Introduction
Pyropia yezoensis, a marine red alga
previously known as Porphyra yezoensis, is a commercially
important seaweed in a number of Asian countries, including Korea,
China, and Japan. P. yezoensis is a crucial source of
bioactive substances that contain large quantities of essential
proteins, vitamins and minerals. P. yezoensis has
potentially beneficial biological effects, including
anti-inflammatory, antioxidant, anticancer, anti-fatigue and
anti-aging (1,2).
Aging is a multifactorial process that is
facilitated by a decline in neuromuscular functions and stress
tolerance, which results in tissue degeneration and malfunction,
most notably in the skeletal muscles. Age-related muscle atrophy
and a reduction in skeletal muscle mass and strength is a condition
known as sarcopenia (from the Greek for ‘lack of flesh’).
Sarcopenia leads to muscle weakness and greatly affects physical
activity and the quality of life of elderly individuals (3,4);
sarcopenia is common in elderly people, and is estimated to occur
in 5–13% of people aged 60–70 years and in 11–50% of those ≥80
years (5). The mechanisms of
sarcopenia development are actively being studied and involve both
intrinsic and extrinsic factors, including the decline of satellite
cell activation, altered hormonal status, contraction-induced
injury, cellular vacuolization, autophagy, apoptosis and increased
oxidative stress (6).
C2C12 mouse skeletal muscle cells are an in
vitro model that is widely used to study the factors that
regulate muscle growth, proliferation and differentiation (7). The differentiation of myotubes can be
induced by 2% fetal bovine serum (FBS). Previous studies have
demonstrated that C2C12 myoblasts cultured in a
growth-factor-deficient state causes the mononucleated myoblasts to
exit the cell cycle, which activates the expression of genes that
promote myoblast fusions and the formation of multinucleated
myotubes (8,9). During this process, changes in cell
shape along with cell fusion occur, and myotube phenotypes can be
observed within 5–6 days. In addition, when muscle fiber formation
(myogenesis) begins, myogenic transcriptional regulatory factors
belonging to the MyoD family, including MyoD, myogenin, Myf4 and
Myf5, are activated (10). Amongst
these, myogenin has an important role within the MyoD family as it
regulates the differentiation of single nucleated myoblasts into
multinucleated myofibers (11).
Glucocorticoids are important in the development of
muscle atrophy in humans and animals (12). In both in vivo and in
vitro experiments, muscle atrophy is induced by synthetic
glucocorticoids such as dexamethasone (DEX) (12). In skeletal muscles, DEX causes a
reduction in protein synthesis and an increase in protein
degradation through the ubiquitin-proteasome pathway (13). The ubiquitin-proteasome pathway has
been revealed to mediate the degradation of short-lived proteins
and long-lived myofibrillar proteins (14). This protein-degradation system
comprises three enzymatic components: the ubiquitin-activating E1
enzyme, the ubiquitin-conjugating E2 enzyme and the
ubiquitin-ligating E3 enzyme. E3 ubiquitin ligases serve a crucial
role in identifying and targeting proteins for proteasomal
degradation (14,15). A previous study characterized two
muscle-specific E3 ubiquitin ligases, muscle RING-finger 1 (MuRF1)
and muscle atrophy F-box (MAFbx; also known as atrogin1), as
markers of skeletal muscle atrophy (16). Levels of MuRF1 and atrogin1/MAFbx
expression are induced early in the atrophy process, prior to the
loss of muscle mass (16). The
expression of atrogin1/MAFbx is controlled by forkhead box O,
whereas MuRF1 transcription is controlled by nuclear factor κB
(17).
The present study investigated the potential
protective effects of the P. yezoensis peptide PYP1-5 on
DEX-induced muscle atrophy in C2C12 mouse skeletal muscle cells,
based on the efficacy of P. yezoensis muscle contraction and
release. The synthetic peptide PYP1-5 corresponds to the 15
N-terminal residues of PYP1 (ALEGGKSSGGGEATRDPEPT) (18). The anti-atrophy potential of PYP1-5
was determined by focusing on its effects on the expression of
atrogin1/MAFbx and MuRF1.
Materials and methods
P. yezoensis peptide synthesis
The N-terminal 15 residues of P. yezoensis
peptide PYP1 (1–15) (D-P-K-G-K-Q-Q- A-I-H-V-A-P-S-F;
designated as PYP1-5) were synthesized by Peptron (Daejeon, Korea).
