Introduction
Renal carcinoma is the 13th most common cancer
worldwide (1). There were ~61,560
new cases of renal cancer and 14,080 deaths due to renal cancer in
2015 in the United States according to statistics by Siegel et
al (2). Renal cell carcinoma
(RCC) accounts for ~3% of all malignancies in adults (3), Clear cell renal cell carcinoma
(CCRCC) is the most common type of RCC, which is responsible for
~75% of all RCC cases worldwide (4). Radical nephrectomy is effective for
curing early and local RCC, however, the response of advanced or
metastatic RCC to chemotherapy or radiotherapy is limited and
individuals have poor prognosis with an average survival of only
6–12 months from the time of diagnosis (5,6).
Therefore, it is important to improve the understanding of the
pathogenesis of aggressive RCC and to select biomarkers in order to
develop effective strategies for the prevention and treatment of
RCC.
Cyclic AMP (cAMP) responsive element-binding protein
(CREB) is a transcription factor that has a critical role in the
regulation of tumorigenesis (7,8). In
response to various signals, on binding to the cAMP-responsive
element (CRE) sequence (TGACGTCA), phosphorylation of CREB (pCREB)
at the Ser133 residue is activated by a number of kinases (9). Activated CREB subsequently regulates
various cell functions by enhancing the expression of target genes
(10–12). Several studies have demonstrated
that overexpression of pCREB promotes tumorigenesis in various
cancer tissues, including acute myeloid leukemia (13,14),
non-small cell lung carcinoma (15,16),
breast (17,18), melanoma (19) and hepatocellular cancers (20). However, the role of CREB in RCC
requires further investigation.
Our previous study demonstrated that pCREB was
upregulated in human renal cancer cell lines and tissues.
Lentiviral vector production was used to knockdown the expression
of pCREB in OS-RC-2 cells and the results demonstrated that
decreasing the level of pCREB inhibited the growth and metastasis
of OS-RC-2 cells (21). However,
although the results are promising, further studies are needed to
confirm this. Therefore, the present study investigated, in
multiple human renal cell carcinoma cell lines, whether pCREB
regulates metastasis via epithelial-mesenchymal transition
(EMT)-associated proteins and matrix metallopeptidase (MMP)-2/9.
The current study additionally aimed to determine whether the human
fibronectin promoter contains a functional CRE that activates
fibronectin transcription upon pCREB binding at the Ser133
site.
Materials and methods
Cell culture
The human renal cancer cell lines (786-Oand ACHN)
and an immortalized proximal tubule epithelial cell line (HK-2)
were obtained from American Type Culture Collection (Manassas, VA,
USA). The human renal cancer cell line OS-RC-2 was obtained from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
The 786-O and OS-RC-2 cell lines were maintained in RPMI-1640
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA), the ACHN
and HK-2 cell lines were maintained in Dulbecco's modified Eagle's
medium (HyClone; GE Healthcare Life Sciences). The medium was
supplemented with 10% fetal bovine serum (FBS; Shanghai ExCell
Biology, Inc., Shanghai, China). All cells were cultured in a
humidified atmosphere at 37°C in 5% CO2 (17).
Transfection of cell lines
Small interfering RNA (siRNA) for CREB (siCREB;
5′-GUCUCCACAAGUCCAAACATT-3′; antisense,
5′-UGUUUGGACUUGUGGAGACTT-3′) and siRNA for negative control (siNC;
sense, 5′-UCCUCCGAACGUGUCACGUTT-3′; antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from Shanghai
GenePharma Co., Ltd (Shanghai, China). A total of 5×105
cells were seeded in 6-well plates for 24 h at 37°C and transfected
with specific siCREB (100 pmol) or control siNC (100 pmol) by using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA, according to the manufacturer's
protocol (22).
Wound healing assay
Cells were added to a 24 well-plate at a
concentration of 0.7×105 cells/well in 1 ml 10%
FBS-containing medium. Cells were cultured for 24 h and reached 70%
confluence at 37°C in 5% CO2. Linear scratches were made
with a micropipette tip across the diameter of the well and
dislodged cells were rinsed with PBS. Cell culture medium was
replaced with fresh, serum-free medium. Images of the scratch were
acquired as baseline and images of the same location were obtained
after 24 h. Experiments were performed in quadruplicate (23).
Cell invasion assay
The cell invasion assay was performed using BD
Matrigel 24-well 8 µm invasion chambers with filters coated with
extracellular matrix on the upper surface (BD Biosciences, Franklin
Lakes, NJ, USA). Briefly, 100 µl serum-free medium containing
1×104 cells from each subgroup were added to the upper
chamber. The 600 µl medium (HyClone; GE Healthcare Life Sciences)
containing 20% FBS (Shanghai ExCell Biology, Inc.) was added to the
lower chamber as a chemoattractant. Cells were allowed to invade
for 24 h at 37°C in 5% CO2. The cells in the upper
chamber were removed using a cotton swab. Cells that had migrated
to the bottom of the membrane were fixed and stained 30 min with
0.1% crystal violet staining solution at room temperature and were
photographed from 5 different microscopic fields (Motic AE31; Motic
China Group Co., Ltd., Xiamen, China). Crystal violet was removed
by addition of 33% acetic acid at 200 µl/well to the 96-well plates
and absorbance was measured at 570 nm using a SpectraMax Plus384;
Molecular Devices LLC (Sunnyvale, CA, USA). Experiments were
performed in triplicate (23).
Western blot analysis
For western blot analysis, 50 µg of each sample were
processed as described (24). CREB
(catalog no. 9197), pCREB (Ser133; catalog no. 9198), N-cadherin
(catalog no. 14215), E-cadherin (catalog no. 3195), β-actin
(catalog no. 8457) and GAPDH (catalog no. 5174) antibodies were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
MMP-2 (catalog no. ab7033), MMP-9 (catalog no. ab137651), vimentin
(catalog no. ab8978) and fibronectin (catalog no. ab2413)
antibodies were obtained from Abcam (Cambridge, UK). All primary
antibodies were diluted at 1:1,000 and were used according the
manufacturer's protocol. Goat anti-rabbit IgG-HRP or goat
anti-mouse IgG-HRP were used (dilution 1;5,000; catalog no
BA1054/BA1050; Wuhan Boster Biological Technology, Ltd., Wuhan,
China) and detected by chemiluminescence. Densitometric analysis
was performed by Tanon GIS version 4.1.2 software (Tanon Science
and Technology Co., Ltd., Shanghai, China).
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed using a
SimpleChIP® Enzymatic Chromatin IP kit (catalog no.
9002; Cell Signaling Technology, Inc.) according to the
manufacturer's protocol. IgG antibody was included in the ChIP
assay kit. pCREB (Ser133) antibody (dilution 1:50; catalog no.
9198) was obtained from Cell Signaling Technology, Inc.
Quantification of immunoprecipitated DNA was performed using
quantitative polymerase chain reaction (qPCR) with LightCycler 480
SYBR-Green I Master (Roche Diagnostics, Indianapolis, IN, USA)
according to the manufacturer's protocol. CREB binds to a target
sequence termed the CRE-site and this serves as a model for the
research of CREB function, and the CREB-binding protein (CRE-BP)
site is the transcription factor CRE-BP binding site. The −449 to
−442 bp (CRE site) fibronectin promoter sequence from the
transcription start site had the following primers:
5′-CCGAAAAAAAGTTGTCTTGCCC-3′ (forward); and
5′-CAGCCGACCGCGCGCCGATTGG-3′ (reverse). The −731 to −722 bp (CRE-BP
site) vimentin promoter sequence from the transcription start site
had the following primers: 5′-TATTGCCGCCAAAGATTCTG-3′ (forward);
and 5′-TACCCTGGTGGAAGTCATTAAAG-3′ (reverse). ChIP results were
calculated and presented as a percentage relative to the input DNA
by the ∆∆Cq method (25,26).
