Introduction
Esophageal squamous cell carcinoma (ESCC), which
begins in flat cells lining the esophagus, is a common esophageal
malignant tumor globally, and particularly in China (1). Alcohol intake and tobacco smoking are
major risk factors for ESCC (2,3). It
is possible to treat small localized ESCC (<2 cm) with surgery,
while larger tumors may be slowed with chemotherapy and radiation
therapy; however, as diagnosis is frequently late the outcomes tend
to be relatively poor (4,5). Therefore, multiple studies have been
performed to study the pathogenesis of ESCC and to identify more
effective molecular methods of diagnosis.
Lee et al (6) suggested that hereditary genetic
polymorphisms of autophagy related 5 and collagen type IV α3 chain
may be prognostic predictors for patients with ESCC. Elevated
expression of AXL receptor tyrosine kinase in esophageal tumor
tissues has been suggested to be associated with increased risk of
death and recurrence of ESCC (7).
Previous reports have identified genetic alternations of oncogenes
and tumor suppressors in ESCC (8,9), but
the genetic and molecular basis for esophageal carcinogenesis
remains largely unknown. Altered gene expression, which is the
result of gene mutation or dramatic changes in gene regulation, is
associated with a variety of diseases. MicroRNAs (miRNAs) act as
oncogenes or tumor suppressors, mediating multiple gene expression
in human cancers (10,11). Previous studies demonstrated that
in ESCC, tumor-suppressive miRNAs of miR-145, miR-133a, miR-133b
(12) and miR-143 (13) inhibited tumor cell proliferation
and invasion. MiR-155 (14),
miR-21 (15) and miR-31 (16) act as oncogenes in ESCC. miRNA
expression is correlated with carcinogenesis, and associated with
prognosis and therapeutic outcomes. Guo et al (17) suggested that high expression of
miR-103/107 was correlated with poor survival of patients with
ESCC. miR-142-3p is a potential prognostic biomarker for ESCC
(18). However, whether these
miRNAs are efficient enough or whether it is possible to use them
to evaluate prognosis in patients with ESCC remains unclear.
Therefore, further studies are required to improve the survival
rates of patients with ESCC.
Using the non-coding RNA (miRNA) profile GSE43732,
Chen et al (19) identified
four miRNA signatures which had good prognostic value, and
investigated the replicability and accuracy of these miRNAs as
predictors. The present study used different methods to screen
prognosis-associated miRNAs, and the results may provide novel
predictable biomarkers and contribute to improved survival in
patients with ESCC.
Materials and methods
Microarray data
The non-coding RNA profile GSE43732, which was
contributed by Chen et al (19), was downloaded from the Gene
Expression Omnibus database (20).
Paired frozen tissues (cancer tissue and adjacent normal tissue)
taken from 119 patients were assessed by microarray for miRNA
expression analysis. This data was obtained based on the platform
of GPL16543 Agilent-038166 cbc_human_miR18 (Cancer Institute and
Hospital, Chinese Academy of Medical Sciences, Beijing, China).
Data preprocessing
Series Matrix Files containing normalized chip
signals were transformed to gene symbols. The aggregate function in
R package (bioconductor.org/packages/2.1/bioc/html/impute.html)
was used to calculate the mean value when multiple probes were
mapped to the same gene. The k-nearest neighbor method (21) from the impute package (22) in R was used with the default
k-value of 10. Quantile normalization of miRNA expression profile
was performed with the preprocessCore package (23) in R. The distribution of miRNA
expression prior to and following normalization was visualized as a
box plot.
Screening for differentially expressed
miRNAs
Differentially expressed miRNAs were screened using
the limma package (24) in R. The
difference of mean expression level between two groups of samples
was examined using the paired Student's t-test in the limma
package. The P-value was adjusted using the Benjamini-Hochberg
method (25) to control the false
discovery rate. Genes with log2 fold change >2 and
adjusted P<0.05 were considered to be differentially expressed
miRNAs.
