Introduction
Human cytomegalovirus (HCMV) infection is a major
cause of high morbidity and mortality in patients that have
undergone allogeneic hematopoietic stem cell transplantation (HSCT)
(1,2). Cellular immunity through
antigen-specific cytotoxic T lymphocytes (CTLs) is involved in
long-term suppression (3,4). Each individual CTL has a specific
complementarity determining region 3 (CDR3) located in the T cell
receptor β variable (TCR BV) region, which occurs as a result of
V(D)J recombination and junctional diversity. During an antiviral
immune response, the interactions between a TCR and its
antigen-specific peptides, which are mediated in part by CDR3,
result in a polyclonal expansion of T cells and clones expressing
different CDR3 sequences (5).
Determining the frequency of specific CDR3 sequences within a
T-cell population may provide an accurate estimation of the extent
of clonal expansion and the function of the expanded populations.
It is well-known that TCRs are closely associated with viral
infections, specifically hepatitis B virus (HBV) and human
immunodeficiency virus (HIV) (6,7). The
occurrence of HCMV reactivation in patients following HSCT is also
well documented (8,9). However, the underlying mechanisms
responsible for this reactivation remain unknown. In the present
study, reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and DNA melting curve analysis were used to evaluate the
distribution of TCR BV CDR3 genes expressed in peripheral blood
mononuclear cells (PBMCs) isolated from patients that had undergone
HSCT. This analysis evaluated the impact of T-cells on HCMV
reactivation beyond T cell clonal expansion, thus providing
molecular evidence that an association exists between HCMV
infection and immune dysregulation in patients following HSCT.
Materials and methods
Subjects
A total of 3 HSCT recipients at The First Affiliated
Hospital, Zhejiang University School of Medicine (Hangzhou, China)
were enrolled in the present study between January 2011 and
December 2012. One healthy donor from the same hospital was
enrolled as a control, in December 2012. Patients with the
following viral infections were excluded from this study: HIV, HBV,
hepatitis A virus, hepatitis C virus, hepatitis D virus, hepatitis
E virus, herpes simplex virus, and Epstein-Barr virus. Recipient 1
was not infected with HCMV, while repeated HCMV reactivation
occurred in recipients 2 and 3. Additional patient information is
listed in Table I. The present
study was approved by the Ethics Committee of the First Affiliated
Hospital at the Medical School of Zhejiang University (Hangzhou,
China). Written, informed consent was obtained from patients
according to the Declaration of Helsinki.
 | Table I.Characteristics of recipients and the
healthy control. |
Table I.
Characteristics of recipients and the
healthy control.
| General
information | Recipient1 | Recipient 2 | Recipient 3 | Healthy control |
|---|
| Sex | Female | Female | Male | Male |
| Age | 31 | 24 | 20 | 25 |
| Underlying
disease | CML | AMMOL(M4) | ALL | None |
| HIV | − | − | − | − |
| HSV | − | − | − | − |
| HAV | − | − | − | − |
| HBV | − | − | − | − |
| HCV | − | − | − | − |
| HDV | − | − | − | − |
| HEV | − | − | − | − |
| EBV | − | − | − | − |
| HCMV-IgG | + | + | + | + |
| Conditioning
regimen |
ARA-C+BUCY+Me-CCNU+ATG |
ARA-C+BUCY+ATG+MeCCNU | BUCY+MeCCNU | N/A |
| Blood type | B/B | B/B | B/B | B |
| HLA | HLA0201 | HLA0201 | HLA0201 | HLA0201 |
| Immunosuppressant
regimen | CSA, MMF, PRED | CSA, MMF, PRED | CSA, MMF, PRED | N/A |
| Antiviral
pretreatment | GCV, ACV | GCV, ACV | GCV, ACV | N/A |
TCR BV CDR3 genes expressed in PBMCs from all
subjects were detected using RT-qPCR and a DNA melting curve
analysis at the third month after transplantation (10,11).
HCMV-pp65, HCMV-immediate early protein (HCMV-IE),
HCMV-immunoglobulin (HCMV-Ig) M and HCMV-IgG were detected on the
same day, and serial analysis of HCMV infection continued monthly
until ~1 year after HSCT.
RNA extraction and cDNA synthesis
A total of 5 ml blood was collected from each
subject, and PBMCs were isolated using a Ficoll-Paque density
gradient technique. Total RNA was extracted using TRNzol reagent
(Tiangen Biotech Co., Ltd., Beijing, China), according to the
manufacturer's protocol. Total RNA was reverse transcribed to cDNA
using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. Sample RNA of 1–5 µg was reverse transcribed with the
OligodT18 primer in a 20 µl reaction volume and stored at −80°C
prior to being used as the template for PCR amplification.