Reverse transcription-qPCR
Using the standard TRIzol protocol (Invitrogen;
Thermo Fisher Scientific, Inc.), total RNA was isolated and
extracted from cells. The amounts of total RNA were quantified
using spectrophotometric measurements. Using a RevertAid First
Strand cDNA Synthesis Kit (K1622; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol, total RNA was reversed
transcribed into cDNA. qPCR was performed using the LightCycler 480
SYBR-Green I Master (Roche Diagnostics). Each standard qPCR
reaction contained the following; Reverse transcriptase product (2
µl); 0.5 µl of each primer (final concentration, 0.1 mM); 12.5 µl
SYBR-Green PCR Master Mix™ (Roche Diagnostics). The thermocycling
conditions included an initial 15 min holding period at 95°C,
followed by a PCR program repeated for 50 cycles: 15 sec at 94°C,
57 sec at 30°C and 30 sec at 70°C. The following primers were used:
MMP-2, 5′-TTGACGGTAAGGACGGACTC-3′ (forward) and
5′-ACTTGCAGTACTCCCCATCG-3′ (reverse); MMP-9,
5′-TTGACAGCGACAAGAAGTGG-3′ (forward) and 5′-CCCTCAGTGAAGCGGTACAT-3′
(reverse); GAPDH, 5′-AAGCCTGCCGGTGACTAAC-3′ (forward) and
5′-GCATCACCCGGAGGAGAAAT-3′ (reverse). GAPDH was used as a
normalization and other gene expression levels were analyzed using
the ∆∆Cq method (25). All samples
were performed in triplicate and, for each reaction, negative
controls without reverse transcriptase or RNA were performed
(12,27).
Statistical analysis
All experiments were repeated 3 times. Using SPSS,
version 18.0 (SPSS, Inc., Chicago, IL, USA), results are presented
as the mean ± standard deviation. One-way analysis of variance and
Fisher's least significant difference tests were performed to
evaluate the differences between groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
Effects of CREB downregulation on cell
migration and invasion
In the present study, to investigate, in multiple
human RCC cell lines, the functional role of the upregulation of
CREB, siCREB was used to knockdown the expression of CREB in 786-O,
OS-RC-2 and ACHN RCC cells, and normal HK-2 cells. To detect the
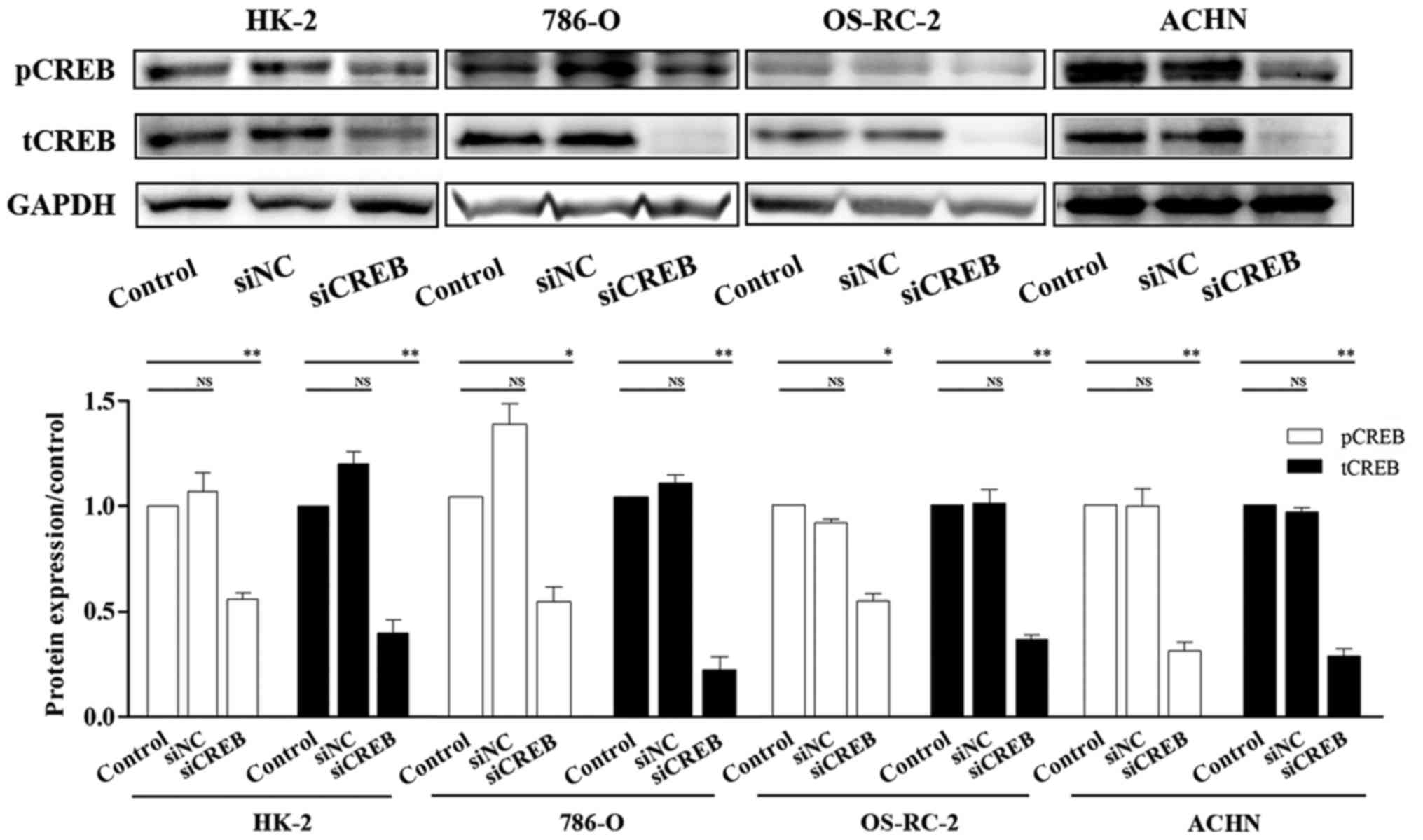
effect of siCREB, the expression of CREB was investigated by
western blotting. The results demonstrated that the protein
expression of CREB and pCREB were significantly downregulated
compared with cells transfected with siNC and control, following
cell transfection with siCREB (Fig.
