Introduction
Thyroid cancer is the most common type of cancer of
the endocrine system, with cases increasing worldwide (1,2).
Thyroid cancer may be classified into numerous types according to
the histopathological characteristics. Medullary thyroid carcinoma
(MTC) is a form of thyroid cancer which originates from the
parafollicular cells of the thyroid (3). It is the third most common type of
thyroid cancer and accounts for ~3% of all thyroid cancer cases.
Approximately 1 in 4 of MTC cases are caused by mutations in the
rearranged during transfection (RET) proto-oncogene (4). The majority of MTC cases are
sporadic, presenting with metastatic disease at diagnosis (5). Nearly all patients with distant
metastases succumb to this disease (6).
Presently, RET mutation have been suggested to be an
indicator of the poor prognosis of MTC; however, this is not
sufficient for understanding the underlying molecular mechanisms of
MTC tumourigenesis. Soh et al (7) demonstrated that vascular endothelial
growth factor receptor 2 was involved in the pathogenesis of MTC
via promotion of pro-invasive and pro-angiogenic phenotypes.
Additionally, dysregulation of the Dickkopf/Wnt signaling pathway
inhibitor 4 has been identified in MTC (8). A previous study revealed that
aberrant expression levels of microRNAs (miRNAs) have a potential
role in tumourigenesis (9), which
may provide novel insight in MTC research. Notably, increasing
evidence has supported the important role of miRNAs in cancers
including thyroid cancer (10,11).
For example, He et al (12)
reported that three miRNAs, including miR-221, −222, and −146, are
overexpressed in papillary thyroid cancer. Furthermore, miR-197 and
−346 are significantly overexpressed in follicular thyroid cancers
(13). Although great advances
have been made in understanding the functions of miRNAs in thyroid
cancers, the underlying molecular mechanisms of this disease remain
to be elucidated.
The present study aimed to use GSE40807 miRNA
microarray data provided by Lassalle et al (14) to identify differentially expressed
miRNAs (DEMs) between human MTC and healthy control tissues.
Subsequently, transcription factor (TF)-miRNA and miRNA-target gene
regulatory networks were constructed. Finally, the target genes of
DEMs were performed functional enrichment analyses to predict their
potential functions that may be associated with MTC. To the best of
our knowledge, this is the first time that the dataset of GSE40807
was analyzed.
Materials and methods
Data source
The GSE40807 miRNA microarray dataset was downloaded
from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) database in the National
Center for Biotechnology Information based on the Agilent-019118
Human miRNA Microarray 2.0 G4470B platform (Agilent Technologies,
Inc., Santa Clara, CA, USA). The dataset included 14 pairs of miRNA
microarrays from human MTC and adjacent healthy tissues.
Data preprocessing and DEM
analysis
The original data were firstly converted into
identifiable expression form using a Linear Models for Microarray
Data (limma) package in R language (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(15). Following this, background
correction and quartile data normalization were performed using a
robust multiarray average algorithm affy package in R (http://www.bioconductor.org/packages/release/bioc/html/affy.html)
(16).
TF-miRNA regulatory association pair
prediction
The TF-miRNA regulatory database (TransmiR;
cmbi.bjmu.edu.cn/transmir) (17) is a valuable resource for the study
of TF-miRNA regulation, which provides an interface for easy
retrieval of TF-miRNA regulatory pairs by searching for a miRNA or
a TF. Currently, TransmiR has curated 735 entries, which includes
201 TFs, 209 miRNAs and 16 organisms from 268 publications. The
present study inputted the obtained DEMs into the database and
extracted regulatory association pairs between DEMs and TFs.
miRNA-target gene regulatory
association pair prediction and TF-miRNA-target gene regulatory
network construction
The starBase v2.0 (starbase.sysu.edu.cn/) database (18) provides certain miRNA-target
regulatory association pairs which are verified by experiments and
predicted by five algorithms including TargetScan (19), miRanda (20), Pictar2 (21), PITA (22) and RNA22 (23). In the present study, miRNA-target
gene regulatory association pairs verified by ≥ one experiment and
predicted by ≥ three algorithms were selected for construction of
the regulatory network.
