2 and Cervista are used for the detection
of HPV DNA using an RNA-DNA hybridization probe and
chemiluminescence or fluorescence respectively for signal
amplification and detection. However, these methods do not detect
non-amplified HPV DNA or identify specific HPV genotypes (5). Target amplification using polymerase
chain reaction (PCR) is the most outstanding target amplification
method, using oligonucleotide primers and thermocycling to amplify
DNA. PCR methods offer high sensitivity and specificity in the
detection of HPV genotypes (6).
However, special devices are required to perform these methods,
which are often time consuming and costly.
Loop-mediated isothermal amplification (LAMP), an
alternative method for nucleotide amplification under isothermal
conditions, has been described previously (7). The LAMP method for HPV genotyping has
been successfully developed using turbidity (8). However, observing turbidity using the
naked eye is not practical and might be difficult, particularly for
the identification of low copy DNA (9).
Lateral flow dipstick (LFD) tests are routinely used
for the detection of biological infectious agents and chemical
contaminants, including bacteria, viruses, toxins, veterinary drugs
and pesticides (10). The
interpretation of LFD tests is easy without external
instrumentation (11). In the
present study, LAMP and LFD methods were combined in order to
develop a simple assay for the detection of HPV16 and HPV18 with
high sensitivity and high specificity and the LAMP-LFD novel assay
was evaluated against the nested PCR assay (a gold standard) using
clinical samples of known HPV genotype.
Materials and methods
Samples and DNA extraction
Clinical samples (142 cervical tissues, including 44
HPV16-positive, 18 HPV18-positive and 80 HPV-negative samples) were
collected from Ubon Ratchathani Cancer Hospital (Ubon Ratchathani,
Thailand) during the year 2015. The human research ethics committee
of Ubon Ratchathani Cancer Hospital approved the protocol
(EC04/2015). DNA was extracted from all clinical samples using the
ExiPrep Dx Viral DNA Kit (Bioneer Corporation, Daejeon, Korea)
according to the manufacturer's protocol, and the initial detection
of DNA samples for high-risk HPV genotyping was performed using
nested PCR according to Sotlar et al (12). All DNA samples were stored at −20°C
until further use.
LAMP primers and LAMP conditions
The LAMP primer sets were obtained from a previous
study (8). These primer sets
(listed in Table I) comprised a
5′biotin-labelled forward inner primer (FIP; termed here as BIP),
outer primers (F3, B3) and loop primers (LF, LB), synthesized at
Pacific Science Co, Ltd. (Bangkok, Thailand). The optimal
conditions for the LAMP method were determined after assessing
varying reaction temperatures and times. The 25 µl reaction mixture
contained 1.6 µM of each inner primer, 0.2 µM of each outer primer,
0.8 µM of each loop primer, 1.4 mM dNTPs (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 0.8 mM betaine (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany), 6 mM MgSO4, 8 units of
Bst 2.0 DNA polymerase (New England BioLabs, Inc., Ipswich, MA,
USA), 1x Bst buffer (New England BioLabs, Inc.) and 3 µl of HPV16
and HPV18 plasmid DNA at 104 copies (provided by
Professor Ethel-Michele de Villiers, The German Cancer Research
Centre, Heidelberg, Germany) (13). The reaction was conducted at
different temperatures (60, 63 and 65°C) for 10, 20, 30, 40, 50 and
60 min.
 | Table I.Primer sets for LAMP and probe
sequences for the LAMP-LFD detection of HPV16 and HPV18. |
Table I.
Primer sets for LAMP and probe
sequences for the LAMP-LFD detection of HPV16 and HPV18.
| Primer | Primer sequence
(5′-3′) | Genome position |
|---|
| HPV16 |
|
|
| F3 |
TCGGTTGTGCGTACAAAG | 756–773 |
| B3 |
AGCCTCTACATAAAACCATCC | 933–913 |
| FIP |
Biotin-TGGGGCACACAATTCCTAGT-CACACACGTAGACATTCGT |
838-819/TTTT/774–792 |
| BIP |
TCAGAAACCATAATCTACCATGGC-ATTACATCCCGTACCCTCTT |
846-869/TTTT/912–893 |
| LF |
CCCATTAACAGGTCTTCCAAAGT | 815–793 |
| LB |
CCTGCAGGTACCAATGGGG | 874–892 |
|
Probe |
FITC-TACCAATGGGGAAGAGGGTA | 882–901 |
| HPV18 |
|
|
| F3 |
TCAGAGGAAGAAAACGATGA | 689–708 |
| B3 |
GTTGCTTACTGCTGGGAT | 912–895 |
| FIP |
Biotin-GCTTCACACTTACAACACATACACAATCAACATTTACCAGCCCG |
798-774/TTTT/726–744 |
| BIP |
TTGAGCTAGTAGTAGAAAGCTCAGCACGGACACACAAAGGACA |
804-828/TTTT/887–870 |
| LF |
ACGTTGTGGTTCGGCTCGT | 763–745 |
| LB |
GCATTCCAGCAGCTGTTTCTGAAC | 842–865 |
|
Probe |
FITC-TGAACACCCTGTCCTTTGTG | 861–880 |
LAMP-LFD assay conditions
The HPV16 and HPV18 LAMP products were hybridized
with the appropriate DNA probe (Pacific Science Co, Ltd.) labelled
with fluorescein isothiocyanate (FITC) at the 5′-end, designed
according to the HPV16 and HPV18 sequences between the LB and B3
primer targets (Table I). The
LAMP-LFD conditions were optimized after assessing various
hybridization times and DNA probe concentrations. The reaction
mixtures containing LAMP product and DNA probe (200 and 20 pmol,
respectively) were incubated at 65°C for 5, 10 or 15 min.