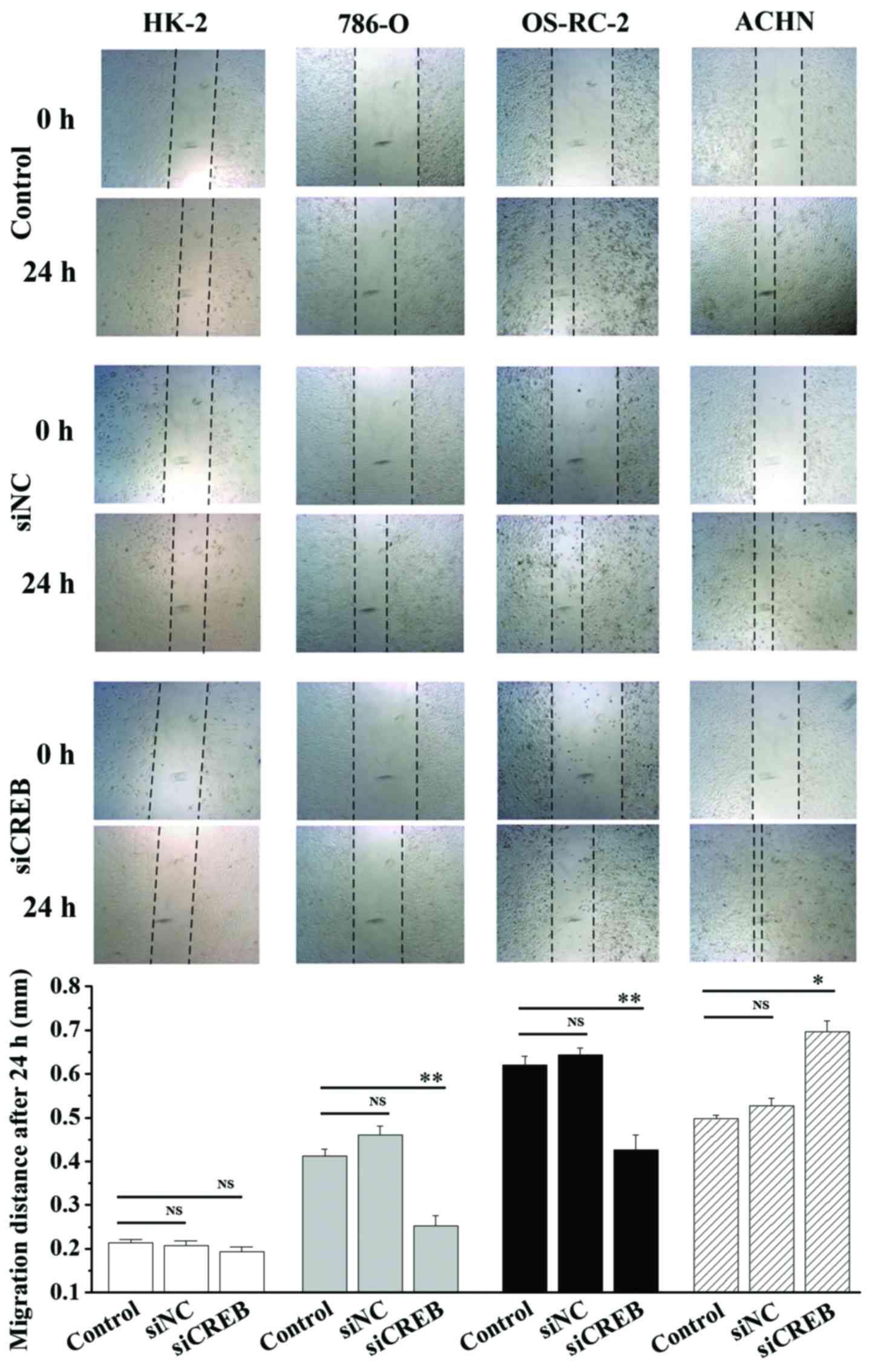
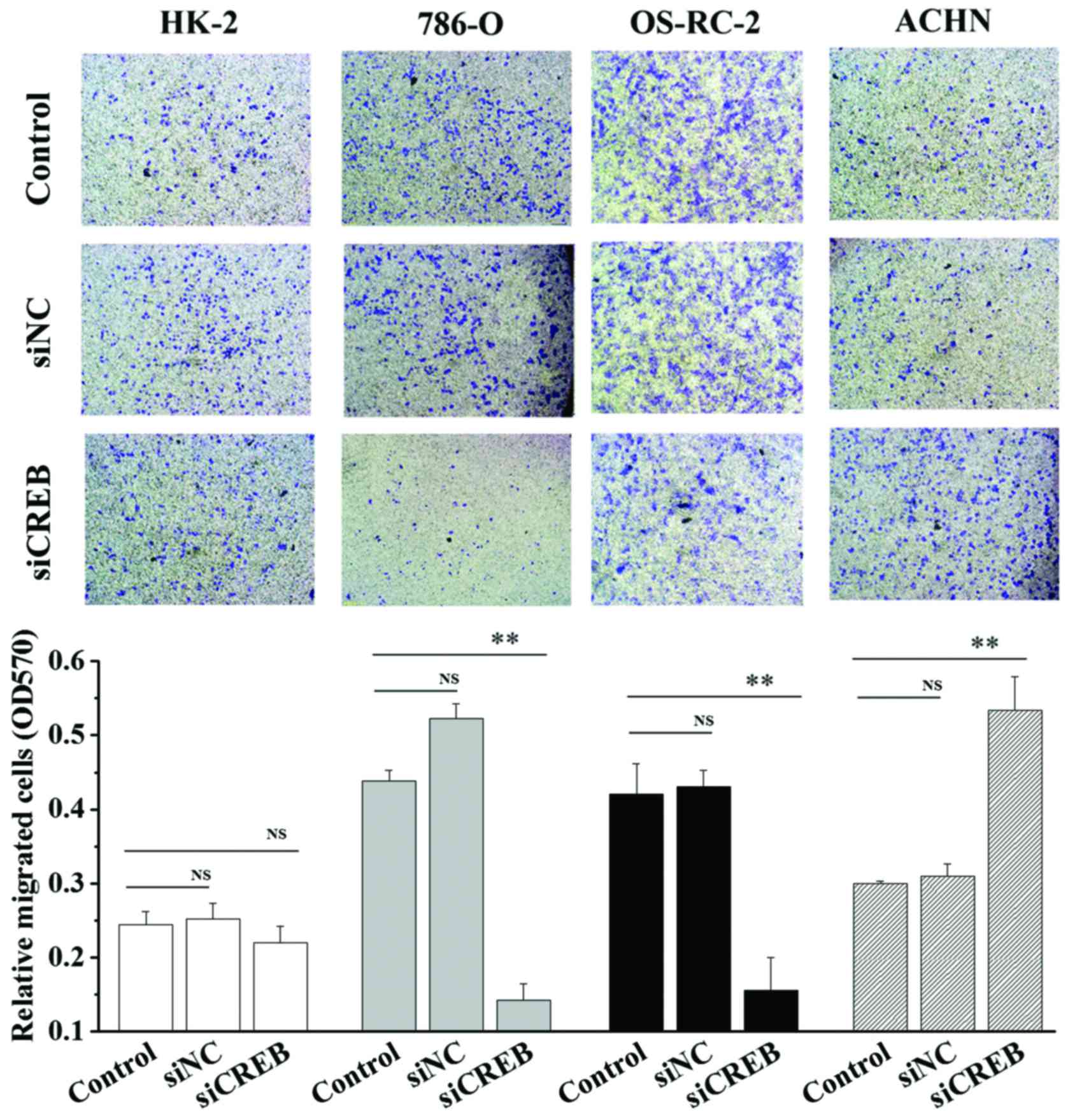
1). In addition, cell migration and invasion ability was
analyzed using wound healing and cell invasion assays. Transfection
with siCREB significantly blocked migration (P<0.01; Fig. 2) and invasion (P<0.01; Fig. 3) in 786-O and OS-RC-2 cells
compared with control, however, significantly increased migration
(P<0.05; Fig. 2) and invasion
(P<0.01; Fig. 3) in ACHN cells
compared with control, and demonstrated no significant effects in
HK-2 cells (Figs. 2 and 3). These results indicate that CREB may
contribute to the migration and invasion phenotype of RCC cells,
and not HK-2 cells, however, results were not consistent between
the3 RCC cell lines.