Based on the predicted TF-miRNA and miRNA-target
gene regulatory association pairs, the TF-miRNA-target gene
regulatory network was constructed using Cytoscape software version
3.2.0 (www.cytoscape.org/) (24). From the network, TFs, miRNAs and
target genes that had higher connective degrees (hub nodes) were
extracted. Hub nodes are small numbers of nodes with numerous
interaction partners, which serve important roles in the network
(25). Thus, these TFs, miRNAs and
target genes might serve roles in MTC.
Functional enrichment analyses
clusterProfiler software (Bioconductor version 3.1;
bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(26) is a package used for gene
classification and enrichment analysis. The Kyoto Encyclopedia of
Genes and Genomes (KEGG; www.genome.ad.jp/kegg/) (27) is a database of biological systems
that collects genomic, chemical and systemic functional
information. To analyzed the potential biological functions of
DEMs, KEGG pathway enrichment analysis was performed for the target
genes of the obtained DEMs based on the clusterProfiler package.
P<0.01 was set as the threshold value.
Statistical analysis
DEMs between MTC and healthy tissues were identified
using the limma (15) package
(Bioconductor version 3.1). Student's t-test in the limma package
was used to compare DEM values, and fold changes (FCs) were
calculated. miRNAs with P<0.01 and |log2FC|≥1 were
selected as DEMs. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEMs
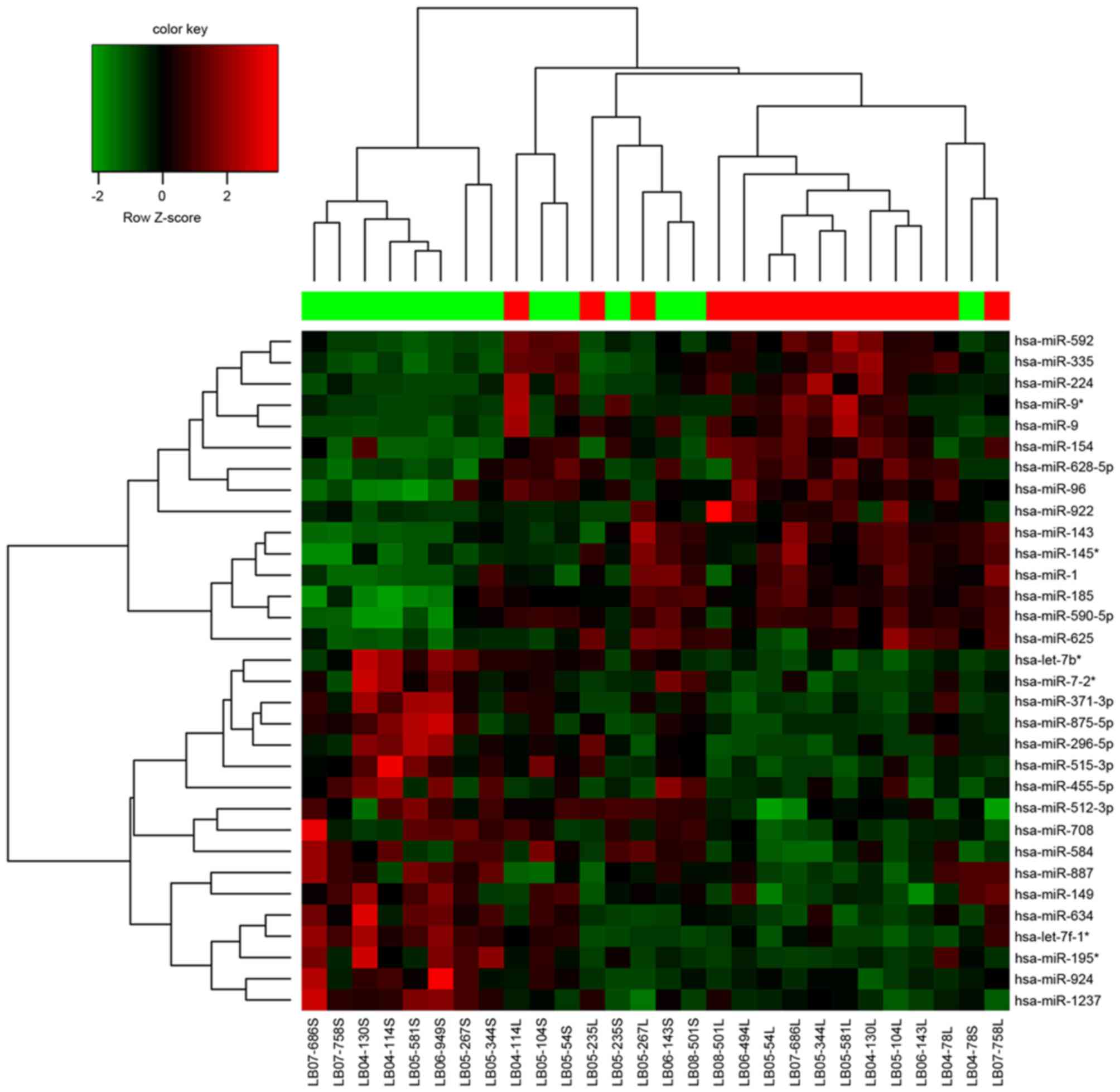
A total of 32 DEMs were identified between MTC and
healthy tissues. Among these DEMs, 15 were upregulated and 17 were
downregulated (Table I).
Hierarchical clustering analysis of these DEMs and samples are
presented in Fig. 1.
 | Table I.Up- and downregulated miRs. |
Table I.
Up- and downregulated miRs.
| miR |
log2FC | P |
|---|
| hsa-miR-9-5p |
1.771272789 | 9.96E-05 |
| hsa-miR-149-5p | −1.285205715 | 0.000830471 |
| hsa-miR-708-5p | −1.14714938 | 0.003865191 |