Subsequently, 8 µl of hybridized product was added to 150 µl of
assay buffer in a new tube (14).
An LFD strip (Milenia HybriDetect; Milenia Biotec GmbH, Giessen,
Germany) was dipped into the reaction mixture for 5 min. A
red-purple line was observed at the control line for all strips,
which confirmed that the test was correctly operated.
Sensitivity of LAMP-turbidity and
LAMP-LFD assays
To detect sensitivity limits, the HPV16 and
HPV18-containing plasmid DNA, varying from 105 to
100 copies, was used as a template for the LAMP
reaction. The plasmids pBR322-HPV16 and pBR322-HPV18 were kindly
provided by Professor Ethel-Michele de Villiers (13). The LAMP products were detected
using LAMP-turbidity (15)
compared with LAMP-LFD.
Specificity of the DNA-probe LFD
assay
The specificity of the DNA probe was examined using
104 copies of HPV16 and HPV18 plasmid DNA for the LAMP
reaction, and subsequently, the LAMP products were detected using
the LFD assay. The negative control was performed using the same
test, but water was added instead of DNA.
Evaluation of clinical samples
All 142 clinical samples were examined using
LAMP-turbidity (15) and LAMP-LFD
assays. The sensitivity and specificity of these assays were
calculated using standard formulae based on the results of nested
PCR.
Use of quantitative PCR (qPCR) to
examine discrepant results
All samples with inconsistent results were further
analysed using qPCR. The set of primers and probes for E2/E6 of
HPV16 and HPV18 were designed according to previous studies
(16,17). qPCR was conducted using an
Exicycler 96 system (Bioneer Corporation), as previously described
(16,17), and 10 µl of the clinical samples
were used as the DNA templates. The standard curves were obtained
from amplification of a dilution series using 108,
107, 106, 105, 104,
103, 102 and 101 copies of HPV16
and HPV18 plasmid DNA.
Results
Design of the LAMP-LFD HPV test
The LAMP-LFD assay was designed to detect
biotin-labelled LAMP products hybridized to a FITC-labelled
specific DNA probe (design explained in the schematic of Fig. 1). Subsequently, the FITC-labelled
specific DNA probe was recognized by a gold-labelled anti-FITC
antibody. This triple-labelled complex was then trapped at a test
line using avidin, generating a red-purple band (positive result).
By contrast, non-LAMP products hybridized with the FITC-labelled
specific probe and bound the gold-labelled anti-FITC antibody, but
did not bind avidin, due to lack of biotin; therefore, this complex