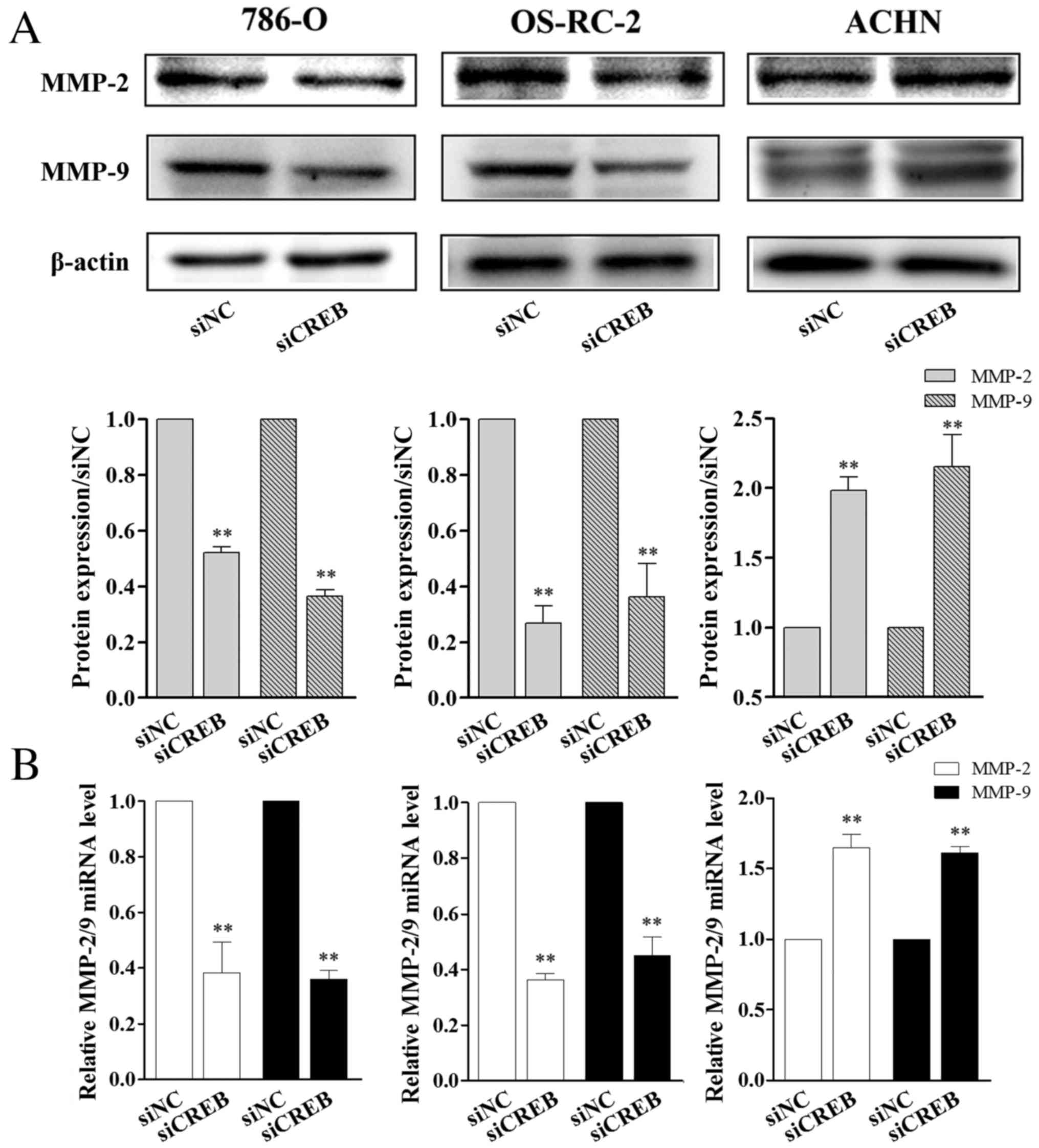
CREB regulates the expression of MMP-2
and MMP-9 in RCC cells
MMPs may disrupt the extracellular matrix to promote
cancer cell mobility, which eventually causes metastasis (28,29).
Among MMPs, MMP-2 and MMP-9 are vital enzymes involved in the
degradation of gelatin, collagen and laminin (30). The expression of numerous genes is
activated by CREB (31).
Therefore, the present study aimed to investigate whether CREB
regulates RCC cell migration and invasion ability by affecting
MMP-2/9 expression. As observed in the results of the wound healing
and cell invasion assay, 786-O and OS-RC-2 cells, following siCREB
transfection, expressed significantly lower levels of MMP-2/9
protein (P<0.01; Fig. 4A) and
mRNA (P<0.01; Fig. 4B) compared
with the siNC group, however, the opposite was observed in ACHN
cells where protein and mRNA levels of MMP-2/9 were significantly
increased compared with the siNC group (P<0.01; Fig. 4). Combined, the results demonstrate
that CREB regulated the migration and invasion of the 3 types of
RCC cells, however, the effects of CREB knockdown were different
between the 3 RCC cell lines.
CREB expression is associated with the
expression of certain EMT markers in RCC cells
The EMT is a highly conserved program necessary for
orchestrating distant cell migration during embryonic development.
A previous report demonstrated a critical role for EMT during the
initial stages of tumorigenesis and later during tumor invasion
(32). Thus, in order to further
investigate the association between CREB and EMT, markers
associated with EMT were assessed using western blot analysis in 3
types of human RCC cell lines. The results demonstrated that
protein levels of N-cadherin and fibronectin were significantly
reduced (P<0.01; Fig. 5), and
E-cadherin protein levels were significantly increased (P<0.01;
Fig. 5) in siCREB-transfected
786-O and OS-RC-2 cells compared with the siNC group, however, the
opposite was observed in ACHN cells, which corresponds with the
results obtained in wound healing and invasion assays. Furthermore,
siCREB transfection did not significantly affect the expression of
vimentin compared with the siNC group in all 3 types of RCC cells
(Fig. 5). These results suggested
that there may be additional mechanisms by which CREB controls RCC
function, however, the present study demonstrated that this does
not occur via vimentin in vitro, as siCREB transfection did
not significantly affect vimentin expression.