PYP1-5 purification was performed using a Prominence
High-Performance Liquid Chromatography apparatus (Shimadzu
Corporation, Kyoto, Japan) with a Capcell Pak C18 column (column
dimensions, 150×4.6 mm; particle size, 2.7 µm; Shiseido Co., Ltd.,
Tokyo, Japan) in 0.1% trifluoroacetic acid (TFA; v/v in water), and
an elution gradient of 10–70% acetonitrile (0–20% acetonitrile for
2 min; 20–50% acetonitrile for 10 min; 50–80% acetonitrile for 2
min) in 0.1% TFA, a flow rate of 1.0 ml/min and UV detection at 220
nm; analysis was conducted with the Class-VP software package,
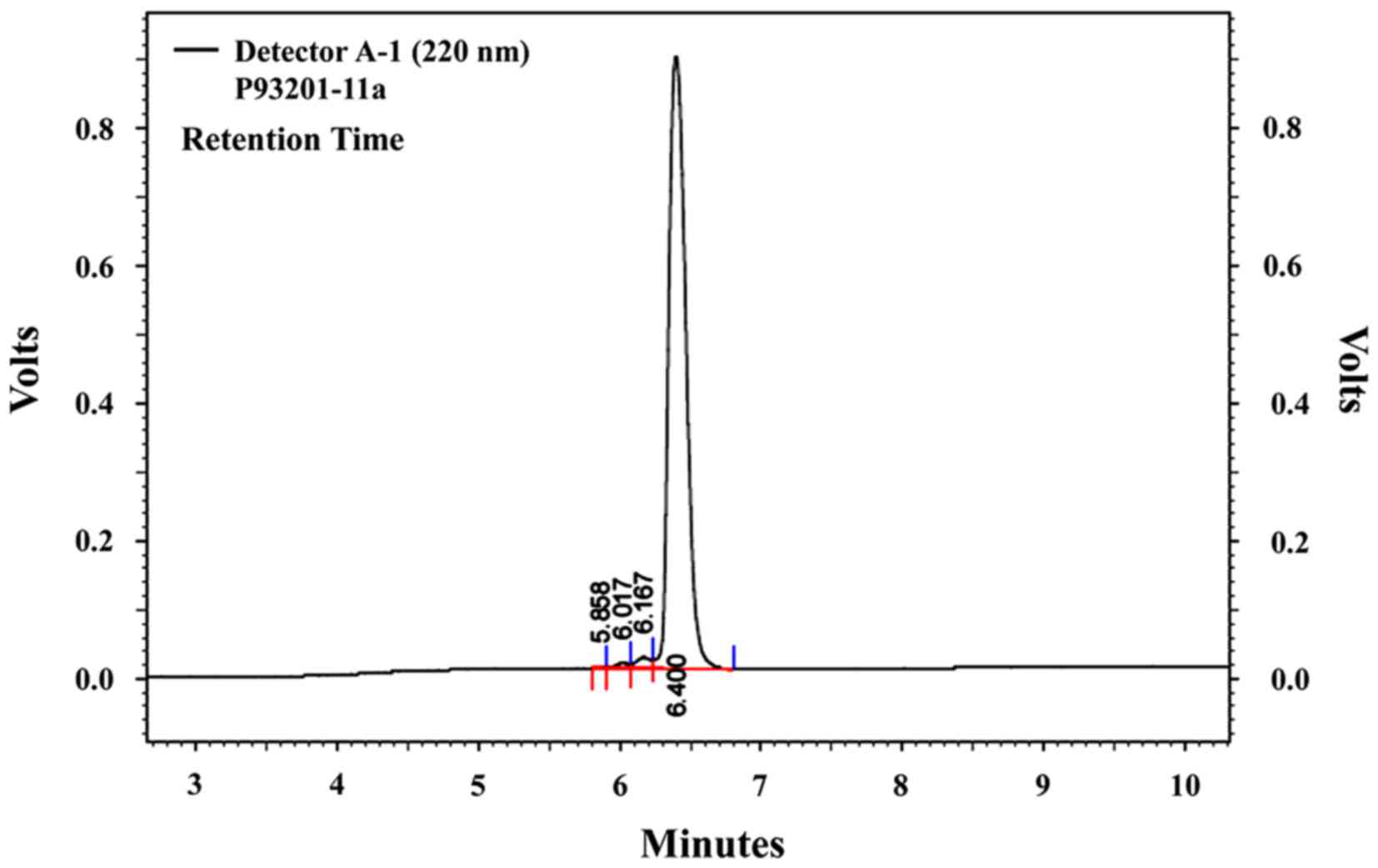
version 6.14 (Shimadzu Corporation). The molecular mass of PYP1-5
was confirmed to be 1,622 Da (Fig.
1) using an HP 110 Series LC/MSD mass spectrometer [ionization
mode, positive; nitrogen flow, 7 l/min; high vacuum, 1.3E-5 torr;
neb press, 40 psi; quadrupole temperature, 100°C; flow rate, 0.4
ml/min (isocratic ANC:DW=8:2, 0.1% TFA); Peptron, Inc., Daejeon,
Korea].
Cell culture
C2C12 mouse skeletal muscle cells (ATCC no.
CRL-1772; American Type Culture Collection, Manassas, VA, USA) were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
at a temperature of 37°C in a humidified atmosphere of 5%
CO2. The cells were cultured to 70–80% confluence in 100
mm dishes, and the medium was replaced every 2 days.
Induction of differentiation
C2C12 myoblasts were grown to 70–80% confluence in
culture dishes at 37°C, trypsinized and seeded (4×104
cells/well) into 6-well culture plates for experiments. Cells were
grown to 70–80% confluence in DMEM supplemented with 10% FBS at
37°C for 24 h, at which time the medium was replaced with DMEM
containing 2% FBS to induce myotube differentiation; the medium was
replaced every 2 days. Cells were allowed to differentiate for 6
days, at which point 90% of the cells had fused into myotubes.
Treatment with DEX and PYP1-5
Following 6 days of differentiation, C2C12 myotubes
were treated with 1, 10, 50 or 100 µM DEX at 37°C for 24 h for
concentration screening. The C2C12 myotubes were subdivided into
four groups: i) the control group, in which cells were incubated in
serum-free medium (SFM; DMEM containing 100 U/ml penicillin and 100
mg/ml streptomycin); ii) the DEX group, in which cells were treated
with 100 µM DEX; iii) the DEX+PYP1-5 group, in which cells were
treated with 100 µM DEX and 500 ng/ml PYP1-5; and iv) the PYP1-5
group, in which cells were treated with 500 ng/ml PYP1-5. All
groups were incubated at 37°C for 24 h, prior to harvesting cells
for experiments.
MTS assay
Cell viability was measured using the CellTiter 96
AQueous Non-Radioactive Cell Proliferation assay (Promega
Corporation, Madison, WI, USA), which is based on the cleavage of
3-(4,5-dimethythiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfonyl)-2H-tetrazolium
(MTS) into a formazan product that is soluble in tissue culture
medium, according to the manufacturer's protocol. Cells
(1.5×104 cells/well) were seeded in 96-well plates in
100 µl DMEM supplemented with 10% FBS and were allowed to attach at
37°C for 24 h. Attached cells were maintained in SFM at 37°C for 4
h and were subsequently treated with or without PYP1-5 (125, 250,
500 and 1,000 ng/ml) for 24 h. MTS solution (10 µl) was added and
the cells were incubated at 37°C for 30 min; the absorbance of each
well was measured at 490 nm using a SpectraMax 340 PC Microplate
Reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Experiments
were performed in triplicate.