moved past the test line and was trapped at the control line
(18).
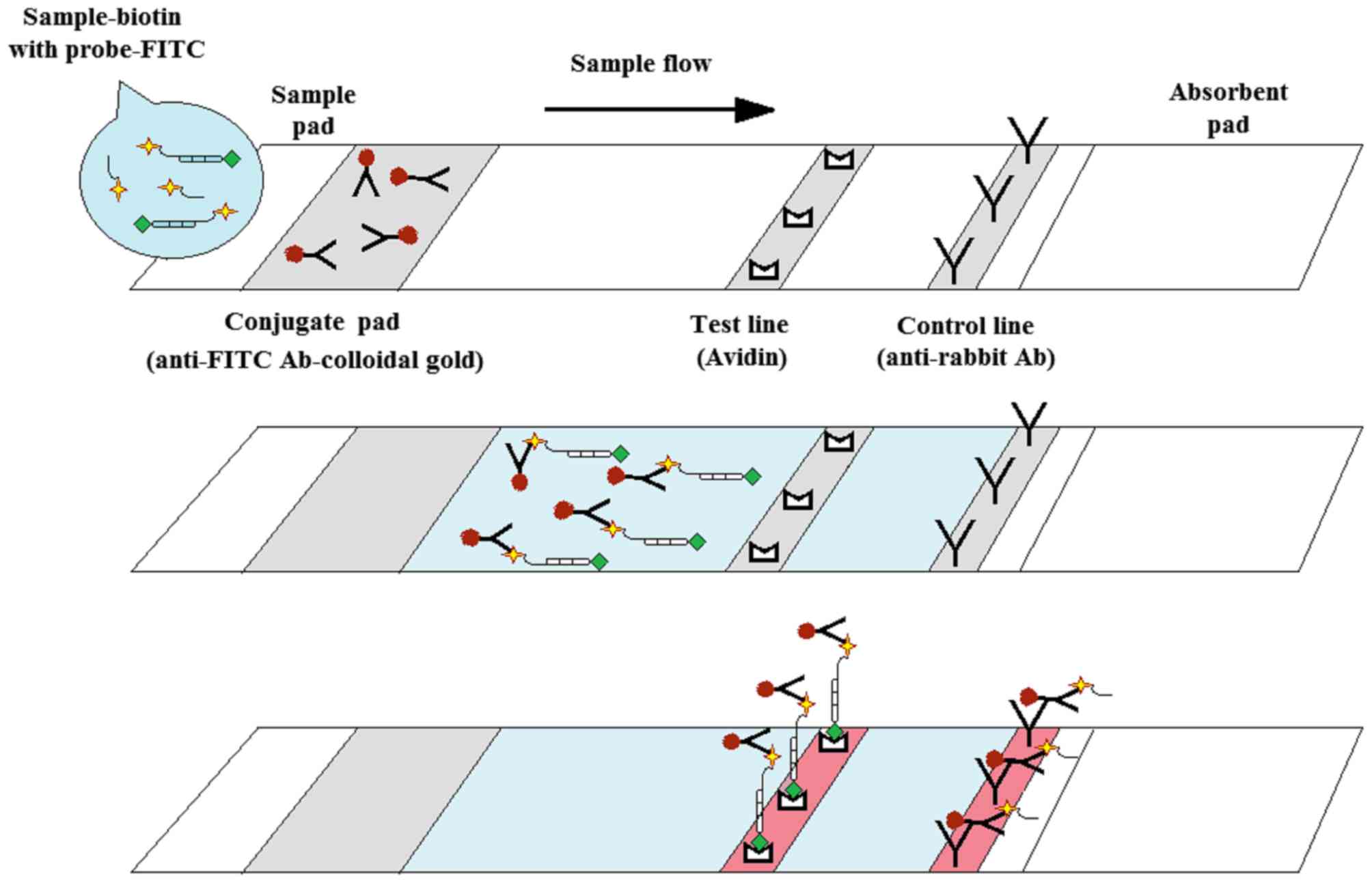 | Figure 1.Schematic description of the LAMP-LFD
assay design for the detection of HPV16- and HPV18-LAMP products.
Following the 30 min LAMP reaction, a FITC-labelled specific probe
was added to the reaction mixture and incubated at 65°C for 15 min.
Subsequently, the hybridized product was added to assay buffer in a
new tube. An LFD strip was then dipped into the reaction mixture
for 5 min. A red-purple band appearing at the control line should
be observed in all strips, indicating that the test was correctly
operated. For a positive result, the triple-labelled complex
(biotin-labelled LAMP product hybridized to FITC-labelled specific
probe combined with gold-labelled anti-FITC antibody) was trapped
at the test line using avidin, appearing as a red-purple band. For
a negative result (no biotin-labelled LAMP product), a double
complex (FITC-labelled specific probe bound by the gold-labelled
anti-FITC antibody) is trapped at the control line using an
anti-rabbit antibody, appearing as a red-purple band. LAMP,
loop-mediated isothermal amplification; LFD, lateral flow dipstick;
HPV, human papillomavirus virus; FITC, fluorescein
isothiocyanate. |
LAMP optimal conditions
The LAMP conditions for HPV16 and HPV18 detection
were optimized by assaying variable temperatures and reaction
duration times, and the turbidity of the amplified product was
observed using the naked eye. The results demonstrated that all
three temperatures tested (60, 63 and 65°C) generated a slightly
different turbidity; however, the highest turbidity was observed at
65°C (data not shown). In addition, the turbidity was initiated at
20 min, but it was difficult to observe under the naked eye.
Therefore, the best set of conditions, 65°C for 30 min, was
selected for the present study.
LAMP-LFD assay conditions
The specific FITC-labelled DNA probes were
hybridized with the HPV16 and HPV18 LAMP products. It was
determined that the best conditions for LAMP product hybridization
were as follows: 20 pmol of DNA probe as hybridization input and
incubation at 65°C for 15 min (data not shown). Subsequently, 8 µl
of hybridization product was added to 150 µl of assay buffer in a
new tube. Next, the LFD strip was dipped into this reaction mixture
for 5 min, and positive result was denoted by the presence of 2
red-purple lines on LFD strips using the naked eye.