| hsa-miR-335-5p |
2.093096976 | 0.003949694 |
| hsa-miR-592 |
1.802025779 | 0.004011795 |
| hsa-miR-875-5p | −1.148677692 | 0.004350708 |
| hsa-miR-455-5p | −1.191505453 | 0.004403134 |
| hsa-miR-590-5p |
1.642054145 | 0.004446886 |
| hsa-miR-96-5p |
2.177657366 | 0.005246821 |
| hsa-miR-584-5p | −1.157995671 | 0.005083235 |
| hsa-miR-922 |
1.022980071 | 0.005377546 |
| hsa-miR-1 |
1.816907981 | 0.005578293 |
| hsa-miR-296-5p | −1.335624531 | 0.00595956 |
| hsa-miR-634 | −1.222619212 | 0.006085225 |
| hsa-miR-224-5p |
1.794623266 | 0.006202874 |
| hsa-miR-185-5p |
1.573515208 | 0.006464766 |
| hsa-miR-628-5p |
1.035790796 | 0.007467156 |
| hsa-miR-924 | −1.075200672 | 0.007467315 |
| hsa-miR-154-5p |
1.230953928 | 0.007599707 |
| hsa-miR-625-5p |
1.081907512 | 0.009088003 |
| hsa-miR-145-3p |
1.656652903 | 0.00054755 |
| hsa-miR-195-3p | −1.337668541 | 0.001551485 |
|
hsa-let-7f-1-3p | −1.15398965 | 0.001765947 |
| hsa-miR-515-3p | −1.033833078 | 0.002354703 |
| hsa-miR-9-3p |
1.917564249 | 0.004292166 |
| hsa-miR-7-2-3p | −1.106382457 | 0.005054543 |
| hsa-miR-143-3p |
1.498076967 | 0.005687211 |
| hsa-miR-887-3p | −1.372273521 | 0.005784544 |
| hsa-miR-512-3p | −1.149225488 | 0.006533263 |
|
hsa-miR-1237-3p | −1.009014763 | 0.006904609 |
|
hsa-miR-371a-3p | −1.099403781 | 0.007869777 |
| hsa-let-7b-3p | −1.362383564 | 0.008104953 |
TF-miRNA regulatory relationship
pairs
From TransmiR, 54 TF-miRNA regulatory association
pairs were extracted, including 33 TFs and 10 miRNAs. Among these
miRNAs, hsa-miR-9-5p was regulated by nine TFs, including nuclear
factor of κ light polypeptide gene enhancer in B-cells 1 (NF-κB1),
and interleukin 1β (IL-1β), and hsa-miR-1 was regulated by eight
TFs, including CCAAT/enhancer binding protein α. Additionally, TFs
of NF-κB1 regulated four DEMs, including hsa-miR-9-5p and −3p, and
v-myc avian myelocytomatosis viral oncogene homolog (MYC) regulated
three DEMs including hsa-miR-195-3p, hsa-let-7b-3p and
hsa-let-7f-1-3p.