Identification of prognosis-related
differentially expressed miRNAs
The high and low critical values of miRNA expression
were derived based on the expression values of miRNAs and the
survival time of patients using receiver operating characteristic
(ROC) curve analysis. The ROC curves were generated and analyzed
using Proc (26), KMsurv (27) and survival package (28) in R. Patients with lower and higher
values compared with the critical values were divided into two
groups. Subsequently, the significant variation of survival time
between these two groups was examined using the Kaplan-Meier (KM)
method and log rank test, with P<0.05 considered to indicate a
statistically significant difference.
Only miRNAs with an area under the curve (AUC) of
≥0.7 were selected. They were subsequently screened again for
miRNAs which were highly expressed in tumor tissues and associated
with worse prognosis than others. Similarly, those miRNAs expressed
at low levels in tumor tissues and associated with a worse
prognosis were also identified.
Construction of the microRNA
regulatory network for prognostic risk
Databases providing information on miRNA-target
genes were used to identify the target genes of differentially
expressed miRNAs. Verification databases included miRNecords
(29) and MiRWalk (30), and predictive databases included
miRanda (31), MirTarget2
(32), PicTar (33), PITA (34) and TargetScan (35). The identified target genes should
be predicted by at least three of these databases. Target genes
obtained from the two types of database were used to construct the
miRNA-regulatory network, which was subsequently visualized using
Cytoscape software (36).
Functional enrichment analysis of
target genes
Gene Ontology (GO, http://www.geneontology.org/) analysis consisting of
molecular function, cell component and biological process (BP), the
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Reactome
database, http://www.reactome.org/ and KEGG
database, http://www.genome.jp/kegg/), and the
Disease Ontology (BMC Genomics 10 Suppl 1:S6) enrichment analyses
(37) for miRNAs-target genes were
performed using the TargetMine (targetmine.nibio.go.jp) (38) online tool. The hypergeometric test
within TargetMine for enrichment results was performed and the
P-value was adjusted with the Holm-Bonferroni correction. An
adjusted P<0.05 was considered to indicate a statistically