Measurement of myotube diameter
Images of myotube cultures were captured with a
phase contrast microscope set to ×100 magnification at 1, 2, 4, 5
and 6 days following induction of differentiation. Images of
myotube cultures were also captured following treatment with 100 µM
DEX and/or 500 ng/ml PYP1-5 for 24 h. A total of 50 myotube
diameters from at least 10 random fields were measured using ImageJ
software (version 4.16; National Institutes of Health, Bethesda,
MD, USA).
Preparation of whole-cell protein
lysates
Cells were allowed to differentiate for 6 days at
37°C, followed by incubation at 37°C for 24 h in either SFM
(control group) or SFM containing 100 µM DEX (DEX group), 100 µM
DEX + 500 ng/ml PYP1-5 (DEX+PYP1-5 group) or 500 ng/ml PYP1-5
(PYP1-5 group). Cells were washed in cold phosphate-buffered saline
(PBS) and lysed with extraction buffer [1% NP-40, 0.25% sodium
deoxycholate, 1 mM ethylene glycol-bis (β-aminoethyl ether)-N,
N,N', N'-tetraacetic acid, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5]
containing protease inhibitors (1 mg/ml aprotinin, 1 mg/ml
leupeptin, 1 mg/ml pepstatin A, 200 mM
Na3VO4, 500 mM NaF and 100 mM
phenylmethylsulfonyl fluoride) on ice. Extracts were centrifuged at
18,341 × g for 10 min at 4°C, and protein levels were
quantified using a Bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The supernatant was then used in western
blotting.
Extraction of nuclear lysates
Cell were treated and harvested as aforementioned,
lysed with hypotonic lysis buffer [25 mM
4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES; pH
7.5), 5 mM EDTA, 5 mM MgCl2 and 5 mM dithiothreitol
(DTT)], and incubated for 15 min on ice. NP-40 (2.5%) was added and
the cells were lysed for an additional 10 min. Nuclei were
collected by centrifugation at 7,310 × g for 15 min at 4°C.
Nuclear proteins were resuspended in extraction buffer (10 mM HEPES
pH 7.9, 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA and 0.2
mM DTT) and incubated for 20 min at 4°C. Extracts were centrifuged
at 18,341 × g for 10 min, and the protein levels were
determined using a Bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The supernatant was used then for western blot
analysis.
Western blot analysis
Proteins (20 µg) were separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA). Membranes were blocked at room temperature for
1 h with 1% bovine serum albumin (BSA) in TBS-T (10 mM Tris-HCl,
150 mM NaCl and 0.1% Tween-20) and then incubated with primary
antibodies against Myogenin, MuRF1, MAFbx and GAPDH (Table I; all purchased from Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at room temperature for 3 h.
Membranes were incubated with secondary horseradish
peroxidase-conjugated rabbit anti-goat IgG, goat anti-mouse IgG
(both Santa Cruz Biotechnology, Inc.) and goat anti-rabbit IgG
(Cell Signaling Technology, Inc., Danvers, MA, USA; Table II) at room temperature for 1 h and
bands were detected using an enhanced chemiluminescence western
blotting kit (Thermo Fisher Scientific, Inc.). Experiments were
performed in triplicate and densitometry analysis was performed
using Multi-Gauge software version 3.0 (Fujifilm Life Science,
Tokyo, Japan).
 | Table I.Primary antibodies used in western
blot analysis. |
Table I.
Primary antibodies used in western
blot analysis.
| Antibody | Catalog no. | Species raised in,
monoclonal or polyclonal | Dilution |
|---|
| Anti-Myogenin | sc-12732 | Mouse monoclonal | 1:1,000 |
| Anti-MuRF1 | sc-27642 | Goat polyclonal | 1:2,000 |
|
Anti-atrogin1/MAFbx | sc-27645 | Goat polyclonal | 1:2,000 |
| Anti-GAPDH | sc-25778 | Rabbit
polyclonal | 1:2,000 |
 | Table II.Secondary antibodies used in western
blot analysis. |
Table II.
Secondary antibodies used in western
blot analysis.
| Antibody | Catalog no. | Dilution |
|---|
| Goat anti-mouse
IgG | sc-2031 | 1:10,000 |
| Rabbit anti-goat
IgG | sc-2768 | 1:10,000 |
| Goat anti-rabbit
IgG | #7074 | 1:10,000 |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from C2C12 cells
(4×104 cells/well) using TRIzol Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). RNA quality was evaluated by
measuring absorbance at 260 and 280 nm to calculate concentration
and to assess RNA purity. The RevoScript Reverse Transcriptase
PreMix (oligo dT) kit (Intron Biotechnology, Inc., Seongnam, Korea)
was used to prepare cDNA, according to the manufacturer's protocol,
and the samples were stored at −50°C. qPCR was conducted in 20 µl
reactions using the QuantiMix SYBR kit (PhilKorea Technology, Inc.,
Daejeon, Korea) and the Eco Real-Time PCR System (Illumina, Inc.,
San Diego, CA, USA), following the manufacturer's protocol.
Oligonucleotide primers for MuRF1, atrogin1/MAFbx and GAPDH are
shown in Table III. qPCR
reactions were incubated for an initial denaturation at 95°C for 10
min, followed by 40 cycles of: 95°C for 15 sec, 55°C for 15 sec,
and 72°C for 15 sec. For each sample, the expression level of
target mRNA was quantified to those of GAPDH mRNA and calculated
using the comparative ΔΔCq method (19). All reactions were performed in
triplicate and repeated in two independent experiments.
 | Table III.Oligonucleotide primer sequences used
in reverse transcription-quantitative polymerase chain
reactions. |
Table III.