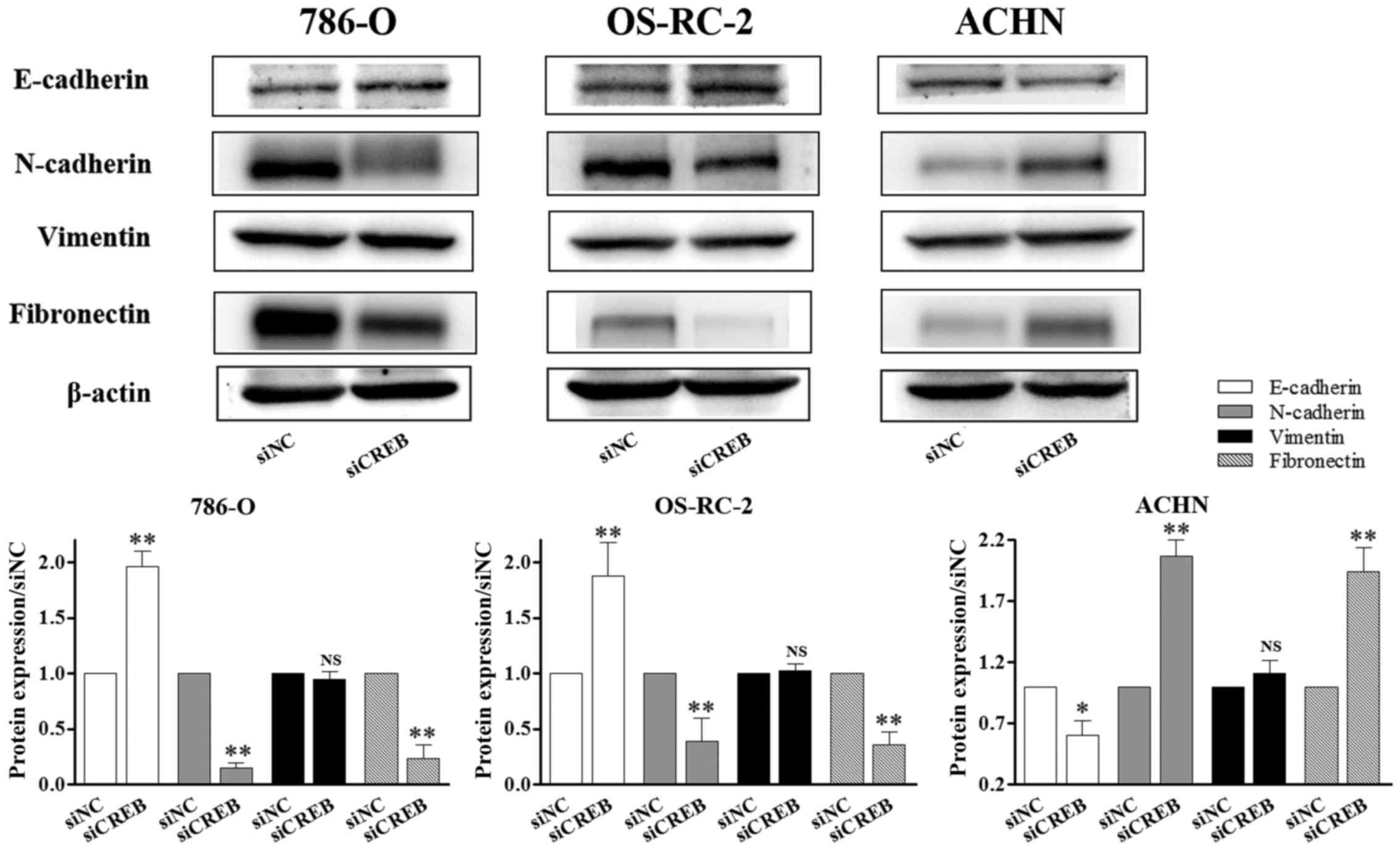 | Figure 5.CREB-mediated RCC cell metastasis
does not occur via vimentin. RCC cell lines were transiently
transfected with siNC or siCREB. The expression of proteins
associated with epithelial-mesenchymal transition was determined by
western blot analysis. N-cadherin and fibronectin expression was
reduced, E-cadherin was increased in 786-O and OS-RC-2 cells
compared with the siNC group, however, the opposite results were
observed in ACHN cells, and the expression of vimentin was not
significantly changed in the 3 types of RCC cells. *P<0.05 and
**P<0.01 vs. siNC group. Results are presented as the mean ±
standard deviation from 3 independent experiments. CREB, cyclic AMP
responsive element-binding protein; RCC, renal cell carcinoma;
siRNA, small interfering RNA; siNC, siRNA negative control; siCREB,
siRNA targeting CREB; NS, not significant. |
The human fibronectin promoter
contains a functional CRE sequence
Using bioinformatics software, the authors
previously demonstrated that that there is a putative CRE sequence
in the promoter of fibronectin, and a putative binding site of
CRE-binding protein is found in the promoter of vimentin (21). The present study aimed to determine
whether there is a direct interaction between CREB and the CREsite
in the fibronectin promoter or the CRE-binding protein site in the
vimentin promoter. The current study employed siCREB, to target
CREB mRNAs, and ChIP assays with the specific pCREB (Ser133)
antibody were performed in 786-O (Fig.
6A), OS-RC-2 (Fig. 6B) and
ACHN (Fig. 6C) cells. Compared
with siNC treatment, siCREB treatment significantly decreased pCREB
binding to the fibronectin promoter CRE sequence in all 3 cell
lines (P<0.05; Fig. 6A-C) and
did not significantly affect pCREB binding to the vimentin promoter
CRE-binding protein site. The results of this experiment increased
evidence for a direct interaction of pCREB (Ser133) with the
fibronectin promoter in RCC, whilst confirming that pCREB (Ser133)
does not target the vimentin promoter.
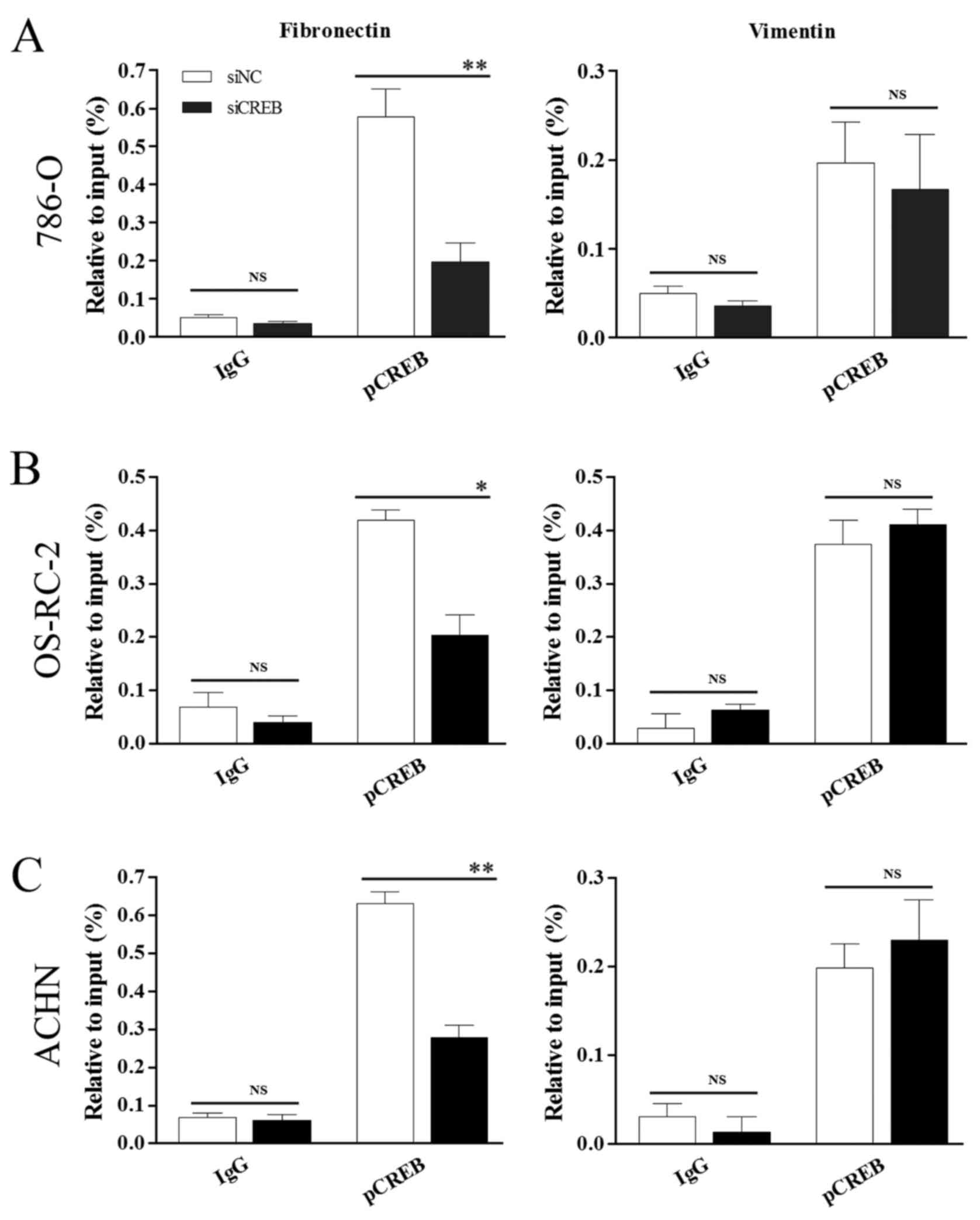 | Figure 6.pCREB directly targets fibronectin by
binding to its promoter and does not control vimentin. Chromatin
immunoprecipitation analysis demonstrated a pCREB interaction with
the CRE located at −449 to −442 in the fibronectin promoter and no
interaction with the CRE-BP site located at −731 to −722 in the
vimentin promoter in (A) 786-O, (B) OS-RC-2 and (C) ACHN cells with
specific pCREB (Ser133) antibody. *P<0.05 and **P<0.01 vs.