significant difference.
Results
Data normalization and differentially
expressed miRNAs
Following normalization, a total of 726 miRNA
expression values were obtained (Fig.
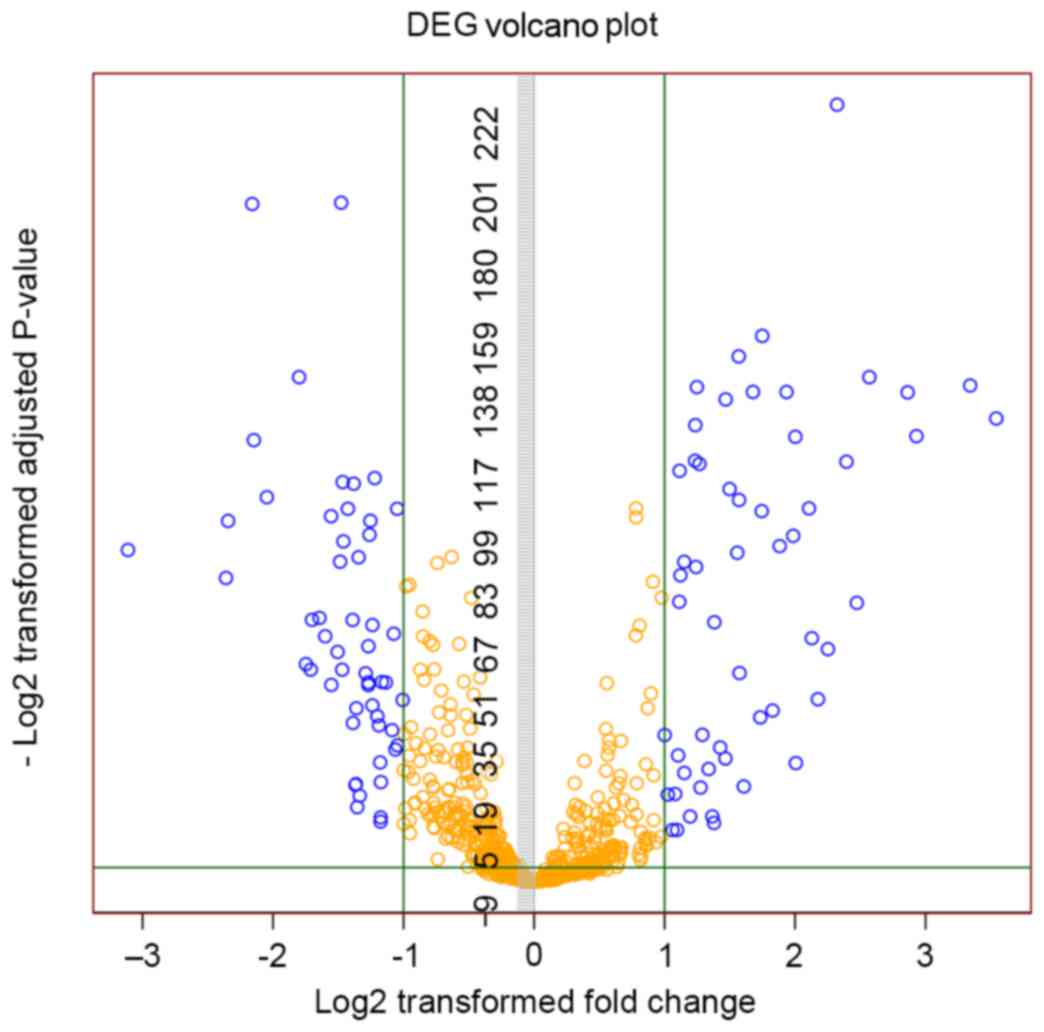
1). A total of 107 differentially expressed miRNAs, including
54 upregulated and 53 downregulated miRNAs, were obtained. The
volcano plot of differentially expressed miRNAs is depicted in
Fig. 2.
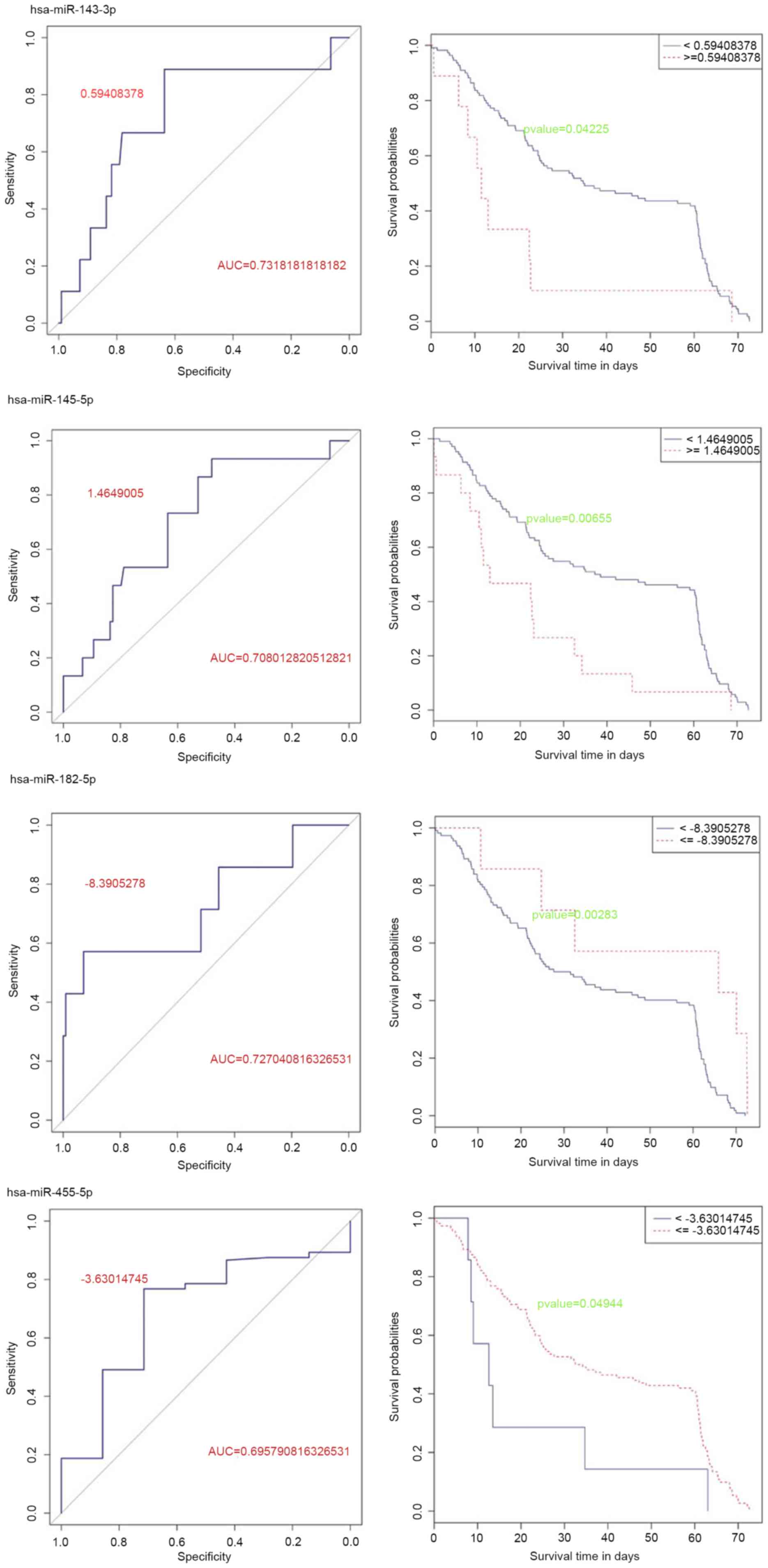
Prognosis related miRNAs
Critical values of the 107 differentially expressed
miRNAs were determined. Kaplan-Meier (KM) curve analysis revealed
that 44 miRNAs had significant effects on prognosis (P<0.05;
data not shown). Among them, 27 miRNAs were upregulated and 17
miRNAs were downregulated. Furthermore, 12 miRNAs, including 9
upregulated and 3 downregulated miRNAs, were screened with AUC
≥0.7. In the end, hsa-miR-143-3p and hsa-miR-145-5p were revealed
to be highly expressed in tumor tissues and were associated with
prognosis (Fig. 3). Meanwhile,
hsa-miR-182-5p and hsa-miR-455-5p, which were also associated with
prognosis, were expressed at low levels in tumor tissues (Fig. 3).
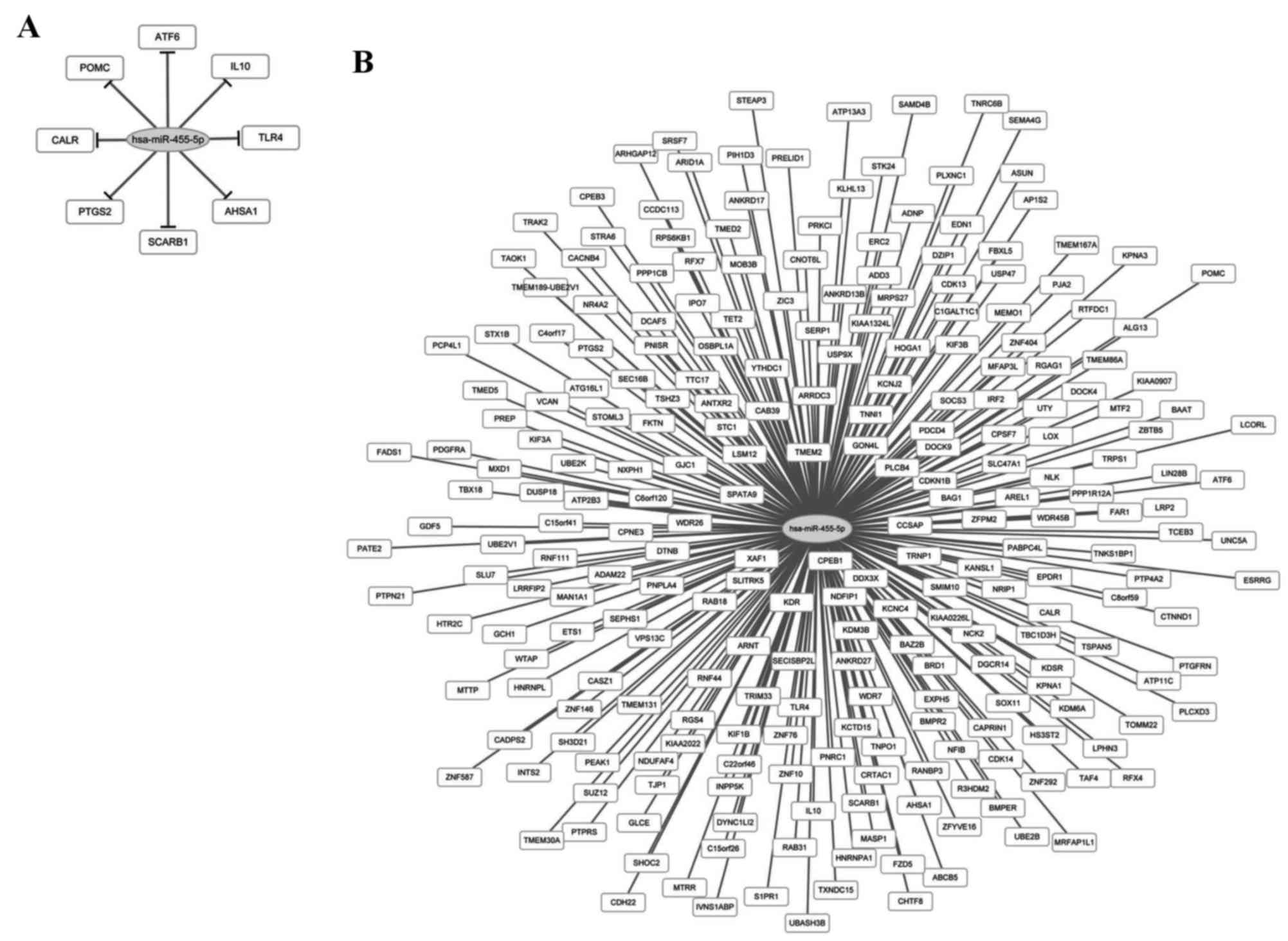
Regulatory network of prognosis
related miRNAs and target genes
A total of 8 target genes of hsa-miR-455-5p were
obtained from the verification database, but not the other three
miRNAs (hsa-miR-143-3p, hsa-miR-145-5p and miR-182-5p). The target
genes were activating transcription factor 6, interleukin 10
(IL10), toll-like receptor 4 (TLR4), activator of
heat shock 90 kDa protein ATPase homolog 1, scavenger receptor
class B member I, prostaglandin-endoperoxide synthase 2,
calreticulin and proopiomelanocortin. From the predicted databases,
a total of 245 target genes including ring finger protein
(RNF) 44/111, lysine demethylase (KDM)
3B/6A and chromosome 15 open reading frame
26/41, and the miRNA regulatory network is depicted
in Fig. 4.
Functional enrichment analysis of
target genes of miRNAs
No pathway was significantly enriched by the 253
target genes of hsa-miR-455-5p; however, two GO-BP terms were
statistically significant (Table
I). These were associated with the macromolecule metabolic
process. The Disease Ontology enrichment results revealed that 15
target genes of hsa-miR-455-5p were enriched in term of melanoma
(Table I).
 | Table I.GO and DO enrichment analyses of
target genes. |
Table I.