Oligonucleotide primer sequences used
in reverse transcription-quantitative polymerase chain
reactions.
| Gene | GenBank accession
no. | Sequence (5′-3′) | Amplicon size
(bp) |
|---|
| MuRF1 | DQ229108.1 | F:
TGTCTGGAGGTCGTTTCCG | 59 |
|
|
| R:
GTGCCGGTCCATGATCACTT |
|
| MAFbx | NM_026346.3 | F:
ATGCACACTGGTGCAGAGAG | 168 |
|
|
| R:
TGTAAGCACACAGGCAGGTC |
|
| GAPDH | NM_008084.3 | F:
ACTCCACTCACGGCAAATTCA | 91 |
|
|
| R:
CGCTCCTGGAAGATGGTGAT |
|
Statistical analysis
Multiple mean values were assessed by analysis of
variance using SPSS version 10.0 software (SPSS Inc., Chicago, IL,
USA) using one-way analysis of variance followed by a Duncan's
multiple range test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Confirmation of C2C12 myotube
differentiation
C2C12 skeletal muscle myoblasts were induced to
differentiate in a mitogen-poor media such as 2% FBS, fusing to
form multinucleate myotubes (20).
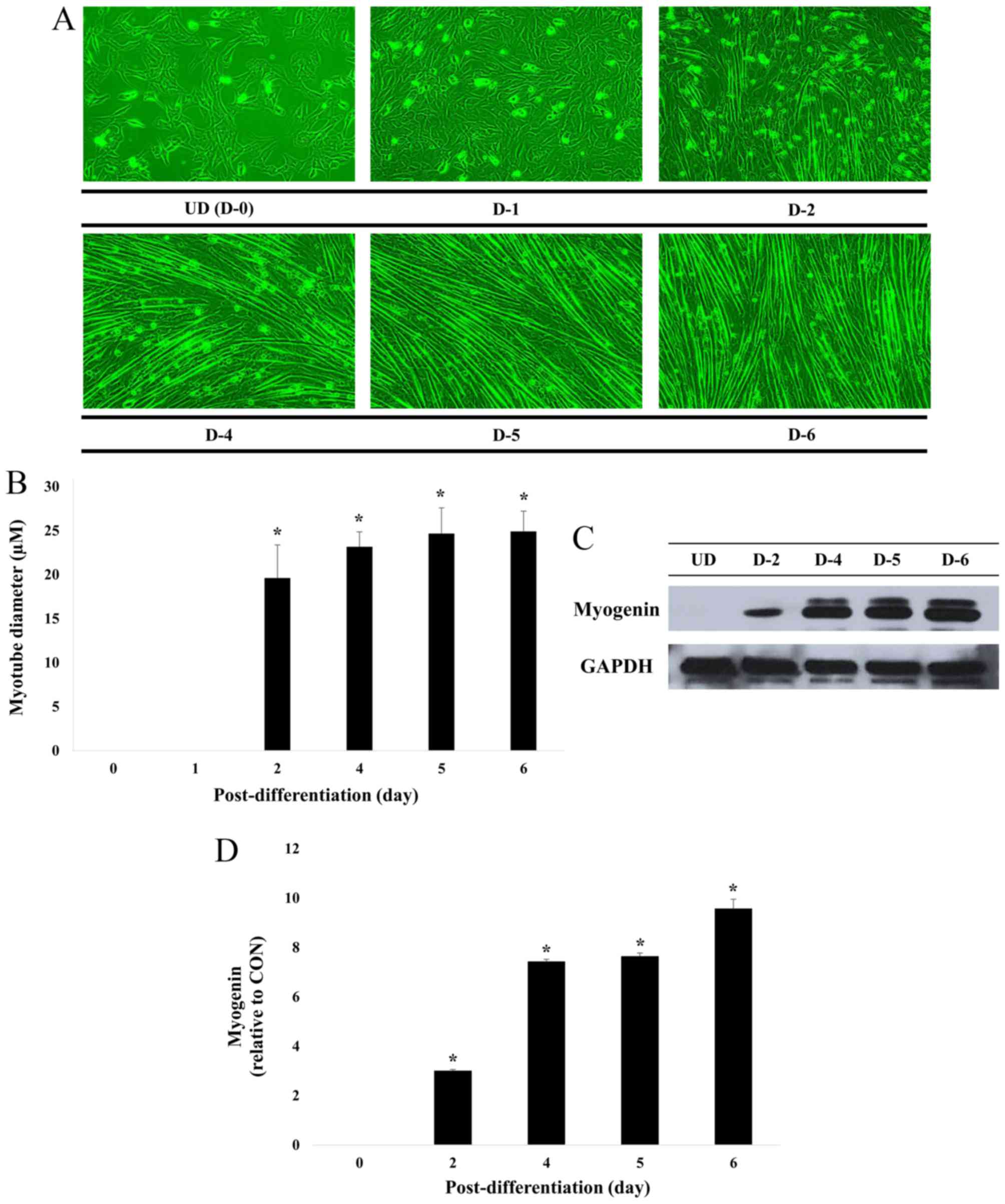
Fig. 2 illustrates that the cell
diameter remained similar following 4, 5 and 6 days of
differentiation. In addition, the level of myogenin expression, a
factor that regulates terminal differentiation of muscle cells, was
similar following 4 and 5 days then increased following 6 days.
These results indicate complete differentiation of myoblasts into
myotubes at this time (6 days).