siNC group. Results are presented as the mean ± standard deviation
from 3 independent experiments. CREB, cyclic AMP responsive
element-binding protein; pCREB, phosphorylated CREB; CRE, cAMP
responsive element; CRE-BP, CRE-binding protein; siRNA, small
interfering RNA; siNC, siRNA negative control; siCREB, siRNA
targeting CREB; IgG, immunoglobulin G; NS, not significant. |
Discussion
CREB has important roles in the regulation of
various cellular functions. It is highly expressed and
constitutively phosphorylated in a number of types of human cancer
(8) and it is widely regarded that
CREB is a proto-oncogenic transcription factor (8,33).
However, whether CREB regulates the metastatic potential of RCC
cells requires further investigation. The present study aimed to
determine the anti-metastatic effect of CREB on human RCC cells by
investigating the regulation of MMP-2/9 expression and the
potential pathways involved in this regulation.
Invasion and migration are two vital features of
metastatic malignancies, and are thought to increase the metastatic
potential of cancer cells (34).
The present study demonstrated that silencing CREB suppressed the
migration and invasion of 786-O and OS-RC-2 cells, whereas it
promoted the migration and invasion of ACHN cells, and had no
effect in HK-2 cells (Figs. 2 and
3). During metastasis, MMPs
facilitate the degradation and invasion of extracellular matrix
components, and participate in the onset and progression of tumors
(35,36). Park et al (37) demonstrated that intracellular
adhesion molecule 3 induced MMP-2 and MMP-9 expression via Akt, and
CREB enhanced the migratory and invasive potential of human
non-small cell lung cancer cells (37). Other previous studies have
identified that, in human osteosarcoma cells, cholangiocarcinoma
cells and macrophages, CREB induced expression of MMPs (38–40).
The results of the present study further demonstrated that, in
786-O and OS-RC-2 cells, CREB knockdown induced reduced MMP-2/9
expression, as determined by western blot and qPCR analysis.
However, CREB silencing increased MMP-2/9 expression in ACHN cells
(Fig. 4). The results of western
blot and qPCR analysis corresponded with the results of wound
healing and invasion assays. The results of the present study
indicated that CREB-induced MMP-2/9 expression may be involved in
tumor metastasis progression.
EMT has been implicated as a potential mechanism of
metastasis (41), it transforms
epithelial tumor cells and confers the mesenchymal characteristics
that facilitate the dissemination of cells, which subsequently
leads to metastasis (42). It has
been previously reported that EMT influences RCC progression
(43). However, the results of the
present study indicate that expression of the mesenchymal markers
N-cadherin and fibronectin, and the epithelial marker E-cadherin,
were altered as a result of CREB silencing in 786-O and OS-RC-2
cells. However, in ACHN cells, siCREB transfection had the opposite
effect on the expression of these proteins (Fig. 5). Furthermore, no changes in the
expression of vimentin protein were detected in the 3 types of RCC
cells (Fig. 5), which is
consistent with the results of our previous study (21). Previously, our laboratory
identified putative CRE sites in the promoter of fibronectin,
however, only a putative binding site of CRE-BP was identified in
the promoter of vimentin (21).
Notably, the present study demonstrated the binding of pCREB to the
promoter sequence of fibronectin and the absence of pCREB binding
to vimentin in 786-O, OS-RC-2 and ACHN cells by ChIP assay
(Fig. 6). We hypothesized that the
decrease in metastatic potential when CREB was suppressed in 786-O
and OS-RC-2 cells is linked to MMP-2/9 and EMT-associated markers,
including fibronectin, E-cadherin and N-cadherin. The results of
the present study were similar to a report by Cho et al
(44), which demonstrated that
γ-ionizing radiation induced increases in CREB-1 led to increased
invasion/migration and EMT in non-small cell lung cancer cells
(44). However, the regulation of
ACHN cell function by CREB requires further investigation and
discussion.
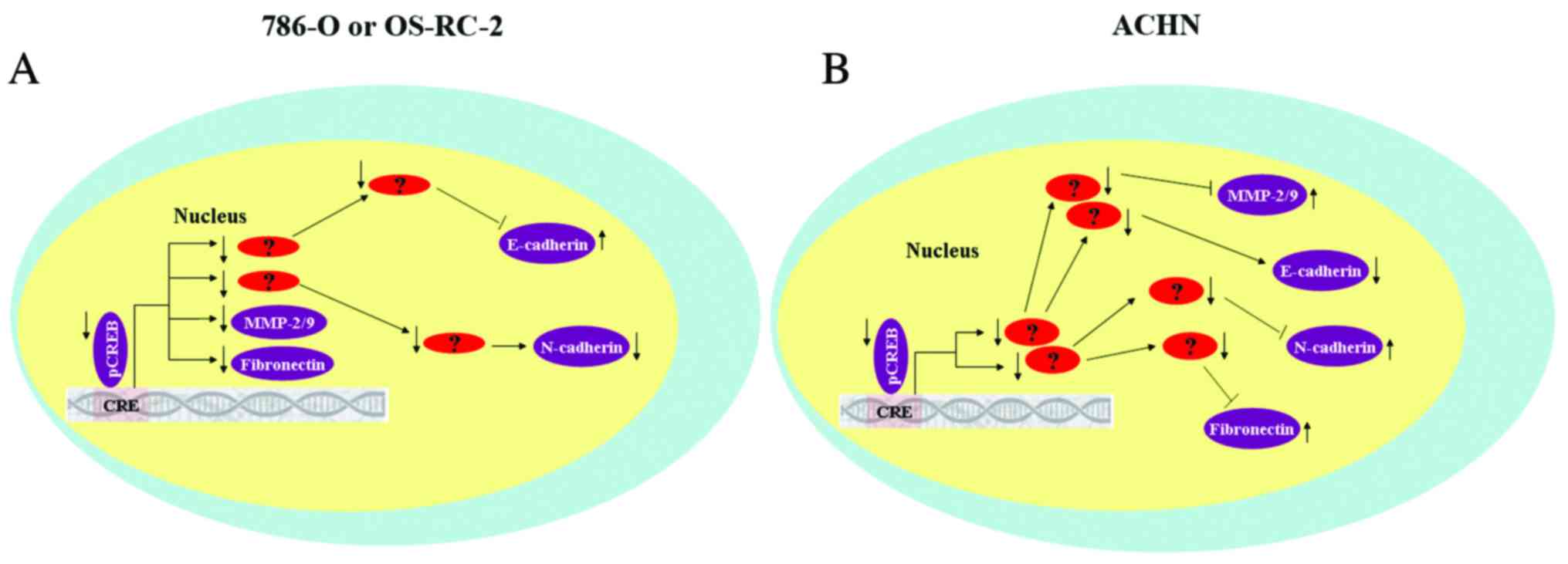
The results of the current study demonstrated that
CREB may regulated metastatic RCC cells by mediating expression of
MMP-2/9 and EMT-associated proteins, however, results were not
consistent between the 3 RCC cell lines. It was hypothesized that
there at least two reasons that may explain this phenomenon.