RT-qPCR amplification of CDR3
cDNA
In this study, we used primers for 24 TCR BV gene
families (Table II) (12). A TransStart TM Green qPCR Super Mix
(Tiangen Biotech Co., Ltd., Beijing, China) was used for qPCR. PCR
reactions contained 0.5 µl reverse primer (TCR BV), 0.5 µl forward
primer (24 TCR BV genes), 1 µl template cDNA, 12.5 µl qPCR Super
Mix (2X), 0.5 µl passive reference (50X), and 10 µl RNase-free
distilled water, to a final volume of 25 µl. Reactions were
performed using an ABI 7500 system and were analyzed with v2.0.6
software (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
reaction parameters were as follows: 2 min at 94°C to activate the
GoTaq DNA polymerase enzyme (Promega Corporation, Madison, WI,
USA), followed by 45 cycles at 94°C for 15 sec, 56.0°C for 25 sec,
72°C for 35 sec, 80°C for 2 sec, and a final extension at 72°C for
8 min. The melting step was performed by slow heating from 75 to
95°C with a ramping rate of 0.2°C/s, during which the fluorescence
signal was continuously measured. Simultaneous amplification of TCR
β chain constant 1 and glyceraldehyde 3-phosphate dehydrogenase
were used as positive controls.
 | Table II.Primers of 24 TCR BV families and
GAPDH. |
Table II.
Primers of 24 TCR BV families and
GAPDH.
| Name of primer | Sequence
(5′-3′) | V region to
CDR3 |
|---|
| BV1 |
gcacaacagttccctgacttgcac | 90
bp |
| BV2 |
tcatcaaccatgcaagcctgacct | 90
bp |
| BV3 |
gtctctagagagaagaaggagcgc | 91
bp |
| BV4 |
acatatgagagtggatttgtcatt | 124 bp |
| BV5.1 |
atacttcagtgagacacagagaaac | 142 bp |
| BV5.2 |
ttccctaactatagctctgagctg | 81
bp |
| BV6 |
aggcctgagggatccgtctc | 85
bp |
| BV7 |
cctgaatgccccaacagctctc | 91
bp |
| BV8 |
atttactttaacaacaacgttccg | 128 bp |
| BV9 |
cctaaatctccagacaaagctcac | 91
bp |
| BV10 |
ctccaaaaactcatcctgtacctt | 83
bp |
| BV11 |
tcaacagtctccagaataaggacg | 97
bp |
| BV12 |
aaaggagaagtctcagat | 118 bp |
| BV13.1 |
caaggagaagtccccaat | 118 bp |
| BV13.2 |
ggtgagggtacaactgcc | 136 bp |
| BV14 |
gtctctcgaaaagagaagaggaat | 91
bp |
| BV15 |
agtgtctctcgacaggcacaggct | 95
bp |
| BV16 |
aaagagtctaaacaggatgagtcc | 139 bp |
| BV17 |
cagatagtaaatgactttcag | 139 bp |
| BV18 |
gatgagtcaggaatgccaaaggaa | 124 bp |
| BV19 |
caatgccccaagaacgcaccctgc | 88
bp |
| BV20 |
agctctgaggtgccccagaatctc | −131
bpa |
| BV21 |
gattcacagttgcctaagga | 121 bp |
| BV22 |
cagagaagtctgaaatattcga | 128 bp |
| BV23 |
gatcgattctcagctcaacag | 103 bp |
| BV24 |
aaagattttaacaatgaagcagac | 133 bp |
| TCR BC1
anti-sense |
ttctgatggctcaaacac |
|
| GAPDH
anti-sense |
aggggtctacatggcaact |
|
| GAPDH sense |
cgaccactttgtcaagctca | 227 bp (predicted
size) |
CDR3 sequencing of monoclonal TCR BV
families
Using this melting curve, PCR products from the TCR
BV gene families that had a single-peak expansion were selected.
Single-peak expansion was defined as ‘monoclonal’, and appeared as
only one main peak in the gene melting spectral pattern (GMSP).
‘Nonskewed’ means polyclonal amplification, which appeared as no
visibly apparent main peak. The PCR products were re-amplified
using GoTaq DNA polymerase (Promega Corporation, Madison, WI, USA)
by touchdown PCR. The parameters were as follows: pre-incubation at
95°C for 2 min, 95°C for 30 sec, 58.5°C for 40 sec and 72°C for 45
sec, and 6 cycles with annealing temperature decreasing 0.5°C per
cycle, followed by 34 cycles of 95°C for 45 sec, 56°C for 45 sec
and 72°C for 45 sec. At the end, a terminal elongation step at 72°C
for 8 min was added. Nested PCR products were sequenced using an
ABI 3730 DNA Sequencer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Samples were sent to Sangon Biotech Co., Ltd.,
Shanghai, China, and sequenced there. Results were analyzed
automatically by a 3730xl DNA Analyzer and the sequencing reagent
was BigDye terminator version 3.1.
Analysis of CDR3 sequences
Chromas software version 2.22 (Technelysium Pty
Ltd., Brisbane, Australia) was used to translate nucleotide
sequences into amino acid sequences (13). When the PCR nucleic acid product
underwent electrophoresis and only one band was present, this
represented direct sequencing. The presence of a thin strip in
addition to a clear band, suggested cloned sequencing. Samples were
sent to Sangon Biotech Co., Ltd., and sequenced there.
Detection of HCMV-pp65 and IE
antigenemia
Peripheral blood samples were collected in
ethylenediaminetetraacetic acid (EDTA)-anticoagulant tubes. To
evaluate pp65 and IE antigenemia a standard two-step
immunohistochemical method was used, as described previously
(14,15). Briefly, 5×104 PBMCs were
fixed on polylysine-coated slides, and incubated with mouse
anti-HCMV (pp65 catalog no. ab53495; IE catalog no. ab53489; 1:100;
Abcam, Cambridge, UK) monoclonal antibodies at 37°C for 30 min, and
horseradish peroxidase-conjugated rabbit anti-mouse IgG polyclonal
antibody (catalog no. ab6728; 1:250; Abcam) at 37°C for 30 min. A
total of 5×104 PBMCs were fixed on one slide, and every
sample was fixed on 2 slides. Results were quantified based on the
average number of brown stained positive cells per 5×104
leukocytes in the 2 slides. Cells were observed under a light
microscope (BH-2; Olympus Corporation, Tokyo, Japan) with
magnification ×100/200/400.