DEX-induced muscle atrophy in C2C12
myotubes
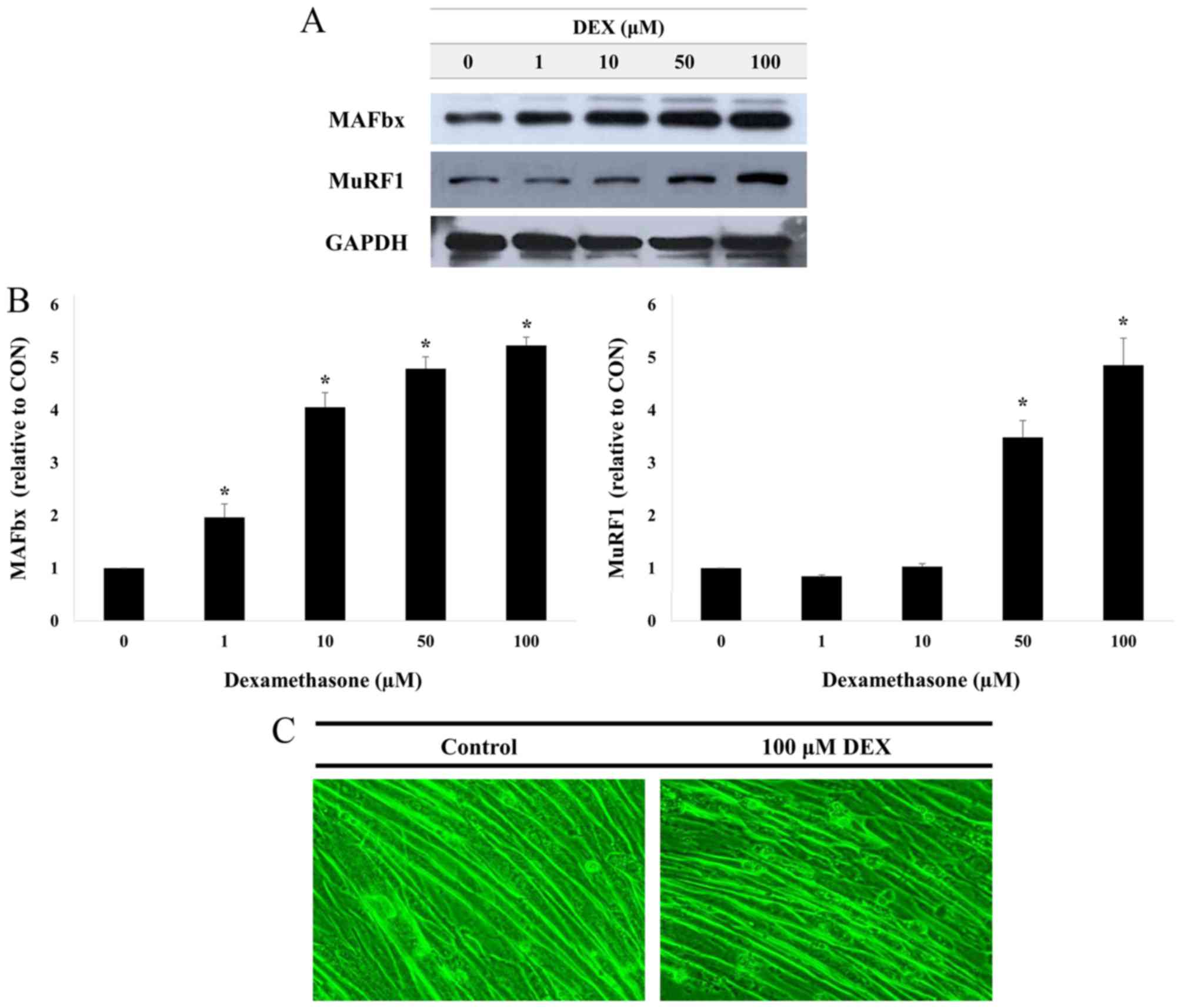
MuRF1 and atrogin1/MAFbx protein expression levels
were measured in C2C12 myotubes treated with 1, 10, 50 or 100 µM
DEX for 24 h (Fig. 3A and B).
Treatment with DEX led to increased protein expression levels for
MuRF1 and atrogin1/MAFbx, and the levels of expression increased in
a dose-dependent manner. These data suggest that DEX may induce
muscle atrophy in C2C12 myotubes. In addition, as shown in Fig. 3C, when compared with the control
group, the C2C12 myotube was markedly atrophied in the 100 µM
DEX-treated group. Therefore, all further experiments were
performed following treatment with 100 µM DEX.
Toxicity of PYP1-5 in C2C12
myoblast
Through mass spectrometry, we identified a 1,622 Da
compound from P. yezoensis, which we named PYP1-5 (Fig. 1). The MTS assay was used to
determine PYP1-5 cytotoxicity to C2C12 myoblasts, which
demonstrated no cytotoxic effects following PYP1-5 treatment for 24
h (Fig. 4). Therefore, all further
experiments were performed 24 h following treatment with 500 ng/ml
PYP1-5, which is the optimum concentration without toxicity as
described by Choi et al (18).