Sensitivity of LAMP-turbidity and
LAMP-LFD assays
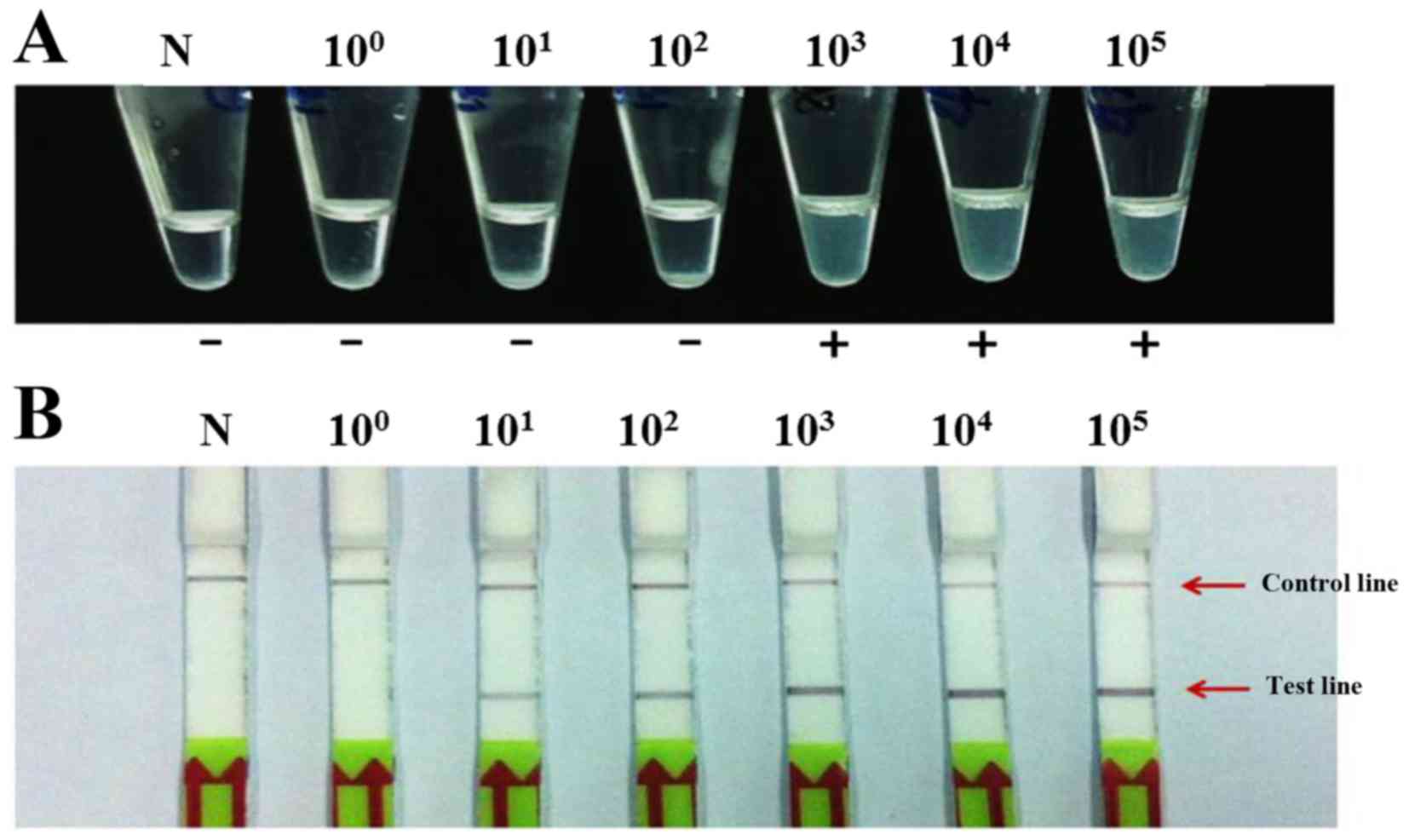
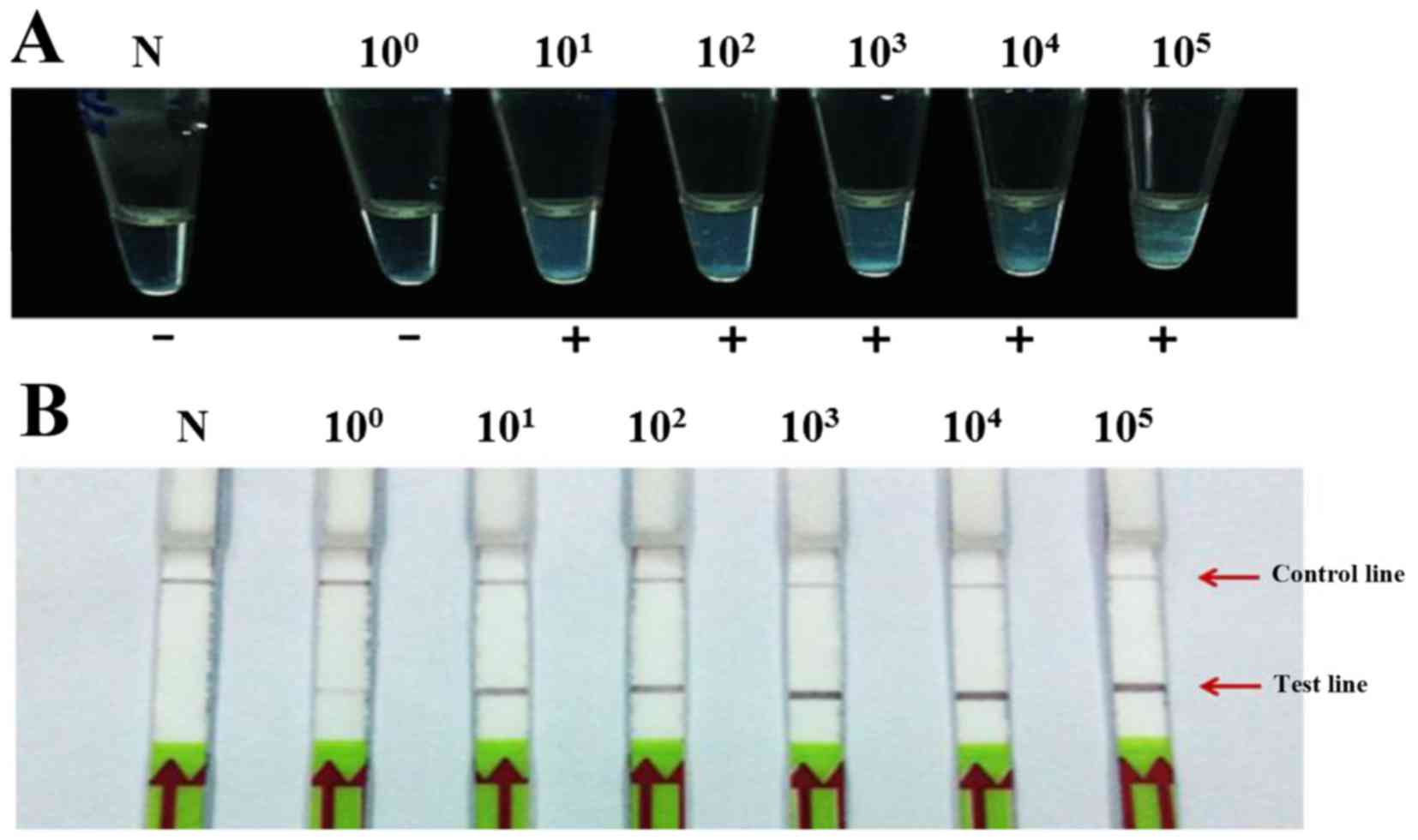
The limits of LAMP-turbidity detection for HPV16 and
HPV18 were 103 and 101 plasmid copies,
respectively (Figs. 2A and
3A, respectively). The LAMP-LFD
assay showed a limit of detection of 101 and
100 copies of HPV16 and HPV18, respectively (Figs. 2B and 3B, respectively). Therefore, the
detection limits of LAMP-LFD were ~100 and 10-fold more sensitive
for the detection of HPV16 and HPV18, respectively, compared with
the LAMP-turbidity method.
Specificity of the DNA-probe LFD
assay
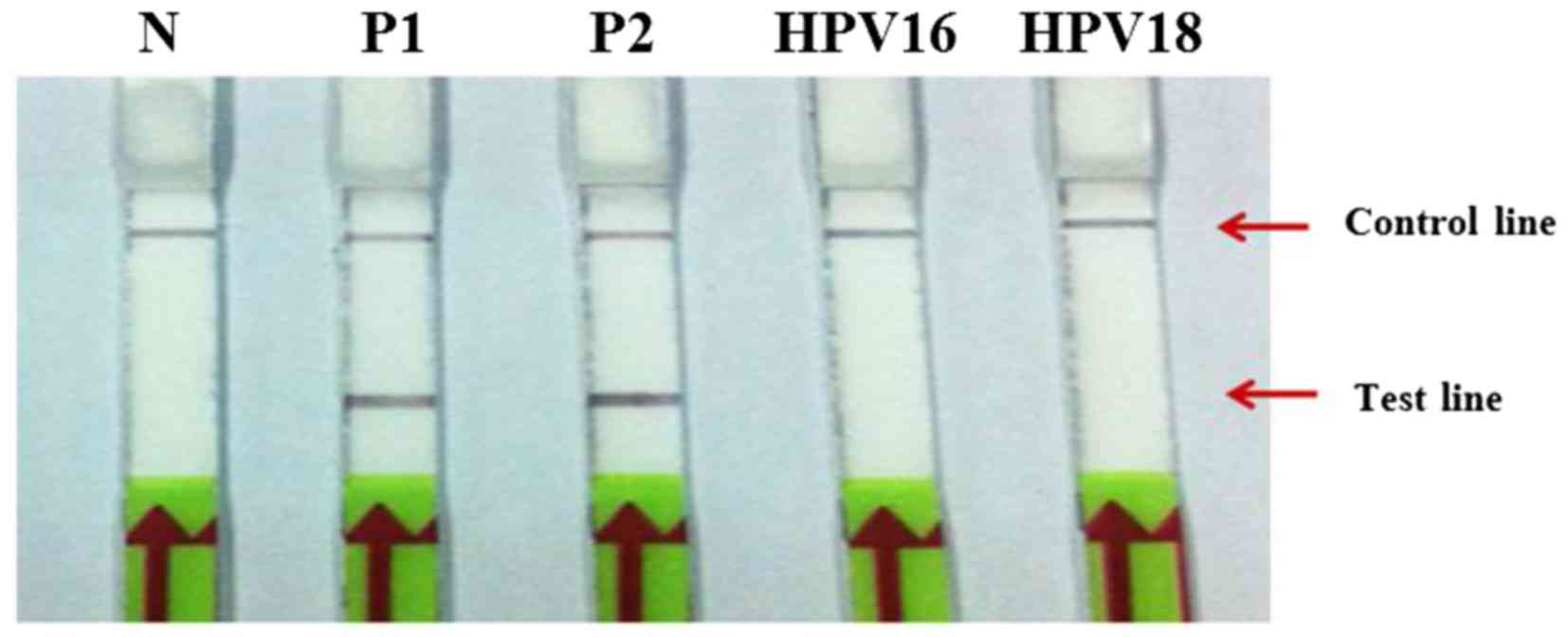
The specificity of the DNA probe was examined using
104 copies of HPV16 and HPV18 as templates for HPV18 and
HPV16 LAMP, respectively. The results revealed no cross-reactivity
between HPV16 and HPV18 using the LAMP-LFD assay (Fig. 4).
Evaluation of LAMP-LFD assay
All 142 clinical samples, including HPV16-positive
(n=44), HPV18-positive (n=18) and HPV-negative (n=80) samples, were