Detection of HCMV-IgG/IgM
Blood samples were collected in EDTA-anticoagulant
tubes. HCMV-antibody serostatus (IgG and IgM) was determined using
enzyme-linked immunosorbent assays (ELISA) according to the
manufacturer's protocols (Dia.Pro Diagnostic Bioprobes s.r.l.,
Milan, Italy).
Statistical analysis
Differences in HCMV antigenemia and the presence of
HCMV-IgG/IgM among the 3 HSCT recipients were examined using
one-way analysis of variance followed by Tukey's test. Graphical
analyses of results were generated using Prism 5 (Graph-Pad, San
Diego, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Frequency of skewed TCR BV gene
families and CDR3 sequences derived from monoclonal TCR BV
expansion in 3 recipients of HSCT
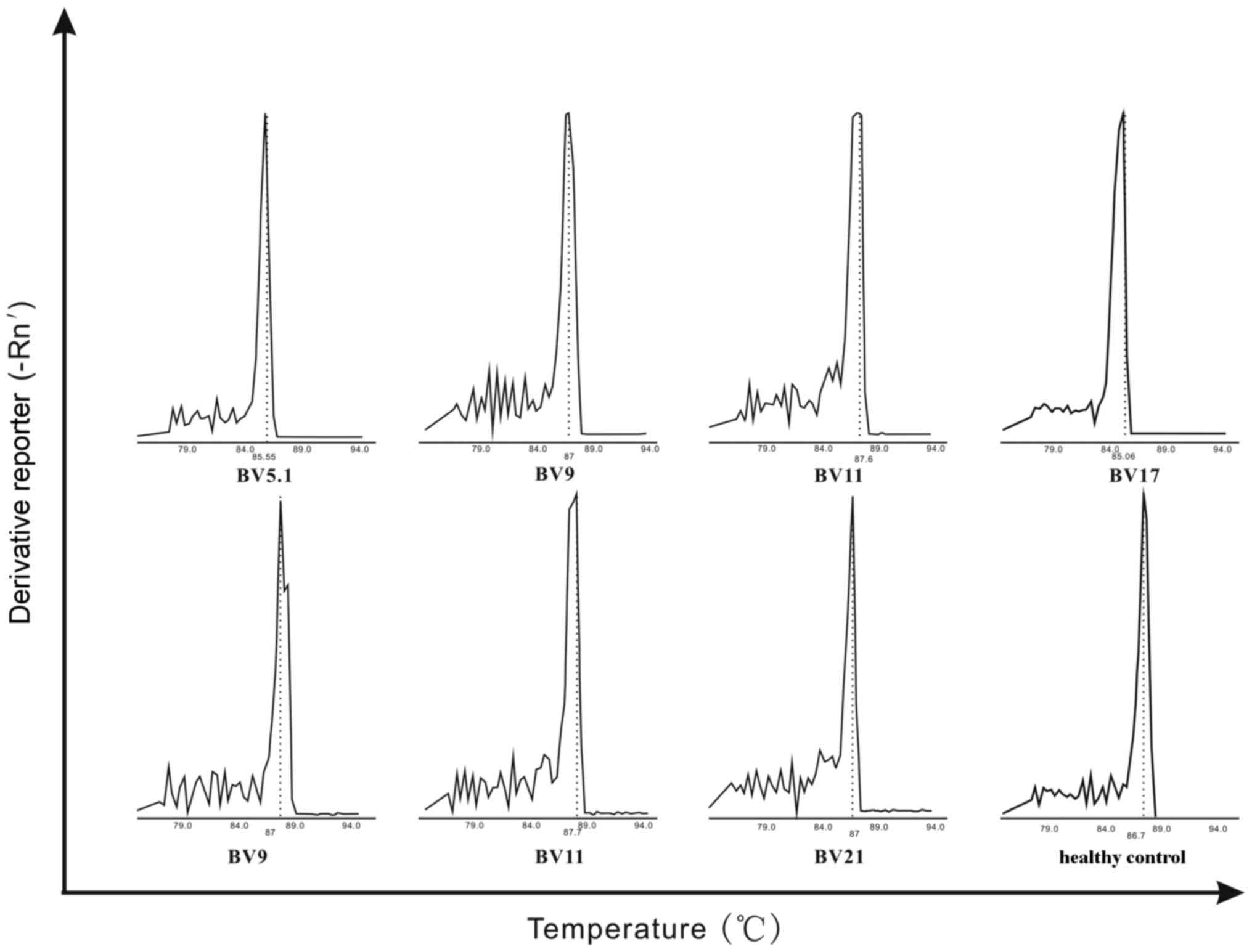
All 3 recipients of HSCT demonstrated preferential
expansion of specific TCR BV gene families. The characteristics of
TCRBV CDR3 expression are visualized in Fig. 1. For recipient 1, 5 monoclonal
peaks were observed and TCR BV5.1 was preferentially expressed. In
recipient 2, 4 monoclonal peaks were observed, including TCR BV9,
TCR BV11, and TCR BV17. In recipient 3, 10 monoclonal peaks were
observed and TCR BV9, TCR BV11, and TCR BV21 were selectively
expressed. Recipients 2 and 3 had 2 gene families in common; TCR
BV9 and TCR BV11 (Fig. 1).