Inhibitory effect of PYP1-5 on
DEX-induced myotube atrophy
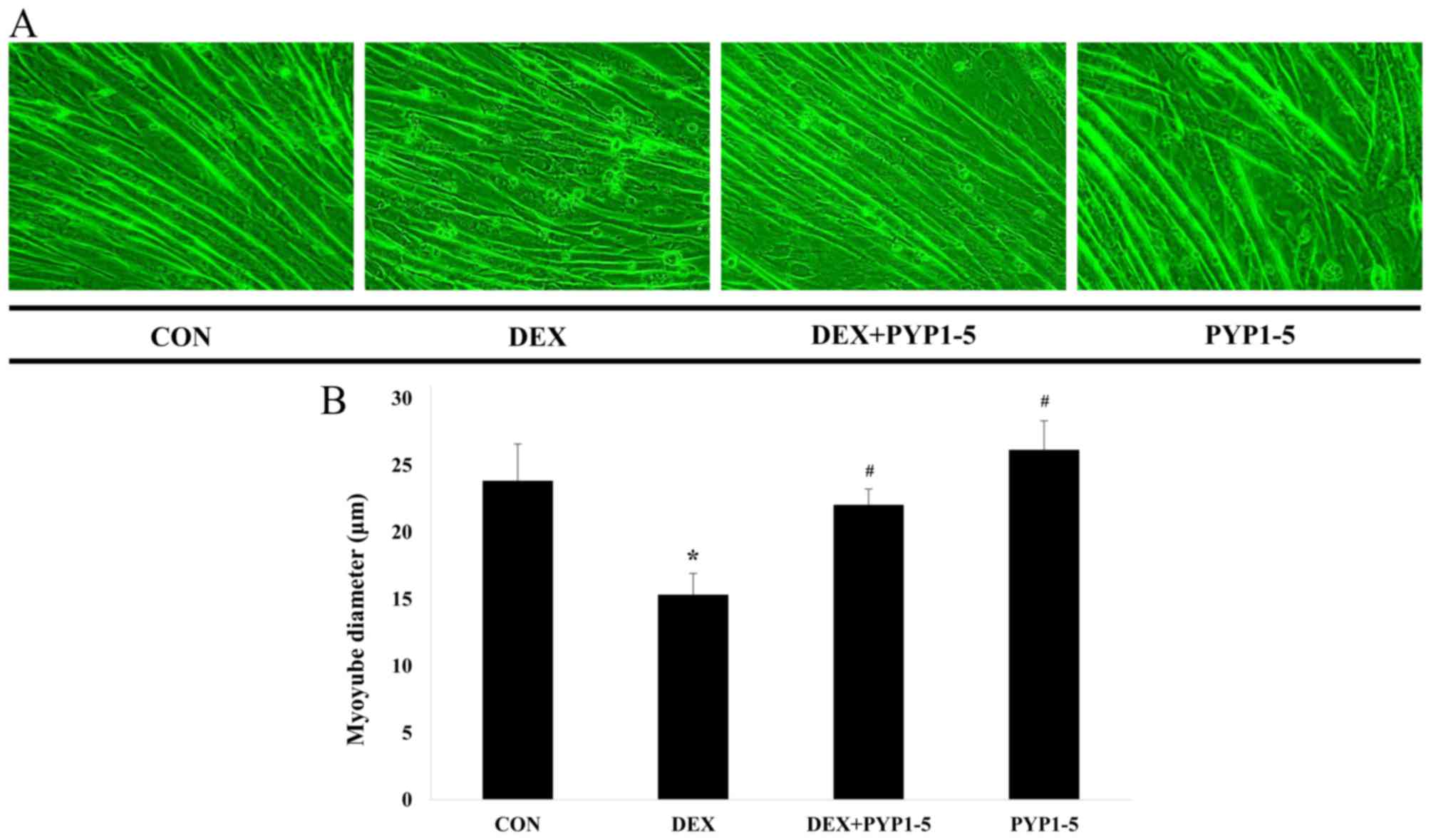
C2C12 myotubes were allowed to differentiate for 6
days, followed by 100 µM DEX and/or 500 ng/ml PYP1-5 treatment for
a further 24 h. As a result, when compared with the control group,
the DEX-treated group exhibited a 36% reduction in cell diameter,
whereas the PYP1-5-treated group exhibited a 10% increase in cell
diameter. In addition, DEX-induced reduction of cell diameter was
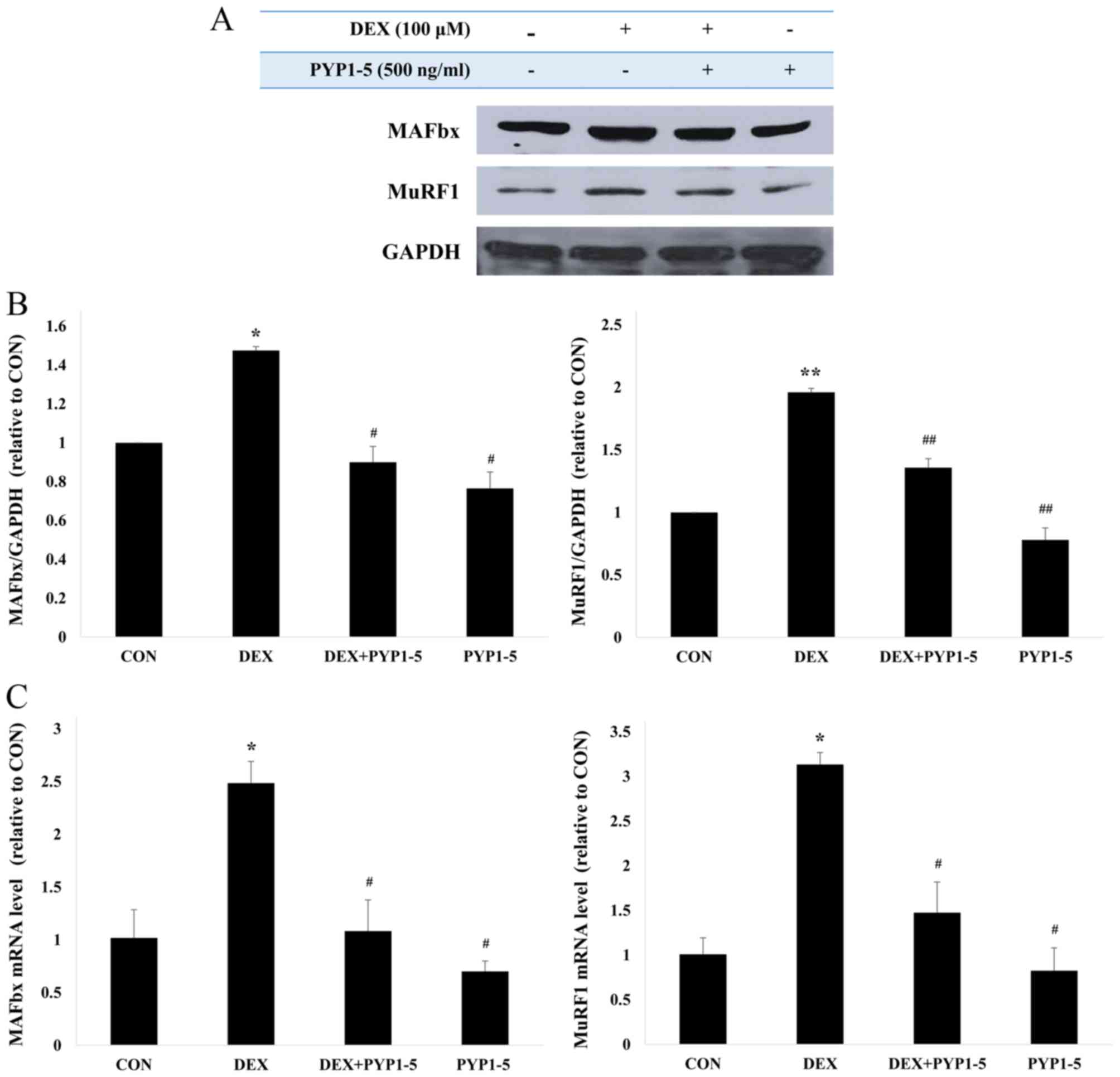
suppressed by co-treatment with PYP1-5 (Fig. 5A and B). The anti-atrophic effects
of PYP1-5 were investigated by examining the protein and mRNA
expression levels of the muscle atrophy markers MuRF1 and
atrogin1/MAFbx in C2C12 myotubes that were treated with 100 µM DEX
and 500 ng/ml PYP1-5 for 24 h (Fig.
6). Cells treated with DEX alone exhibited a marked increase in
the mRNA and protein expression levels of MuRF1 and atrogin1/MAFbx
compared with cells in the untreated control group (Fig. 6). The DEX-induced upregulation of
MuRF1 and atrogin1/MAFbx was suppressed by co-treatment with 500
ng/ml PYP1-5. Therefore, PYP1-5 may be able to inhibit atrophy and
induce hypertrophy pathways in C2C12 myotubes.
Discussion
Skeletal muscle atrophy refers to the decline in
muscle mass and strength that may occur as a result of various
conditions, including denervation, injury, glucocorticoid
treatment, starvation, cancer, joint immobilization, sepsis and
aging (21). The present study
focused on the progression of muscle atrophy caused by aging.
In the present study, anti-atrophic effects of the
P. yezoensis peptide PYP1-5 were investigated in C2C12
myotubes. C2C12 mouse skeletal muscle cells are commonly used as a
model for myotube differentiation to examine the signaling pathways
involved in muscle atrophy. The effects of the PYP1-5 on the muscle
atrophy in C2C12 myotubes were determined by first culturing C2C12
myoblasts in media containing 2% FBS for 6 days to induce
differentiation. Following differentiation, muscle atrophy was
induced by treating the cells with DEX. DEX treatment has been
previously demonstrated to reduce muscle mass, largely owing to the
breakdown of muscle proteins due to upregulated catabolism by the