examined by the LAMP turbidity assay compared with the novel
LAMP-LFD assay. The results from the LAMP-turbidity assay revealed
that 44 and 18 samples were positive for HPV16 and HPV18,
respectively, while the results from the LAMP-LFD assay revealed
that 57 samples and 27 samples were positive for HPV16 and HPV18,
respectively (Table II).
Therefore, a higher number of samples gave positive readings with
the novel LAMP-LFD assay developed in the present study than with
the nested PCR, gold standard method.
 | Table II.Comparison of LAMP-turbidity and
LAMP-LFD assays for HPV16 and HPV18 detection. |
Table II.
Comparison of LAMP-turbidity and
LAMP-LFD assays for HPV16 and HPV18 detection.
|
| HPV16 | HPV18 |
|---|
|
|
|
|
|---|
| Results | Nested PCR | LAMP-turbidity | LAMP-LFD | Nested PCR | LAMP-turbidity | LAMP-LFD |
|---|
| Positive | 44 | 44 | 57 | 18 | 18 | 27 |
|
Negativea | 40 | 40 | 27 | 40 | 40 | 31 |
| Total | 84 | 84 | 84 | 58 | 58 | 58 |
qPCR of discrepant results
The samples with inconsistent results between
LAMP-LFD assay and the gold standard nested PCR method were further
analysed using qPCR. The limits of qPCR detection for HPV16 and 18
were 103 copies (data not shown). When the 22 samples
that exhibited inconsistent results between the LAMP-LFD assay and
the gold standard method were repeated using qPCR, the results
demonstrated that 13 of 13 samples and 7 of 9 samples were positive
by qPCR for HPV16 and HPV18, respectively, while 2 samples
generated negative results by qPCR (Tables III and IV).
 | Table III.Analysis of clinical sample
discrepant results by quantitative PCR. |
Table III.
Analysis of clinical sample
discrepant results by quantitative PCR.
|
| HPV16 | HPV18 |
|---|
|
|
|
|
|---|
| Results | LAMP-LFD | Real time PCR | LAMP-LFD | Real time PCR |
|---|
| Positive | 13 | 13 | 9 | 7 |
| Negative | 0 | 0 | 0 | 2 |
| Total | 13 | 13 | 9 | 9 |
 | Table IV.qPCR confirmation of the LAMP-LFD
discrepant results obtained using the gold standard method. |
Table IV.
qPCR confirmation of the LAMP-LFD
discrepant results obtained using the gold standard method.