A single peak in a GMSP indicates monoclonal
expansion of a particular TCR BV clone, and this was verified by
direct sequencing. Representative amino acid sequences of the TCR
BV CDR3 in PBMCs from all recipients are listed in Table III. All recipients had a CDR3
sequence length of 5–12 amino acids. The amino acid sequences of
TCR BV9 (QVRGGTDTQ) and TCR BV11 (VATDFQ) were similar between
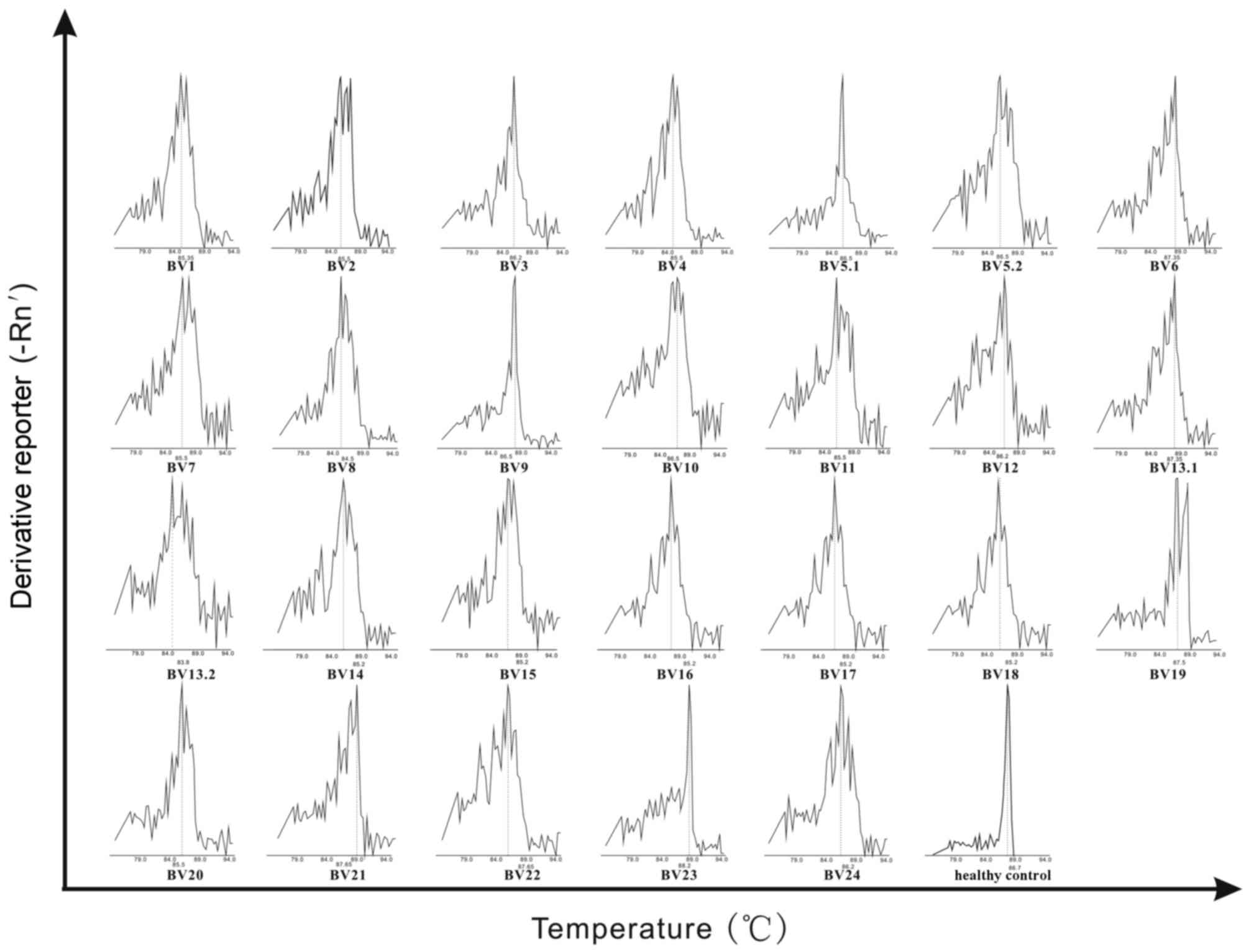
recipients 2 and 3. The healthy control expressed a non-skewed TCR
BV repertoire. Representative GMSPs for non-skewed and
oligoclonally expanded TCRBV gene families are visualized in
Fig. 2.
 | Table III.Representative amino acid sequences
of monoclonal TCRBV populations from 3 recipients of allogeneic
hematopoietic stem cell transplantation. |
Table III.