ubiquitin-proteasome system (22,23).
Administration of high concentrations of DEX has been revealed to
cause muscle atrophy in animals and humans (24). Results from the present study
demonstrated that C2C12 myotubes respond to increasing
concentrations of DEX (1–100 µM) in a dose-dependent manner, as
revealed by the increased expression of MuRF1 and atrogin1/MAFbx
protein levels. Prior to determining the protective effects of
PYP1-5 on muscle atrophy, the potential toxicity of PYP1-5 on C2C12
myoblasts was determined using an MTS assay, which revealed that
PYP1-5 treatment had no cytotoxic effects on C2C12 myoblasts. We
next assessed the protective effects of PYP1-5 by measuring myotube
diameter. Compared with the control group, the DEX-treated group
exhibited a 36% reduction in cell diameter, whereas the
PYP1-5-treated group exhibited a 10% increase in cell diameter.
Subsequently, the effects of PYP1-5 on C2C12 myotube atrophy were
investigated by examining the expression levels of muscle atrophy
markers MuRF1 and atrogin1/MAFbx, as they are upregulated in a
number of catabolic conditions (25). Glucocorticoids such as DEX have
been demonstrated to promote myosin heavy chain degradation through
the activation of the E3 ligase MuRF1 (26). Therefore, the downregulation of
MuRF1 and atrogin1/MAFbx expression may inhibit muscle atrophy.
Western blot and RT-qPCR analyses in the present study revealed
that PYP1-5 treatment led to a decrease in the mRNA and protein
expression levels of MuRF1 and atrogin1/MAFbx, suggesting that
PYP1-5 may potentially be used in anti-atrophy functional
foods.
In conclusion, the present study provides important
new information about the influence of PYP1-5 on DEX-induced muscle
atrophy. The results provide molecular evidence that the
anti-atrophic effects of PYP1-5 are due to the downregulated
expression of the muscle-specific E3 ubiquitin ligases MuRF1 and
atrogin1/MAFbx. Future studies on the anti-atrophic effects of
PYP1-5 are required, and should involve identifying the signaling
pathways that may be associated with the anti-atrophic effects,
such as insulin-like growth factor 1/Akt and myostatin.
Acknowledgements
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant no.
2012R1A6A1028677).