GO and DO enrichment analyses of
target genes.
| Category | Term | P-value | Count |
|---|
| GO-BP | Macromolecule
metabolic process [GO:0043170] | 0.00763 | 117 |
| GO-BP | Cellular
macromolecule metabolic process [GO:0044260] | 0.00860 | 109 |
| DO term | Melanoma
[DOID:1909] | 0.00215 | 15 |
Discussion
The poor prognosis of ESCC frequently results in
severe adverse effects to patients. Novel, effective molecular
markers for ESCC are, therefore, urgently required. In the present
study, several miRNAs associated with prognosis of ESCC were
discovered. miRNAs including miR-143-3p and miR-145-5p were
upregulated, while miR-182-5p and miR-455-5p were downregulated in
ESCC tissues. Previous research has demonstrated the involvement of
miR-143-3p (39) and miR-145-5p in
ESCC (40,41), which were recorded in the MalaCards
database; however, not the downregulated miRNAs miR-182-5p and
miR-455-5p. miR-182-5p and miR-455-5p may be novel miRNAs
associated with ESCC.
The association between miR-182 and tumors has been
widely studied. Segura et al (42) demonstrated that abnormal miR-182
expression represses forkhead box O3 and microphthalmia-associated
transcription factors to promote melanoma metastasis, and
contribute to renal cell carcinoma proliferation (43). miR-182 may be a prognostic marker
for glioma progression and its expression was associated with
patient survival (44). In the
present study, miR-182-5p was revealed to be under expressed in
tumor tissues and was associated with prognosis. Therefore,
miR-182-5p may be a novel miRNA associated with the prognosis and
survival rate of ESCC.
miR-455-5p was another downregulated miRNA
identified in the present study. The expression pattern and its
association with prognosis was similar to that of miR-182-5p.
Swingler et al (45)
hypothesized that miR-455 may regulate apoptosis in certain mouse
embryo tissue. Besides, decreased expression of miR-455-5p has been
demonstrated to be correlated with vascular invasion and poor
overall survival, and to be involved in endometrial serous
adenocarcinomas progression (46).
Based on these previous findings and the results of the present
study, miR-455-5p may be a novel miRNA participating in the
development and prognosis of ESCC. In addition, multiple target
genes were identified. IL-10 (47) and TLR4 (48), which were overexpressed in the
results of the present study, were demonstrated to be involved in
ESCC. Among the predicted target genes, no genes recorded in the
MalaCards database were associated with ESCC, but the homologous
genes including RNF6 and KDM4C were illustrated in
this database. Furthermore, the Disease Ontology enrichment results
demonstrated that the target genes were tumor-associated. In
addition, IL-10 and TLR4, as well as multiple other
target genes of miR-455-5p, were revealed to be enriched in
macromolecule biosynthesis and metabolism, which is associated with
tumor initiation and progression (49,50).
Cellular metabolic perturbation is a crucial hallmark of cancer,
and is associated with angiogenesis, evasion of apoptosis,
metastasis and avoidance of immune detection (51). Macromolecule biosynthesis and
metabolism are critical to supplying enough carbohydrates,
proteins, lipids and nucleic acids to support the rapid cell
division that occurs within a tumor (52). Therefore, miR-455-5p may be another
novel miRNA that participates in the progression of ESCC and is
associated with its prognosis.
In conclusion, in addition to the two known miRNAs
(miR-143-3p and miR-145-5p), two novel tumor suppressor miRNAs,
miR-182-5p and miR-455-5p, were demonstrated to be associated with
ESCC progression. As multiple target genes of miR-455-5p were
identified, it may be involved in complicated regulatory mechanisms
and be involved in ESCC. Although there were no identified target
genes for miR-182-5p, it may also be important in ESCC and warrants
of further research. Whether the two miRNAs are effective targets
for ESCC requires confirmation and further study in future
experiments.
References
|
1
|
Lin D, Meng X, Xu L, Ding L, Garg M, Yang
H, Liu L, Hao J, Wang M, Nagata Y, et al: Comprehensive molecular
characterization of esophageal squamous cell carcinoma. Cancer Res.