Representative amino acid sequences
of monoclonal TCRBV populations from 3 recipients of allogeneic
hematopoietic stem cell transplantation.
| Recipient | Primer BV | Vβ | CDR3 | Jbeta |
|---|
| 1 | BV5.1 | SALYLCASS | SPRDRGYGDTQ | YFGPGTRLTVLED |
| 2 | BV9 | SAVYFCASS | QVRGGTDTQ | YFGPGTRLTVLET |
|
| BV11 | TSQHFCASS | VATDEQ | FFGPGTRLTVLED |
|
| BV17 | TAFYLCASS | IGQGNTEA | FFGQGTRLTV |
| 3 | BV9 | SAVYFCASS | QVRGGTDTQ | YFGPGTRLTVLED |
|
| BV11 | TSQYLCASS | VATDEQ | FFGPGTWLTVLED |
|
| BV21 | SAVYLCASS | GMIGRLTDTQ | YFGPGTRLTVLED |
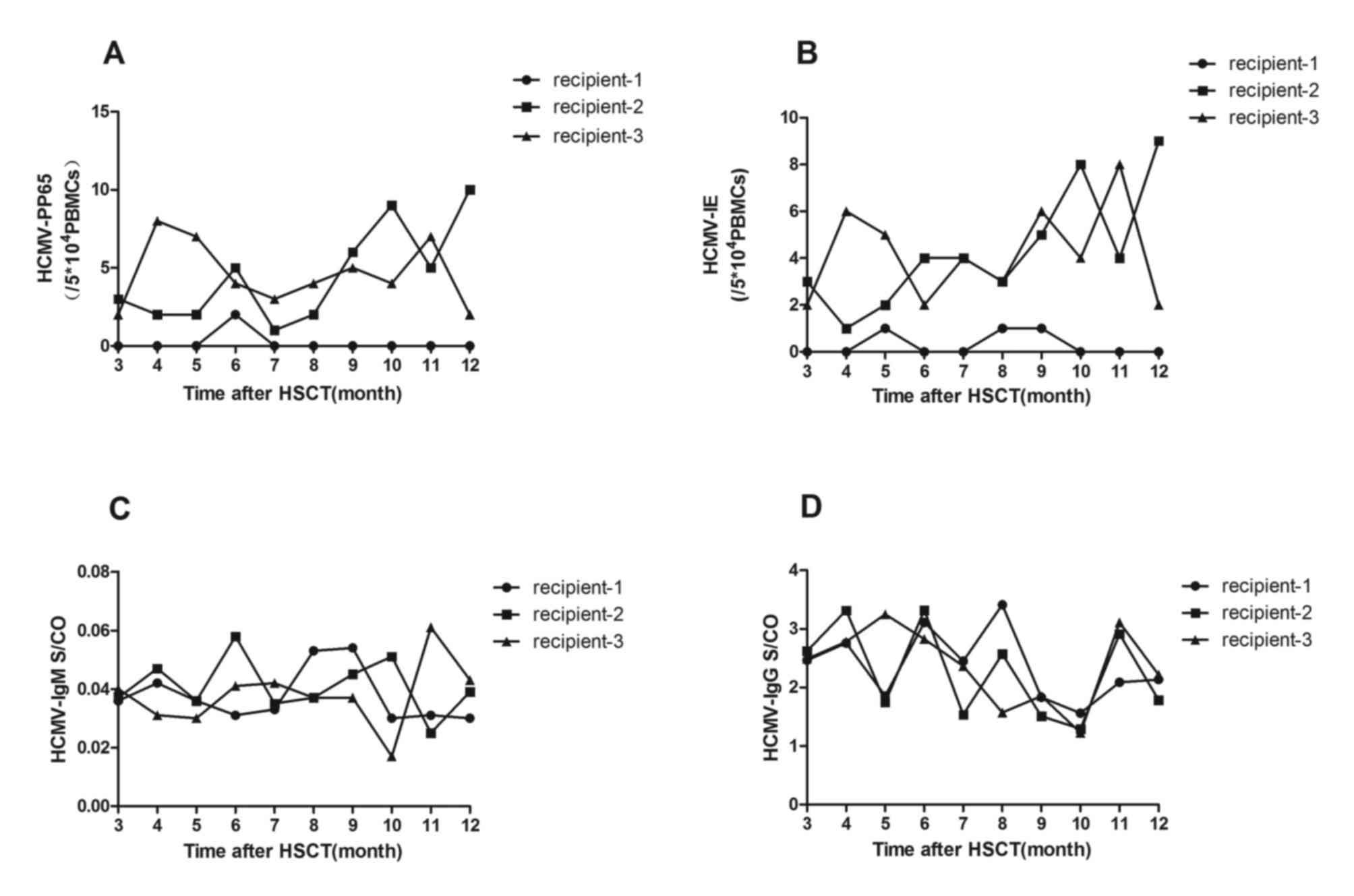
Detection of pp65 and IE
antigenemia
In all transplant recipients, pp65 and IE
antigenemia were monitored ~10 times for up to one year following
HSCT. Samples obtained from recipient 1 were typically pp65- and
IE-negative, while recipients 2 and 3 consistently tested positive
for pp65 (Fig. 3A) and IE
antigenemia (Fig. 3B). For
recipient 2, the mean number of pp65-positive cells was
4.5/5×104 PBMCs (range, 1–10 positive
cells/5×104 PBMCs), and the mean number of IE-positive
cells was 4.3/5×104 PBMCs (range, 1–9 positive
cells/5×104 PBMCs). For recipient 3, a mean number of
pp65-positive cells of 4.6/5×104 PBMCs (range, 2–8
positive cells/5×104 PBMCs) and a mean number of
IE-positive cells of 4.2/5×104 PBMCs (range, 2–8
positive cells/5×104 PBMCs) was observed.