miRNA-target gene regulatory
association pairs and TF-miRNA-target gene regulatory network
construction
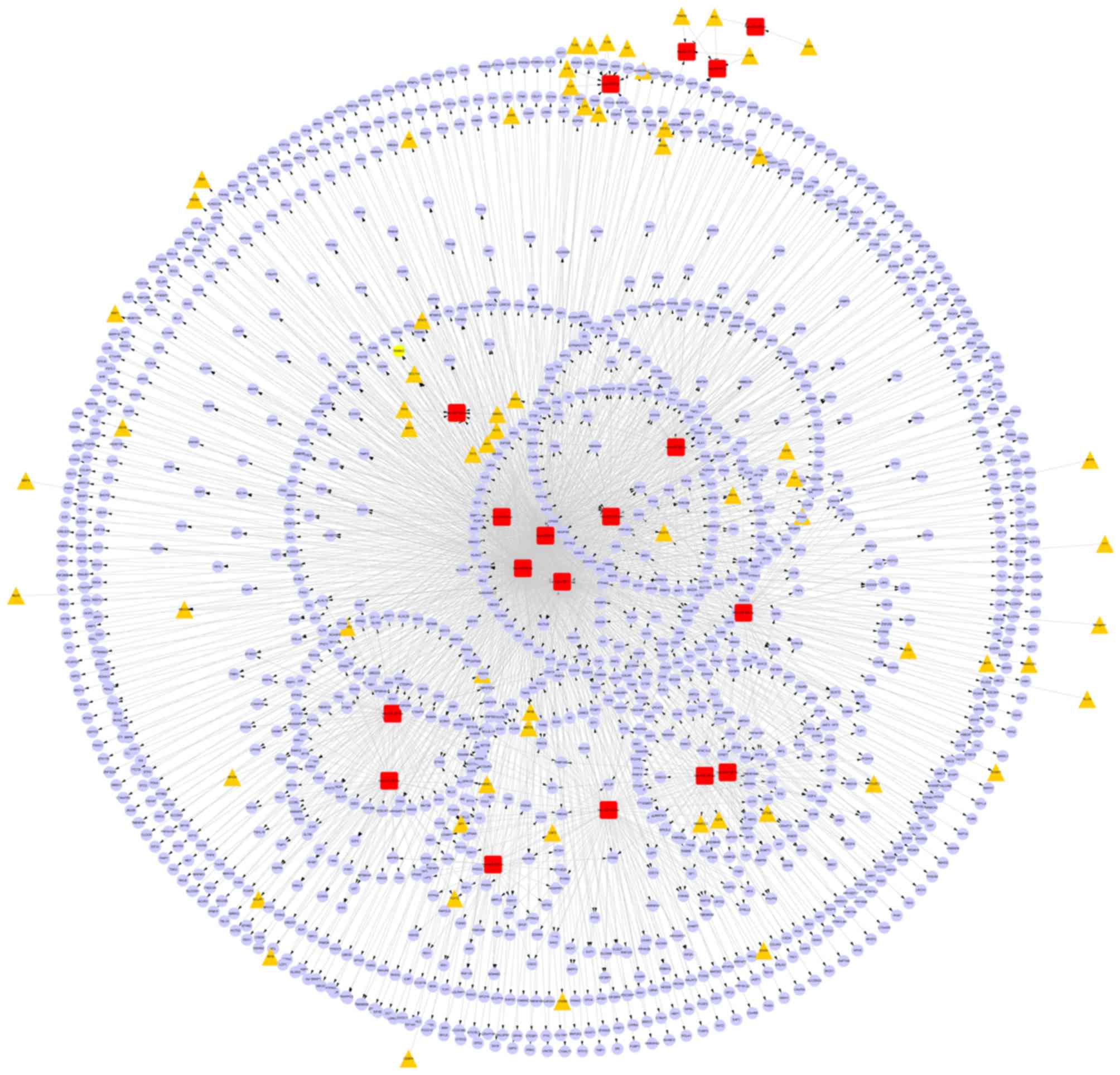
From starBase, 1654 miRNA-target gene regulatory
association pairs were obtained, including 12 DEMs and 1338 target
genes. Among the 12 DEMs, hsa-miR-1, hsa-miR-9-5p, hsa-miR-96-5p
and hsa-miR-590-5p had the top four highest connective degrees
(feature miRNAs). Additionally, seven target genes that were
regulated by at least four DEMs were identified (Table II).