74:22252014. View Article : Google Scholar
|
|
2
|
Lee CH, Lee JM, Wu DC, Hsu HK, Kao EL,
Huang HL, Wang TN, Huang MC and Wu MT: Independent and combined
effects of alcohol intake, tobacco smoking and betel quid chewing
on the risk of esophageal cancer in Taiwan. Int J Cancer.
113:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Znaor A, Brennan P, Gajalakshmi V, Mathew
A, Shanta V, Varghese C and Boffetta P: Independent and combined
effects of tobacco smoking, chewing and alcohol drinking on the
risk of oral, pharyngeal and esophageal cancers in Indian men. Int
J Cancer. 105:681–686. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stahl M, Mariette C, Haustermans K,
Cervantes A and Arnold D; ESMO Guidelines Working Group, :
Oesophageal cancer: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 24 Suppl
6:vi51–vi56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JM, Yang PW, Chiang TH, Huang YC and
Hsieh CY: The genetic polymorphisms of ATG5 and COL4A3 are
associated with the prognosis of patients with esophageal squamous
cell carcinoma. Cancer Res. 74:28592014. View Article : Google Scholar
|
|
7
|
Yang PW, Hsieh MS, Huang YC, Chiang TH and
Lee JM: AXL receptor tyrosine kinase is associated with the
prognosis of patients with esophageal squamous cell carcinoma.
Cancer Res. 74:44052014. View Article : Google Scholar
|
|
8
|
Mandard AM, Hainaut P and Hollstein M:
Genetic steps in the development of squamous cell carcinoma of the
esophagus. Mutation Research/Reviews in Mutation Research.
462:335–342. 2000. View Article : Google Scholar
|
|
9
|
Ma S, Bao JY, Kwan PS, Chan YP, Tong CM,
Fu L, Zhang N, Tong AH, Qin YR, Tsao SW, et al: Identification of
PTK6, via RNA sequencing analysis, as a suppressor of esophageal
squamous cell carcinoma. Gastroenterology. 143:675–686. e12. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Y, Schetter AJ, Yang GB, Nguyen G,
Mathé EA, Li P, Cai H, Yu L, Liu F, Hang D, et al: microRNA and
inflammatory gene expression as prognostic marker for overall
survival in esophageal squamous cell carcinoma. Int J Cancer.
132:2901–2909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nohata N, Hanazawa T, Kikkawa N, Mutallip
M, Sakurai D, Fujimura L, Kawakami K, Chiyomaru T, Yoshino H,
Enokida H, et al: Tumor suppressive microRNA-375 regulates oncogene
AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J Hum
Genet. 56:595–601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang
G, Liu Q and Du J: MicroRNA-143 functions as a tumor suppressor in
human esophageal squamous cell carcinoma. Gene. 517:197–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Cheng C, Yuan X, He JT, Pan QH
and Sun FY: microRNA-155 acts as an oncogene by targeting the tumor
protein 53-induced nuclear protein 1 in esophageal squamous cell
carcinoma. Int J Clin Exp Pathol. 7:6022014.PubMed/NCBI
|
|
15
|
Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H,
Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, et al: Role of
microRNA-21 and effect on PTEN in Kazakh's esophageal squamous cell
carcinoma. Mol Biol Rep. 38:3253–3260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo
L and Lu SH: The oncogenetic role of microRNA-31 as a potential
biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond).
121:437–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo Y, Chen Z, Zhang L, Zhou F, Shi S,
Feng X, Li B, Meng X, Ma X, Luo M, et al: Distinctive microRNA
profiles relating to patient survival in esophageal squamous cell
carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF,
Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM and Xu LY: MiR-142-3p as
a potential prognostic biomarker for esophageal squamous cell
carcinoma. J Surg Oncol. 105:175–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou
F, Shi S, Feng X, Sun N, Yao R, et al: MiRNA expression profile
reveals a prognostic signature for esophageal squamous cell
carcinoma. Cancer Lett. 350:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barrett T and Edgar R: Gene expression
omnibus: Microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Altman NS: An introduction to kernel and
nearest-neighbor nonparametric regression. The American
Statistician. 46:175–185. 1992. View Article : Google Scholar
|
|
22
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: Impute: Imputation for microarray data. R package version.