Detection of HCMV serostatus (IgG and
IgM)
Over the course of 1 year following HSCT,
HCMV-specific IgG and IgM were investigated on 10 separate
occasions for all 3 recipients, during detection of pp65 and IE
antigens. During the present study, all recipients remained
HCMV-IgG-positive and HCMV-IgM-negative.
Analysis of HCMV-pp65 and -IE
antigenemia levels
The levels of HCMV-pp65 and IE antigenemia were
evaluated using a standard two-step immunohistochemical method on
60 samples from 3 recipients following HSCT. All recipients were
tested on 10 separate occasions, up to one year following HSCT and
all data of HCMV-pp65 or HCMV-IE collected during this period were
involved. Statistically significant differences were observed in
the levels of HCMV-pp65 (P<0.05; Fig. 3A) and -IE (P<0.05; Fig. 3B) antigenemia between recipients 1
and 2. This result was also observed between recipients 1 and 3
(P<0.05 and P<0.05, respectively; Fig. 3A and B, respectively). However,
there was no significant difference in the levels of pp65 and IE
antigenemia between recipients 2 and 3 (P>0.05 and P<0.05,
respectively; Fig. 3A and B,
respectively).
Comparison of the HCMV serostatus (IgG
and IgM) among recipients of HSCT
HCMV-specific-IgG and IgM was detected using ELISA
on 60 samples from 3 patients 1 year following HSCT. Each recipient
was tested 10 times on separate occasions. During the study, all
recipients remained HCMV-specific-IgG-positive and IgM-negative. No
significant difference between HCMV-IgG and IgM optical
density/cut-off among the 3 recipients was observed.
(P>0.05).
Comparison of HCMV antigenemia levels
between TCR BV9/TCR BV11-positive recipients and the TCR BV9/TCR
BV11-negative recipient
Recipients who preferentially expressed TCR BV9 and
TCR BV11 were defined as TCR BV9+ and TCR
BV11+, respectively. Among the 3 recipients enrolled,
recipients 2 and 3 were TCR BV9+/TCR BV11+.
‘QVRGGTDTQ’ was the conserved amino acid sequence in TCR BV9 CDR3
and ‘VATDFQ’ was observed in TCR BV11 CDR3 (Table III). Two recipients exhibited
pp65 and IE antigenemia, while being simultaneously positive for
HCMV-IgG and negative for HCMV-IgM. Recipient 1 was TCR
BV9−/TCR BV11−, HCMV-IgG+ and
HCMV-IgM−, but free of pp65 and IE antigenemia (Table IV). To determine whether HCMV
reactivation was associated with a specific amino acid sequence of
TCR BV CDR3, HCMV antigenemia status were compared between the TCR
BV9+/TCR BV11+ recipients and the TCR
BV9−/TCR BV11− recipient (Table IV). This revealed that HCMV
reactivation may be associated with TCR BV9 and TCR BV11, and a
specific amino acid sequence of TCR BV CDR3 may be involved in HCMV
infection.
 | Table IV.HCMV antigenemia status and TCR
BV9/BV11 expression among the 3 recipients of allogeneic
hematopoietic stem cell transplantation. |
Table IV.
HCMV antigenemia status and TCR
BV9/BV11 expression among the 3 recipients of allogeneic
hematopoietic stem cell transplantation.
|
| HCMV
antigenemia |
|---|
|
|
|
|---|
| Recipient | TCR BV9 | TCR BV11 | HCMV-pp65 | HCMV-IE |
|---|
| 1 | − | − | − | − |
| 2 | + | + | + | + |
| 3 | + | + | + | + |
Discussion
Although HCMV reactivation is commonly observed
following immune dysfunction from HSCT, the molecular mechanisms
that drive this phenomenon remain unknown. T cell immune responses
induced by viral antigens are involved in the inflammatory process
of stemming viral infections. CTLs are involved in HCMV control and
pathogenesis (16). In PBMCs,
>95% of the T cells are αβ+ (17). During T cell development, the TCR β
chain undergoes rearrangement earlier than the α chain, according
to rules of allelic exclusion. Therefore, analysis of the TCRBV
CDR3 gene may be beneficial in determining the clonality of a
particular T cell response, and therefore be used as a marker for
the functional status of T cells (18). Studies focused on TCR BV gene
families may provide novel insights and a solid foundation for the
prevention, diagnosis, and treatment of viral infections (19).
The TCR BV gene family demonstrated a diverse,
non-skewed expansion in PBMCs derived from the healthy control.
However, particular TCR BV families were preferentially expressed
or biased, and emerged as a group of monoclonal (oligoclonal) T
cells in the PBMCs of all 3 recipients of HSCT. Due to the
long-term use of systemic steroids, the T cell response to HCMV is
impaired and HCMV reactivation often occurs in patients that have
undergone HSCT (20,21). This was determined by the
restricted use and oligoclonal expression of TCR BV families
following transplantation. The TCR BV gene rearrangement was random
without antigen stimulation (22).
TCR gene transfer was developed as a promising means of generating
large numbers of T cells of a given antigen specificity and
functional avidity ex vivo. This technique has demonstrated
significant potential for clinical use (23–26).