 | Table II.Feature miRNAs and genes in the
miR-target gene regulatory network. |
Table II.
Feature miRNAs and genes in the
miR-target gene regulatory network.
| Node | Number |
|---|
| hsa-miR-9-5p | 326 |
| hsa-miR-96-5p | 297 |
| hsa-miR-1 | 246 |
| hsa-miR-590-5p | 146 |
| CRIM1 |
5 |
| KIF1B |
4 |
| NR4A3 |
4 |
| RNF111 |
4 |
| TNPO1 |
4 |
| FNDC3B |
4 |
| BCL11A |
4 |
Furthermore, based on the constructed miRNA-target
gene and TF-miRNA regulatory networks, a TF-miRNA- target gene
regulatory network was constructed using Cytoscape software. In the
network, there were 1654 miRNA-target gene and 54 TF-miRNA
regulatory relationship pairs (Fig.
2).
Functional enrichment analyses
Among the 12 DEMs in the miRNA-target gene
regulatory network, the target genes of hsa-miR-1, hsa-miR-9-5p,
hsa-miR-96-5p and hsa-miR-590-5p were demonstrated to be enriched
in the KEGG pathways, including the mitogen activated protein
kinase (MAPK) signaling pathway, pathways in cancer, and during
focal adhesion (Table III).
 | Table III.Enriched signaling pathways involving
differentially expressed miRs. |
Table III.
Enriched signaling pathways involving
differentially expressed miRs.
| miR | Pathway
description |
|---|
| hsa-miR-1 | Neurotrophin
signaling pathway |
|
| Renal cell
carcinoma |
|
| Axon guidance |
|
| MAPK signaling
pathway |
|
| Pathways in
cancer |
| hsa-miR-9-5p | Neurotrophin
signaling pathway |
|
| Bacterial invasion
of epithelial cells |
|
| Focal adhesion |
|
| MAPK signaling
pathway |
|
| Endocytosis |
| hsa-miR-96-5p | GnRH signaling
pathway |
|
| ErbB signaling
pathway |
|
| Prostate
cancer |
|
| Neurotrophin
signaling pathway |
|
| Axon guidance |
| hsa-miR-590-5p | MAPK signaling
pathway |
Discussion
Patients with progressive MTC have limited treatment
options (28). Thus, understanding
the underlying molecular mechanism of carcinogenesis may facilitate
diagnosis and therapy options of this disease. In the present
study, 15 upregulated and 17 downregulated DEMs were identified. In
the constructed TF-miRNA regulatory network, hsa-miR-9-5p was
regulated by 9 TFs and hsa-miR-1 was regulated by 8 TFs. The TFs of
NF-κB1 and MYC regulated 4 and 3 DEMs, respectively. Additionally,
the above two miRNAs served key roles in the miRNA-target gene
regulatory network. Their target genes were primarily enriched in
the MAPK signaling pathway and during focal adhesion. These miRNAs
and signaling pathways may be important biomarkers for MTC
diagnosis and treatment.