| Clinical no. | LAMP-LFD
results | qPCR results | Viral load (copy/ng
DNA) |
|---|
| 1 | HPV 16 | HPV 16 | 9.49 |
| 2 | HPV 16 | HPV 16 | 4.45 |
| 3 | HPV 16 | HPV 16 | 11.85 |
| 4 | HPV 16 | HPV 16 | 3.09 |
| 5 | HPV 16 | HPV 16 | 2.71 |
| 6 | HPV 16 | HPV 16 | 11.4 |
| 7 | HPV 16 | HPV 16 | 18.07 |
| 8 | HPV 16 | HPV 16 | 10.92 |
| 9 | HPV 16 | HPV 16 | 6.59 |
| 10 | HPV 16 | HPV 16 | 50.43 |
| 11 | HPV 16 | HPV 16 | 31.09 |
| 12 | HPV 16 | HPV 16 | 18.82 |
| 13 | HPV 16 | HPV 16 | 12.48 |
| 14 | HPV18 | Negative | Not detected |
| 15 | HPV18 | HPV18 | 25.98 |
| 16 | HPV18 | Negative | Not detected |
| 17 | HPV18 | HPV18 | 4.18 |
| 18 | HPV18 | HPV18 | 51.15 |
| 19 | HPV18 | HPV18 | 5.43 |
| 20 | HPV18 | HPV18 | 9.13 |
| 21 | HPV18 | HPV18 | 63.13 |
| 22 | HPV18 | HPV18 | 117.42 |
Discussion
LAMP is an alternative method for nucleotide
amplification (7). This fast,
simple and inexpensive amplification method can be performed within
1 h under isothermal conditions, and visualization of LAMP
amplification products can be detected using the naked eye through
several methods, such as turbidity, fluorescence and colour change
(19). However, specific and
nonspecific products cannot be separated using these detection
methods. Therefore, to avoid false-positive results, LAMP products
can be hybridized to specific probes (20) and subsequently detected using LFD.
LFD is sufficient for the detection of hybridized LAMP products
(21,22), and the results are easy to read
without the use of carcinogens, such as ethidium bromide. LAMP
detection methods have been successfully developed for HPV16 and
HPV18 using visual turbidity and gel electrophoresis, and these
assays can be completed within 70 and 115 min, respectively
(15). In the present study, the
same LAMP primer sets were used as previously published, but
LAMP-LFD combined detection provided higher sensitivity and was
completed within a shorter time period compared with a previous
report (8).
The detection limits of LAMP-turbidity for HPV16 and
HPV18 were 103 and 101 copies, respectively,
and the assay was completed in 30 min. The detection limits of
LAMP-LFD for HPV16 and HPV18 were 101 and 100
copies, respectively, and the assay was completed in 45 min.
Therefore, the sensitivity of LAMP-LFD was higher than
LAMP-turbidity, and the signal for LAMP-LFD was easy to read using
the naked eye.
LAMP-turbidity and LAMP-LFD were further evaluated
using 142 clinical samples (Table
II), and the results revealed that the sensitivity and
specificity of LAMP-turbidity for HPV16 and HPV18 detection were
100%. The sensitivity of LAMP-LFD for HPV16 and HPV18 was 100%,
while the specificity was 67.5 and 77.5%, respectively. The
decrease in LAMP-LFD specificity may be due to the fact that 22 of
80 HPV-negative samples generated positive results with the
LAMP-LFD method. Therefore, those 22 samples were further analysed
using qPCR (Tables III and
IV). The results demonstrated
that all 13 samples that generated positive HPV16 results with
LAMP-LFD were also demonstrated HPV16-positive by qPCR. However,
out of the 9 samples that were positive for HPV18 by LAMP-LFD, only
7 were also HPV18-positive by qPCR. Further analysis of the
remaining 2 samples that generated negative results by qPCR
revealed that the detection limit of the qPCR, although 10 µl of
clinical sample was used as DNA template, increased ~3.3-fold,
potentially reflecting a low concentration of HPV viral DNA.
Therefore, detection of HPV16 and HPV18 using LAMP-LFD demonstrated
higher sensitivity compared with nested PCR and did not require two
reaction steps for PCR cycling (13). However, to avoid
cross-contamination during the detection of the DNA products, the
use of uracil DNA glycosylase has been recommended (23). In conclusion, LAMP-LFD is a rapid
and simple method for the highly sensitive and specific detection
of HPV16 and HPV18. Thus, LAMP-LFD might be useful as an HPV16 and
HPV18 diagnostic tool in local hospitals or field studies.
Acknowledgements
This work was financially supported by grants from
the Centre for Research and Development of Medical Diagnostic
Laboratories, Faculty of Associated Medical Sciences and HPV &
EBV and Carcinogenesis Research Group, Khon Kaen University,
Thailand (grant no. KKU-2556).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chansaenroj J, Junyangdikul P, Chinchai T,
Swangvaree S, Karalak A, Gemma N and Poovorawan Y: Large scale
study of HPV genotypes in cervical cancer and different cytological
cervical specimens in Thailand. J Med Virol. 86:601–607. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suthipintawong C, Siriaunkgul S,
Tungsinmunkong K, Pientong C, Ekalaksananan T, Karalak A, Kleebkaow
P, Vinyuvat S, Triratanachat S, Khunamornpong S and Chongsuwanich
T: Human papilloma virus prevalence, genotype distribution, and
pattern of infection in Thai women. Asian Pac J Cancer Prev.