In the present study, TCR β chain sequences and the
presence of HCMV infection were analyzed in recipients of HSCT. A
relationship between HCMV reactivation and TCR BV families was
observed. TCR BV9 and TCR BV11-positive recipients had HCMV
antigenemia. ‘QVRGGTDTQ’ was the most common amino acid sequence
observed for TCR BV9 CDR3, and ‘VATDFQ’ was observed for TCR BV11
CDR3. The explanation for this finding may be the multifarious
usage of distinctive TCR BV families. Conserved sequences of the
TCR BV repertoire were diverse without peptide stimulation, but
became more restrictive following stimulation by the HCMV peptide.
The clinical course of HCMV reactivation was affected by the usage
of TCR BV9 and TCR BV11 in recipients of HSCT. Expression of TCR
BV9 and TCR BV11 may be associated with HCMV antigenemia, and may
be involved in the immune response. The amino acid sequences
‘QVRGGTDTQ’ and ‘VATDFQ’ may be beneficial for eliciting an
anti-viral response, as well as contributing to HCMV clearance. TCR
BV with the sequences ‘QVRGGTDTQ’ and ‘VATDFQ’ may, therefore, be a
risk factor for HCMV reactivation.
Although the number of patients that underwent HSCT
enrolled in the present study was relatively small, each of the
patients underwent detailed longitudinal analysis with 10 separate
follow-ups over the course of 1 year. In total, >120 samples
were analyzed. In addition, the contents of the present study are
only a part of longer-term research work. Another challenge
regarding the methodology is that the controls would have been
improved had they been HCMV-negative patients that underwent HSCT.
However, ~100% Han Chinese people are HCMV-IgG positive (27), and HCMV-IgG negative HSCT
recipients were not identified. Therefore, such negative controls
were not used in the present study.
Given that immune reconstitution must start from the
beginning for a patient who has undergone HSCT, these patients are
at high risk for HCMV reactivation. The results from the present
study provide a link between HCMV reactivation and immune
homeostasis, and thus help to establish a prophylaxis and diagnosis
of HCMV reactivation following HSCT. Assessment of clonal diversity
of TCR against HCMV may provide important insights into the
molecular basis of T cell immunodominance. However, further
investigation is necessary to address this issue in recipients of
HSCT.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30872239) and the
Zhejiang Provincial Natural Science Foundation of China (grant no.
LY14H190002).
Glossary
Abbreviations
Abbreviations:
|
TCR BV
|
T-cell receptor β variable
|
|
CDR3
|
complementarity determining region
3
|
|
HSCT
|
allogeneic hematopoietic stem cell
transplantation
|
|
HCMV
|
human cytomegalovirus
|
|
GMSP
|
gene melting spectral pattern
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
CTLs
|
cytotoxic T lymphocytes
|
|
HBV
|
hepatitis B virus
|
|
HIV
|
human immunodeficiency virus
|
References
|
1
|
Gratama JW and Cornelissen JJ: Diagnostic
potential of tetramer-based monitoring of cytomegalovirus-specific
CD8+ T lymphocytes in allogeneic stem cell
transplantation. Clin Immunol. 106:29–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mori T and Kato J: Cytomegalovirus
infection/disease after hematopoietic stem cell transplantation.
Int J Hematol. 91:588–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moins-Teisserenc H, Busson M, Scieux C,
Bajzik V, Cayuela JM, Clave E, de Latour RP, Agbalika F, Ribaud P,
Robin M, et al: Patterns of cytomegalovirus reactivation are
associated with distinct evolutive profiles of immune
reconstitution after allogeneic hematopoietic stem cell
transplantation. J Infect Dis. 198:818–826. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gamadia LE, Rentenaar RJ, Baars PA,
Remmerswaal EB, Surachno S, Weel JF, Toebes M, Schumacher TN, ten
Berge IJ and van Lier RA: Differentiation of
cytomegalovirus-specific CD8(+) T cells in healthy and
immunosuppressed virus carriers. Blood. 98:754–761. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alli R, Zhang ZM, Nguyen P, Zheng JJ and
Geiger TL: Rational design of T cell receptors with enhanced
sensitivity for antigen. PLoS One. 6:e180272011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bohne F, Chmielewski M, Ebert G, Wiegmann
K, Kürschner T, Schulze A, Urban S, Krönke M, Abken H and Protzer
U: T cell redirected against hepatitis B virus surface proteins
eliminate infected hepatocytes. Gastroenterology. 134:239–247.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sommermeyer D, Neudorfer J, Weinhold M,
Leisegang M, Engels B, Noessner E, Heemskerk MH, Charo J, Schendel
DJ, Blankenstein T, et al: Designer T cells by T cell receptor
replacement. Eur J Immunl. 36:3052–3059. 2006. View Article : Google Scholar
|
|
8
|
Torelli GF, Lucarelli B, Iori AP, De
Propris MS, Capobianchi A, Barberi W, Valle V, Iannella E, Natalino
F, Mercanti C, et al: The immune reconstitution after an allogeneic
stem cell transplant correlates with the risk of graft-versus-host
disease and cytomegalovirus infection. Leuk Res. 35:1124–1126.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abu-Khader A and Krause S: Rapid
monitoring of immune reconstitution after allogeneic stem cell
transplantation-a comparison of different assays for the detection
of cytomegalovirus-specific T cells. Eur J Haematol. 91:534–545.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang JZ, Li MW, Wang JG, Lu HF, Yao XS, He
JQ and Li LJ: Rapid detection of clonal expansion of T-cell
receptor-beta gene in patients with HBV using the real-time PCR
with DNA melting curve analysis. Hepatol Res. 40:407–414. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang J, He J, Lu H, Wei L, Li S, Wang B,
Diao H and Li L: Molecular features of the complementarity
determining region 3 motif of the T cell population and subsets in
the blood of patients with chronic severe hepatitis B. J Transl
Med. 9:2102011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arenz M, zum Büschenfelde KH Meyer and
Löhr HF: Limited T cell receptor Vbeta-chain repertoire of
liver-infiltrating T cells in autoimmune hepatitis. J Hepatol.