2001.
|
|
23
|
Bolstad B: PreprocessCore: A collection of
pre-processing functions. R package version 1.20.0. 2013.
|
|
24
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society. Series
B (Methodological). 57:289–300. 1995.
|
|
26
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
KMsurv: Data sets from Klein and
Moeschberger, . 1997, Survival Analysis. R package version 0.1–5,
2012. https://cran.r-project.org/web/packages/KMsurv/KMsurv.pdfFebuary
19–2015
|
|
28
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox model. New York, NY: Springer;
2000, View Article : Google Scholar
|
|
29
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X and El Naqa IM: Prediction of both
conserved and nonconserved microRNA targets in animals.
Bioinformatics. 24:325–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schriml LM, Arze C, Nadendla S, Chang YW,
Mazaitis M, Felix V, Feng G and Kibbe WA: Disease Ontology: A
backbone for disease semantic integration. Nucleic Acids Res.
40:D940–D946. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen YA, Tripathi LP and Mizuguchi K:
TargetMine, an integrated data warehouse for candidate gene
prioritisation and target discovery. PLoS One. 6:e178442011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q,
Dan ZL, Tian DA and Zhang P: MicroRNA-143 suppresses gastric cancer
cell growth and induces apoptosis by targeting COX-2. World J
Gastroenterol. 19:7758–7765. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF,
Huang Q, Fang GQ and Li EM: MiRNA profile in esophageal squamous
cell carcinoma: Downregulation of miR-143 and miR-145. World J
Gastroenterol. 17:79–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu R, Liao J, Yang M, Sheng J, Yang H,
Wang Y, Pan E, Guo W, Pu Y, Kim SJ and Yin L: The cluster of
miR-143 and miR-145 affects the risk for esophageal squamous cell
carcinoma through co-regulating fascin homolog 1. PLoS One.
7:e339872012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:pp. 1814–1819.
2009; View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang L, Mao P, Song L, Wu J, Huang J, Lin
C, Yuan J, Qu L, Cheng SY and Li J: miR-182 as a prognostic marker
for glioma progression and patient survival. Am J Pathol.
177:29–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Swingler TE, Wheeler G, Carmont V, Elliott
HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP,
Hajihosseini MK, Münsterberg A, et al: The expression and function
of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum.
64:1909–1919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gholamin M, Moaven O, Memar B, Farshchian
M, Naseh H, Malekzadeh R, Sotoudeh M, Rajabi-Mashhadi MT, Forghani
MN, Farrokhi F and Abbaszadegan MR: Overexpression and interactions
of interleukin-10, transforming growth factor beta, and vascular
endothelial growth factor in esophageal squamous cell carcinoma.
World J Surg. 33:1439–1445. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sheyhidin I, Nabi G, Hasim A, Zhang RP,
Ainiwaer J, Ma H and Wang H: Overexpression of TLR3, TLR4, TLR7 and
TLR9 in esophageal squamous cell carcinoma. World J Gastroenterol.
17:3745–3751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Baxter LT and Jain RK: Transport of fluid
and macromolecules in tumors: III. Role of binding and metabolism.
Microvasc Res. 41:5–23. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Masoudi-Nejad A and Asgari Y: Metabolic
cancer biology: Structural-based analysis of cancer as a metabolic
disease, new sights and opportunities for disease treatment.
Journal. 30:21–29. 2015.
|
|
52
|
Ward PS and Thompson CB: Metabolic
reprogramming: A cancer hallmark even warburg did not anticipate.
Cancer cell. 21:297–308. 2012. View Article : Google Scholar : PubMed/NCBI
|