In the miRNA-target gene regulatory network,
hsa-miR-1 was upregulated, and its target genes, including
MAPK1, were enriched in numerous signaling pathways
associated with cancer, including MAPK. MAPKs are a family of
protein kinases whose functions are conserved during evolution from
unicellular organisms (29). The
MAPK signaling pathway consists of numerous key signaling
components and phosphorylation events which control multiple
fundamental cell processes including proliferation, differentiation
and apoptosis (30). This
signaling pathway has been frequently identified in activated in
human cancers, which leads to malignant phenotypes including
autonomous cell proliferation (31). Notably, Zatelli et al
(32) demonstrated that the growth
of the TT MTC cell line depends on activation of the MAPK signaling
pathway, which suggests its role in MTC. In addition, MAPK
signaling is important in regulating cytokine signaling pathways
(33). Cytokines are released in
response to inflammation and immunity, and have important roles in
cancer development and progression (34). It has been reported that
undifferentiated thyroid cancer cells secrete cytokines (35). Taken together, the MAPK signaling
pathway may serve important roles in MTC via hsa-miR-1 and its
target gene MAPK1.
In addition to hsa-miR-1, hsa-miR-9-5p upregulated
in the miRNA-target gene regulatory network, and was regulated by 9
TFs in the TF-miRNA regulatory network, including NF-κB1 and IL-1ß.
NF-κB is a transcription regulator activated by various intra- and
extracellular stimuli. A previous study demonstrated that NF-κB1
regulates the expression of genes involved in numerous processes,
including proliferation and apoptosis (36). Inappropriate activation of NF-κB
has been associated with numerous inflammatory diseases, whereas
persistent inhibition of NF-κB may lead to delayed cell growth
(37). NF-κB has been demonstrated
to be associated with the development of colorectal (38), breast (39), bladder (40), prostate (41) and advanced thyroid (36) cancers. On the other hand, IL-1β, a
member of the IL-1 cytokine family, is an important mediator of the
inflammatory response. A previous study reported that inflammation
is a critical component of tumor progression (42). Zeki et al (43) suggested that IL-1 regulates G1 cell
cycle progression and arrest in papillary thyroid carcinoma cells.
Therefore, NF-κB1, IL-1ß and their regulated DEM hsa-miR-9-5p may
serve important roles in MTC progression.
Additionally, TFs of MYC were demonstrated to
regulate 3 DEMs including hsa-miR-195-3p, hsa-let-7b-3p and
hsa-let-7f-1-3p. MYC is a multifunctional, nuclear phosphoprotein
which serves roles in cell cycle progression, apoptosis and
cellular transformation (44). The
MYC gene has been widely implicated in numerous human
cancers (45,46). Khosla et al (47) revealed that MYC mRNA expression
levels increased in apoptotic TT cells, suggesting its role in MTC.
Its regulated miRNA hsa-miR-195-3p has been identified to be
abnormally expressed in a variety of cancers. For example, levels
were upregulated in breast cancer and downregulated in gastric,
hepatocellular and bladder cancers (48–50).
The roles of hsa-let-7b-3p and hsa-let-7f-1-3p in cancer remain to
be elucidated; thus, it was hypothesized that these miRNAs may be
involved in MTC via regulation from the MYC gene. Taken
together, MYC and its regulated DEMs, hsa-miR-195-3p, hsa-let-7b-3p
and hsa-let-7f-1-3p, may serve important roles in the development
of MTC.
The present study identified numerous key miRNAs and
TFs that may be associated with MTC using comprehensive
bioinformatics methods. However, no experiments with tissues or
cells were performed to validate the expression levels of these
miRNAs and TFs; a key limitation of this study. Additionally, there
were only 14 pairs of miRNA microarrays in the dataset. Further
studies with experimental validations and more samples are required
to validate these observations.
In conclusion, the results of the present study
indicated that the DEMs hsa-miR-1, hsa-miR-9-5p and hsa-miR-195-3p
may have the potential to be used as diagnostic and therapeutic
targets of MTC. Additionally, hsa-miR-1 and its target gene
MAPK1 may serve a role in MTC, involving in MAPK signaling
pathway. Additionally, TFs of IL-1ß and MYC may be implicated in
the development of MTC.
Acknowledgements
The present study was supported by the Beijing
Municipal Science & Technology Commission (grant no.
Z141107002514003), the Beijing Municipal Administration of
Hospitals Clinical Medicine Development of Special Funding Support
(grant no. XMLX201311) and the National Natural Science Foundation
of China (grant no. 81473499).