Firstly, expression of CREB in the 3 RCC cell lines is different.
Compared with normal renal cells (HK-2), expression of total CREB
is upregulated in 786-O cells, and is marginally decreased in
OS-RC-2 cells and partially downregulated in ACHN cells. In
addition, CREB mediated expression of MMP-2/9 and EMT-associated
protein may control tumor metastasis via different signaling
pathways (Fig. 7). Further studies
are required to determine the precise molecular mechanisms by which
inhibition of CREB expression advanced cancer cell metastasis, and,
conversely, the mechanisms by which the interruption of CREB
signaling inhibited cancer cell metastasis.
In conclusion, the present study demonstrated that
CREB may act as a critical factor that promotes 786-O and OS-RC-2
cell metastasis by regulating the expression of MMP-2/9 and
EMT-associated proteins. It was indicated that this role may be
different in ACHN cells, and pCREB did not control vimentin
expression in any of the3 RCC cell lines. Understanding the
mechanisms by which CREB affects metastatic RCC via the MMP-2/9 and
EMT may aid in the development of novel strategies for the
treatment of patients with acquired or innate resistance to cancer
treatments.
Acknowledgements
The current study was supported by grants from
Scientific Plan of Medical and Health of Zhejiang Province (grant
nos. 2015RCB024 and 2017KY604) and Ningbo Natural Science
Foundation (grant no. 2016A610018).
References
|
1
|
Ljungberg B, Campbell SC, Choi HY, Jacqmin
D, Lee JE, Weikert S and Kiemeney LA: The epidemiology of renal
cell carcinoma. Eur Urol. 60:615–621. 2011. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar
|
|
3
|
Mastoraki A, Mastoraki S, Tsikala-Vafea M,
Papanikolaou IS, Lazaris A, Smyrniotis V and Arkadopoulos N:
Prognostic benefit of surgical management of renal cell carcinoma
invading the inferior vena cava. Indian J Surg Oncol. 8:14–18.
2017. View Article : Google Scholar
|
|
4
|
Fu L, Minton DR, Zhang T, Nanus DM and
Gudas LJ: Genome-wide profiling of TRACK kidneys shows similarity
to the human ccRCC transcriptome. Mol Cancer Res. 13:870–878. 2015.
View Article : Google Scholar :
|
|
5
|
Liang L, Li L, Zeng J, Gao Y, Chen YL,
Wang ZQ, Wang XY, Chang LS and He D: Inhibitory effect of silibinin
on EGFR signal-induced renal cell carcinoma progression via
suppression of the EGFR/MMP-9 signaling pathway. Oncol Rep.
28:999–1005. 2012.
|
|
6
|
Eggener SE, Yossepowitch O, Pettus JA,
Snyder ME, Motzer RJ and Russo P: Renal cell carcinoma recurrence
after nephrectomy for localized disease: Predicting survival from
time of recurrence. J Clin Oncol. 24:3101–3106. 2006. View Article : Google Scholar
|
|
7
|
Jean D and Bar-Eli M: Regulation of tumor
growth and metastasis of human melanoma by the CREB transcription
factor family. Mol Cell Biochem. 212:19–28. 2000. View Article : Google Scholar
|
|
8
|
Xiao X, Li BX, Mitton B, Ikeda A and
Sakamoto KM: Targeting CREB for cancer therapy: Friend or foe. Curr
Cancer Drug Targets. 10:384–91. 2010. View Article : Google Scholar :
|
|
9
|
Sakamoto KM and Frank DA: CREB in the
pathophysiology of cancer: Implications for targeting transcription
factors for cancer therapy. Clin Cancer Res. 15:2583–2587. 2009.
View Article : Google Scholar :
|
|
10
|
Shaywitz AJ and Greenberg ME: CREB: A
stimulus-induced transcription factor activated by a diverse array
of extracellular signals. Annu Rev Biochem. 68:821–861. 1999.
View Article : Google Scholar
|
|
11
|
Huang S, Ren Y, Wang P, Li Y, Wang X,
Zhuang H, Fang R, Wang Y, Liu N, Hehir M and Zhou JX: Transcription
factor CREB is involved in CaSR-mediated cytoskeleton gene
expression. Anat Rec (Hoboken). 298:501–512. 2015. View Article : Google Scholar
|
|
12
|
Zhuang H, Meng X, Li Y, Wang X, Huang S,
Liu K, Hehir M, Fang R, Jiang L, Zhou JX, et al: Cyclic AMP
responsive element-binding protein promotes renal cell carcinoma
proliferation probably via the expression of spindle and
kinetochore-associated protein 2. Oncotarget. 7:16325–16337.
2016.
|
|
13
|
Cho EC, Mitton B and Sakamoto KM: CREB and
leukemogenesis. Crit Rev Oncog. 16:37–46. 2011. View Article : Google Scholar :
|
|
14
|
van der Sligte NE, Kampen KR, ter Elst A,
Scherpen FJ, Meeuwsen-de Boer TG, Guryev V, Van Leeuwen FN,
Kornblau SM and de Bont ES: Essential role for cyclic-AMP
responsive element binding protein 1 (CREB) in the survival of
acute lymphoblastic leukemia. Oncotarget. 6:14970–14981. 2015.
View Article : Google Scholar :
|
|
15
|
Peng B, Lei N, Chai Y, Chan EK and Zhang
JY: CIP2A regulates cancer metabolism and CREB phosphorylation in
non-small cell lung cancer. Mol Biosyst. 11:105–114. 2015.
View Article : Google Scholar
|
|
16
|
Seo HS, Liu DD, Bekele BN, Kim MK, Pisters
K, Lippman SM, Wistuba II and Koo JS: Cyclic AMP response
element-binding protein overexpression: A feature associated with
negative prognosis in never smokers with non-small cell lung
cancer. Cancer Res. 68:6065–6073. 2008. View Article : Google Scholar :
|
|
17
|
Singh R, Shankar BS and Sainis KB:
TGF-β1-ROS-ATM-CREB signaling axis in macrophage mediated migration
of human breast cancer MCF7 cells. Cell Signal. 26:1604–1615. 2014.
View Article : Google Scholar
|
|
18
|
Zhang S, Chen L, Cui B, Chuang HY, Yu J,
Wang-Rodriguez J, Tang L, Chen G, Basak GW and Kipps TJ: ROR1 is
expressed in human breast cancer and associated with enhanced
tumor-cell growth. PLoS One. 7:e311272012. View Article : Google Scholar :
|
|
19
|
Liu YL, Lai F, Wilmott JS, Yan XG, Liu XY,
Luan Q, Guo ST, Jiang CC, Tseng HY, R A Scolyer, et al: Noxa
upregulation by oncogenic activation of MEK/ERK through CREB
promotes autophagy in human melanoma cells. Oncotarget.