28:70–77. 1988. View Article : Google Scholar
|
|
13
|
Yang J, He J, Huang H, Ji Z, Wei L, Ye P,
Xu K and Li L: Molecular characterization of T cell receptor beta
variable in the peripheral blood T cell repertoire in subjects with
active tuberculosis or latent tuberculosis infection. BMC Infect
Dis. 13:4232013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan J, Zhang X, Chen XM, Gao HN, Yang MF,
Zhao H, Hu JH and Ma WH: Monitoring of human cytomegalovirus
glycoprotein B genotypes using real-time quantitative PCR in
immunocompromised Chinese patients. J Virol Methods. 160:74–77.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Huang YP, Gao HN, Yang MF, Zhao
H, Hu JH, Chen XM, Ma WH and Fan J: Quantification of
cytomegalovirus glycoprotein Bn DNA in hematopoietic stem cell
transplant recipients by real-time PCR. PLoS One. 7:e512242012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu A, Ma Y, Wu W, Chen X, Huang Y, Hu J,
Liang H, Wang H, Yang R and Fan J: Evaluation of human
cytomegalovirus-specific CD8+ T-cells in allogeneic haematopoietic
stem cell transplant recipients using pentamer and
interferon-γ-enzyme-linked immunospot assays. J Clin Virol.
58:427–431. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fields BA, Ober B, Malchiodi EL, Lebedeva
MI, Braden BC, Ysern X, Kim JK, Shao X, Ward ES and Mariuzza RA:
Crystal structure of the V alpha domain of a T cell antigen
receptor. Science. 270:1821–1824. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van de Berg PJ, Van Stijn A, Ten Berge IJ
and van Lier RA: A fingerprint left by cytomegalovirus infection in
the human T cell compartment. J Clin Virol. 41:213–217. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakasone H, Tanaka Y, Yamazaki R, Terasako
K, Sato M, Sakamoto K, Yamasaki R, Wada H, Ishihara Y, Kawamura K,
et al: Single-cell T-cell receptor-β analysis of
HLA-A*2402-restricted CMV-pp65-specific cytotoxic T-cells in
allogeneic hematopoietic SCT. Bone Marrow Transplant. 49:87–94.
2104. View Article : Google Scholar
|
|
20
|
Engstrand M, Lidehall AK, Totterman TH,
Herrman B, Eriksson BM and Korsgren O: Cellular responses to
cytomegalovirus in immunosuppressed patients: Circulating
CD8+ T cells recognizing CMVpp65 are present but display
functional impairment. Clin Exp Immunol. 132:96–104. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ozdemir E, St John LS, Gillespie G,
Rowland-Jones S, Champlin RE, Molldrem JJ and Komanduri KV:
Cytomegalovirus reactivation following allogeneic stem cell
transplantation is associated with the presence of dysfunctional
antigen-specific CD8+ T cells. Blood. 100:3690–3697. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matsumoto Y, Matsuo H, Sakuma H, Park IK,
Tsukada Y, Kohyama K, Kondo T, Kotorii S and Shibuya N: CDR3
spectratyping analysis of the TCR repertoire in myasthenia gravis.
J Immunol. 176:5100–5107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frankel TL, Burns WR, Peng PD, Yu Z,
Chinnasamy D, Wargo JA, Zheng Z, Restifo NP, Rosenberg SA and
Morgan RA: Both CD4 and CD8 T cells mediate equally effective in
vivo tumor treatment when engineered with a highly avid TCR
targeting tyrosinase. J Immunol. 184:5988–5998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ray S, Chhabra A, Chakraborty NG, Hegde U,
Dorsky DI, Chodon T, von Euw E, Comin-Anduix B, Koya RC, Ribas A,
et al: MHC-Irestricted melanoma antigen specific TCR-engineered
human CD4+ T cells exhibit multifunctional effector and helper
responses, in vitro. Clin Immunol. 136:338–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kerkar SP, Sanchez-Perez L, Yang S, Borman
ZA, Muranski P, Ji Y, Chinnasamy D, Kaiser AD, Hinrichs CS,
Klebanoff CA, et al: Genetic engineering of murine CD8+ and CD4+ T
cells for preclinical adoptive immunotherapy studies. J Immunother.
34:343–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang QJ, Hanada K, Feldman SA, Zhao Y,
Inozume T and Yang JC: Development of a genetically-modified novel
T-cell receptor for adoptive cell transfer against renal cell
carcinoma. J Immunol Methods. 366:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan J, Meng XQ, Yang MF, Zhou L, Chen XM,
Hu MJ, Fan WW, Ma WH and Li LJ: Association of cytomegalovirus
infection with human leukocyte antigen genotypes in recipients
after allogeneic liver transplantation. Hepatobiliary Pancreat Dis
In. 5:34–38. 2006.
|