References
|
1
|
Byeon Y, Lee H Yool, Choi DW and Back K:
Chloroplast-encoded serotonin N-acetyltransferase in the red alga
Pyropia yezoensis: Gene transition to the nucleus from
chloroplasts. J Exp Bot. 66:709–717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee MK, Kim IH, Choi YH, Choi JW, Kim YM
and Nam TJ: The proliferative effects of Pyropia yezoensis peptide
on IEC-6 cells are mediated through the epidermal growth factor
receptor signaling pathway. Int J Mol Med. 35:909–914.
2015.PubMed/NCBI
|
|
3
|
Wenz T, Rossi SG, Rotundo RL, Spiegelman
BM and Moraes CT: Increased muscle PGC-1alpha expression protects
from sarcopenia and metabolic disease during aging. Proc Natl Acad
Sci USA. 106:pp. 20405–20410. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Doherty TJ: Invited Review: Aging and
sarcopenia. J Appl Physiol (1985). 95:1717–1727. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marzetti E and Leeuwenburgh C: Skeletal
muscle apoptosis, sarcopenia and frailty at old age. Exp Gerontol.
41:1234–1238. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dupont-Versteegden EE: Apoptosis in muscle
atrophy: Relevance to sarcopenia. Exp Gerontol. 40:473–481. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Artaza JN, Bhasin S, Mallidis C, Taylor W,
Ma K and Gonzalez-Cadavid NF: Endogenous expression and
localization of myostatin and its relation to myosin heavy chain
distribution in C2C12 skeletal muscle cells. J Cell Physiol.
190:170–179. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aoki MS, Miyabara EH, Soares AG, Saito ET
and Moriscot AS: mTOR pathway inhibition attenuates skeletal muscle
growth induced by stretching. Cell Tissue Res. 324:149–156. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weintraub H: The MyoD family and
myogenesis: Redundancy, networks, and thresholds. Cell.
75:1241–1244. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Melo F, Carey DJ and Brandan E:
Extracellular matrix is required for skeletal muscle
differentiation but not myogenin expression. J Cell Biochem.
62:227–239. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
te Pas MF, Soumillion A, Harders FL,
Verburg FJ, van den Bosch TJ, Galesloot P and Meuwissen TH:
Influences of myogenin genotypes on birth weight, growth rate,
carcass weight, backfat thickness, and lean weight of pigs. J Anim
Sci. 77:2352–2356. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sandri M, Sandri C, Gilbert A, Skurk C,
Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH and Goldberg
AL: Foxo transcription factors induce the atrophy-related ubiquitin
ligase atrogin-1 and cause skeletal muscle atrophy. Cell.
177:399–412. 2004. View Article : Google Scholar
|
|
13
|
Zhao W, Qin W, Pan J, Wu Y, Bauman WA and
Cardozo C: Dependence of dexamethasone-induced Akt/FOXO1 signaling,
upregulation of MAFbx, and protein catabolism upon the
glucocorticoid receptor. Biochem Biophys Res Commun. 378:668–672.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glass DJ: Skeletal muscle hypertrophy and
atrophy signaling pathways. Int J Biochem Cell Biol. 37:1974–1984.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hershko A and Ciechanover A: The ubiquitin
system. Annu Rev Biochem. 67:425–479. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lecker SH, Jagoe RT, Gilbert A, Gomes M,
Baracos V, Bailey J, Price SR, Mitch WE and Goldberg AL: Multiple
types of skeletal muscle atrophy involve a common program of
changes in gene expression. FASEB J. 18:39–51. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng SJ and Yu LJ: Oxidative stress,
molecular inflammation and sarcopenia. Int J Mol Sci. 11:1509–1526.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi YH, Yamaguchi K, Oda T and Nam TJ:
Chemical and mass spectrometry characterization of the red alga
Pyropia yezoensis chemoprotective protein (PYP): Protective
activity of the N-terminal fragment of PYP1 against
acetaminophen-induced cell death in Chang liver cells. Int J Mol
Med. 35:271–276. 2015.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cooper ST, Maxwell AL, Kizana E, Ghoddusi
M, Hardeman EC, Alexander IE, Allen DG and North KN: C2C12
co-culture on a fibroblast substratum enables sustained survival of
contractile, highly differentiated myotubes with peripheral nuclei
and adult fast myosin expression. Cell Motil Cytoskeleton.
58:200–211. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gomes MD, Lecker SH, Jagoe RT, Navon A and
Goldberg AL: Atrogin1, a muscle-specific F-box protein highly
expressed during muscle atrophy. Proc Natl Acad Sci USA. 98:pp.
14440–14445. 2001; View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Auclair D, Garrel DR, Zerouala A Chaouki
and Ferland LH: Activation of the ubiquitin pathway in rat skeletal
muscle by catabolic doses of glucocorticoids. Am J Physiol.
272:C1007–C1016. 1997.PubMed/NCBI
|
|
23
|
Jackman RW and Kandarian SC: The molecular
basis of skeletal muscle atrophy. Am J Physiol. 287:C834–C843.
2004. View Article : Google Scholar
|
|
24
|
Mitch WE and Goldberg AL: Mechanisms of
muscle wasting. The role of the ubiquitin-proteasome pathway. N
Engl J Med. 335:1897–1905. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krawiec BJ, Nystrom GJ, Frost RA,
Jefferson LS and Lang CH: AMP-activated protein kinase agonists
increase mRNA content of the muscle-specific ubiquitin ligases
MAFbx and MuRF1 in C2C12 cell. Am J Physiol Endocrinol Metab.
292:E1555–E1567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clarke BA, Drujan D, Willis MS, Murphy LO,
Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Laltres E
and Glass DJ: The E3 ligase MuRF1 degrades myosin heavy chain
protein in dexamethasone-treated skeletal muscle. Cell Metab.
6:376–385. 2007. View Article : Google Scholar : PubMed/NCBI
|