12:853–856. 2011.PubMed/NCBI
|
|
4
|
Saslow D, Solomon D, Lawson HW, Killackey
M, Kulasingam SL, Cain J, Garcia FA, Moriarty AT, Waxman AG, Wilbur
DC, et al: American cancer society, American society for colposcopy
and cervical pathology and American society for clinical pathology
screening guidelines for the prevention and early detection of
cervical cancer. CA Cancer J Clin. 62:147–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arney A and Bennett KM: Molecular
diagnostics of human papillomavirus. Labmedicine. 41:523–530.
2010.
|
|
6
|
Zaravinos A, Mammas IN, Sourvinos G and
Spandidos DA: Molecular detection methods of human papillomavirus
(HPV). Int J Biol Marker. 24:215–222. 2009. View Article : Google Scholar
|
|
7
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saetiew C, Limpaiboon T, Jearanaikoon P,
Daduang S, Pientong C, Kerdsin A and Daduang J: Rapid detection of
the most common high-risk human papillomaviruses by loop-mediated
isothermal amplification. J Virol Methods. 178:22–30. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fischbach J, Xander NC, Frohme M and
Glokler JF: Shining a light on LAMP assays-a comparison of LAMP
visualization methods including the novel use of berberine.
Biotechniques. 58:189–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Posthuma-Trumpie GA, Korf J and van
Amerongen A: Lateral flow (immuno)assay: Its strengths, weaknesses,
opportunities and threats. A literature survey. Anal Bioanal Chem.
393:569–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yetisen AK, Akram MS and Lowe CR:
Paper-based microfluidic point-of-care diagnostic devices. Lab
Chip. 13:2210–2251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sotlar K, Diemer D, Dethleffs A, Hack Y,
Stubner A, Vollmer N, Menton S, Menton M, Dietz K, Wallwiener D, et
al: Detection and typing of human papillomavirus by e6 nested
multiplex PCR. J Clin Microbiol. 42:3176–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Villiers EM: Papillomavirus and HPV
typing. Clin Dermatol. 15:199–206. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kiatpathomchai W, Jaroenram W, Arunrut N,
Jitrapakdee S and Flegel TW: Shrimp Taura syndrome virus detection
by reverse transcription loop-mediated isothermal amplification
combined with a lateral flow dipstick. J Virol Methods.
153:214–217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumvongpin R, Jearanaikool P, Wilailuckana
C, Sae-ung N, Prasongdee P, Daduang S, Wongsena M, Boonsiri P,
Kiatpathomchai W, Swangvaree SS, et al: High sensitivity,
loop-mediated isothermal amplification combined with colorimetric
gold-nanoparticle probes for visual detection of high risk human
papillomavirus genotypes 16 and 18. J Virol Methods. 234:90–95.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peitsaro P, Johansson B and Syrjänen S:
Integrated human papillomavirus type 16 is frequently found in
cervical cancer precursors as demonstrated by a novel quantitative
real-time PCR technique. J Clin Microbiol. 40:886–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Damay A, Didelot-Rousseau MN, Costes V,
Konate I, Ouedraogo A, Nagot N, Foulongne V, Van de Perre P, Mayaud
P and Segondy M: Viral load and physical status of human
papillomavirus (HPV) 18 in cervical samples from female sex workers
infected with HPV 18 in Burkina Faso. J Med Virol. 81:1786–1791.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wanga X, Tenga D, Guana Q, Tiana F and
Wang J: Detection of roundup ready soybean by loop-mediated
isothermal amplification combined with a lateral-flow dipstick.
Food Cont. 29:213–220. 2013. View Article : Google Scholar
|
|
19
|
Notomi T, Mori Y, Tomita N and Kanda H:
Loop-mediated isothermal amplification (LAMP): Principle, features,
and future prospects. J Microbiol. 53:1–5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mori Y, Hirano T and Notomi T: Sequence
specific visual detection of LAMP reactions by addition of cationic
polymers. BMC Biotechnol. 6:32006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kaewphinit T, Arunrut N, Kiatpathomchai W,
Santiwatanakul S, Jaratsing P and Chansiri K: Detection of
Mycobacterium tuberculosis by using loop-mediated isothermal
amplification combined with a lateral flow dipstick in clinical
samples. Biomed Res Int. 2013:9262302013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khunthong S, Jaroenram W, Arunrut N,
Suebsing R, Mungsantisuk I and Kiatpathomchai W: Rapid and
sensitive detection of shrimp yellow head virus by loop-mediated
isothermal amplification combined with a lateral flow dipstick. J
Virol Methods. 188:51–56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kil EJ, Kim S, Lee YJ, Kang EH, Lee M, Cho
SH, Kim MK, Lee KY, Heo NY, Choi HS, et al: Advanced loop-mediated
isothermal amplification method for sensitive and specific
detection of Tomato chlorosis virus using a uracil DNA glycosylase
to control carry-over contamination. J Virol Methods. 213:68–74.
2015. View Article : Google Scholar : PubMed/NCBI
|