5:11237–11251. 2014. View Article : Google Scholar :
|
|
20
|
Kovach SJ, Price JA, Shaw CM, Theodorakis
NG and McKillop IH: Role of cyclic-AMP responsive element binding
(CREB) proteins in cell proliferation in a rat model of
hepatocellular carcinoma. J Cell Physiol. 206:411–419. 2006.
View Article : Google Scholar
|
|
21
|
Wang X, Ren Y, Zhuang H, Meng X, Huang S,
Li Y, Hehir M and Wang P: Decrease of phosphorylated proto-oncogene
CREB at Ser 133 site inhibits growth and metastatic activity of
renal cell cancer. Expert Opin Ther Targets. 19:985–95. 2015.
View Article : Google Scholar
|
|
22
|
Zi Y, Zhao W, Zhou J, He H and Xie M:
Silencing of TMSG1 enhances metastasis capacity by targeting
V-ATPase in breast cancer. Int J Clin Exp Pathol. 8:1312–1320.
2015.
|
|
23
|
Xia M, Yao L, Zhang Q, Wang F, Mei H, Guo
X and Huang W: Long noncoding RNAHOTAIR promotes metastasis of
renal cell carcinoma by up-regulating histone H3K27demethylase
JMJD3. Oncotarget. Feb 3–2017.(Epub ahead of print).
|
|
24
|
Wang P, Yan H and Li JC: CREB-mediated
Bcl-2 expression in trichosanthin-induced Hela cell apoptosis.
Biochem Biophys Res Commun. 363:101–105. 2007. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Jeon SH, Chae BC, Kim HA, Seo GY, Seo DW,
Chun GT, Yie SW, Eom SH and Kim PH: The PKA/CREB pathway is closely
involved in VEGF expression in mouse macrophages. Mol Cells.
23:23–29. 2007.
|
|
27
|
Khoufache K, Bazin S, Girard K,
Guillemette J, Roy MC, Verreault JP, Al-Abed Y, Foster W and Akoum
A: Macrophage migration inhibitory factor antagonist blocks the
development of endometriosis in vivo. PLoS One. 7:e372642012.
View Article : Google Scholar :
|
|
28
|
Moss LA Shuman, Jensen-Taubman S and
Stetler-Stevenson WG: Matrix metalloproteinases: Changing roles in
tumor progression and metastasis. Am J Pathol. 181:1895–1889. 2012.
View Article : Google Scholar :
|
|
29
|
Hofmann UB, Houben R, Bröcker EB and
Becker JC: Role of matrix metalloproteinases in melanoma cell
invasion. Biochimie. 87:307–314. 2005. View Article : Google Scholar
|
|
30
|
Cheng HL, Hsieh MJ, Yang JS, Lin CW, Lue
KH, Lu KH and Yang SF: Nobiletin inhibits human osteosarcoma cells
metastasis by blocking ERK and JNK-mediated MMPs expression.
Oncotarget. 7:35208–35223. 2016.
|
|
31
|
Melnikova VO, Mourad-Zeidan AA, Lev DC and
Bar-Eli M: Platelet-activating factor mediates MMP-2 expression and
activation via phosphorylation of cAMP-response element-binding
protein and contributes to melanoma metastasis. J Biol Chem.
281:2911–2922. 2006. View Article : Google Scholar
|
|
32
|
Taparra K, Tran PT and Zachara NE:
Hijacking the hexosamine biosynthetic pathway to promote
EMT-mediated neoplastic phenotypes. Front Oncol. 6:852016.
View Article : Google Scholar :
|
|
33
|
Kinjo K, Sandoval S, Sakamoto KM and
Shankar DB: The role of CREB as a proto-oncogene in hematopoiesis.
Cell Cycle. 4:1134–1335. 2005. View Article : Google Scholar
|
|
34
|
Ma X, Gu L, Li H, Gao Y, Li X, Shen D,
Gong H, Li S, Niu S, Zhang Y, et al: Hypoxia-induced overexpression
of stanniocalcin-1 is associated with the metastasis of early stage
clear cell renal cell carcinoma. J Transl Med. 13:562015.
View Article : Google Scholar :
|
|
35
|
Li H, Zhang K, Liu LH, Ouyang Y, Bu J, Guo
HB and Xiao T: A systematic review of matrix metalloproteinase 9 as
a biomarker of survival in patients with osteosarcoma. Tumour Biol.
35:5487–5491. 2014. View Article : Google Scholar
|
|
36
|
Willis AL, Sabeh F, Li XY and Weiss SJ:
Extracellular matrix determinants and the regulation of cancer cell
invasion stratagems. J Microsc. 251:250–60. 2013. View Article : Google Scholar
|
|
37
|
Park JK, Park SH, So K, Bae IH, Yoo YD and
Um HD: ICAM-3 enhances the migratory and invasive potential of
human non-small cell lung cancer cells by inducing MMP-2 and MMP-9
via Akt and CREB. Int J Oncol. 36:181–192. 2010.
|
|
38
|
Yang SF, Lee WJ, Tan P, Tang CH, Hsiao M,
Hsieh FK and Chien MH: Upregulation of miR-328 and inhibition of
CREB-DNA-binding activity are critical for resveratrol-mediated
suppression of matrix metalloproteinase-2 and subsequent metastatic
ability in human osteosarcomas. Oncotarget. 6:2736–2753. 2015.
View Article : Google Scholar
|
|
39
|
Sun B, Rong R, Jiang H, Zhang H, Wang Y,
Bai X, Zhang M, Ma J, Xia S, Shu W, et al: Prostaglandin E2
receptor EP1 phosphorylate CREB and mediates MMP2 expression in
human cholangiocarcinoma cells. Mol Cell Biochem. 378:195–203.
2013. View Article : Google Scholar
|
|
40
|
Lee DK, Park EJ, Kim EK, Jin J, Kim JS,
Shin IJ, Kim BY, Lee H and Kim DE: Atorvastatin and simvastatin,
but not pravastatin, up-regulate LPS-induced MMP-9 expression in
macrophages by regulating phosphorylation of ERK and CREB. Cell
Physiol Biochem. 30:499–511. 2012. View Article : Google Scholar
|
|
41
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar
|
|
42
|
Ni D, Ma X, Li HZ, Gao Y, Li XT, Zhang Y,
Ai Q, Zhang P, Song EL, Huang QB, et al: Downregulation of FOXO3a
promotes tumor metastasis and is associated with metastasis-free
survival of patients with clear cell renal cell carcinoma. Clin
Cancer Res. 20:1779–1790. 2014. View Article : Google Scholar
|
|
43
|
O'Mahony FC, Faratian D, Varley J, Nanda
J, Theodoulou M, Riddick AC, Harrison DJ and Stewart GD: The use of
automated quantitative analysis to evaluate
epithelial-to-mesenchymal transition associated proteins in clear
cell renal cell carcinoma. PLoS One. 7:e315572012. View Article : Google Scholar :
|
|
44
|
Cho JH, Hong WG, Jung YJ, Lee J, Lee E,
Hwang SG, Um HD and Park JK: Gamma-Ionizing radiation-induced
activation of the EGFR-p38/ERK-STAT3/CREB-1-EMT pathway promotes
the migration/invasion of non-small cell lung cancer cells and is
inhibited by podophyllotoxin acetate. Tumour Biol. 37:7315–7325.
2016. View Article : Google Scholar
|