References
|
1
|
He W, Qi B, Zhou Q, Lu C, Huang Q, Xian L
and Chen M: Key genes and pathways in thyroid cancer based on gene
set enrichment analysis. Oncol Rep. 30:1391–1397. 2013.
|
|
2
|
Geraldo MV and Kimura ET: Integrated
analysis of thyroid cancer public datasets reveals role of
post-transcriptional regulation on tumor progression by targeting
of immune system mediators. PLoS One. 10:e01417262015. View Article : Google Scholar :
|
|
3
|
Hu MI, Vassilopoulou-Sellin R, Lustig R
and Lamont JP: Thyroid and parathyroid cancers. Cancer Management:
A Multidisciplinary Approach. 11:2008.
|
|
4
|
Lodish MB and Stratakis CA: RET oncogene
in MEN2, MEN2B, MTC and other forms of thyroid cancer. Expert Rev
Anticancer Ther. 8:625–632. 2008. View Article : Google Scholar :
|
|
5
|
Santarpia L, Calin GA, Adam L, Ye L, Fusco
A, Giunti S, Thaller C, Paladini L, Zhang X, Jimenez C, et al: A
miRNA signature associated with human metastatic medullary thyroid
carcinoma. Endocr Relat Cancer. 20:809–823. 2013. View Article : Google Scholar
|
|
6
|
Hofstra R, Stelwagen T, Stulp RP, de Jong
D, Hulsbeek M, Kamsteeg EJ, van den Berg A, Landsvater RM, Vermey
A, Molenaar WM, et al: Extensive mutation scanning of RET in
sporadic medullary thyroid carcinoma and of RET and VHL in sporadic
pheochromocytoma reveals involvement of these genes in only a
minority of cases. J Clin Endocrinol Metab. 81:2881–2884. 1996.
View Article : Google Scholar
|
|
7
|
Soh EY, Duh QY, Sobhi SA, Young DM,
Epstein HD, Wong MG, Garcia YK, Min YD, Grossman RF, Siperstein AE
and Clark OH: Vascular endothelial growth factor expression is
higher in differentiated thyroid cancer than in normal or benign
thyroid. J Clin Endocrinol Metab. 82:3741–3747. 1997. View Article : Google Scholar
|
|
8
|
Maliszewska A, Leandro-Garcia LJ,
Castelblanco E, Macià A, de Cubas A, Goméz-López G, Inglada-Pérez
L, Álvarez-Escolá C, De la Vega L, Letón R, et al: Differential
gene expression of medullary thyroid carcinoma reveals specific
markers associated with genetic conditions. Am J Pathol.
182:350–362. 2013. View Article : Google Scholar
|
|
9
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar :
|
|
10
|
Pallante P, Visone R, Croce CM and Fusco
A: Deregulation of microRNA expression in follicular cell-derived
human thyroid carcinomas. Endocr Relat Cancer. 17:F91–F104. 2010.
View Article : Google Scholar
|
|
11
|
Visone R, Pallante P, Vecchione A,
Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V,
Borbone E, et al: Specific microRNAs are downregulated in human
thyroid anaplastic carcinomas. Oncogene. 26:7590–7595. 2007.
View Article : Google Scholar
|
|
12
|
He H, Jazdzewski KW, Li W, Liyanarachchi
S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et
al: The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:pp. 19075–19080. 2005; View Article : Google Scholar :
|
|
13
|
Weber F, Teresi RE, Broelsch CE, Frilling
A and Eng C: A limited set of human microRNA is deregulated in
follicular thyroid carcinoma. J Clin Endocrinol Metab.
91:3584–3591. 2006. View Article : Google Scholar
|
|
14
|
Lassalle S, Zangari J, Popa A, Ilie M,
Hofman V, Long E, Patey M, Tissier F, Belléannée G, Trouette H, et
al: MicroRNA-375/SEC23A as biomarkers of the in vitro efficacy of
vandetanib. Oncotarget. 7:30461–30478. 2016.
|
|
15
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; New York, ΝΥ: pp. 397–420.
2005, View Article : Google Scholar
|
|
16
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar
|
|
17
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38(Database issue): D119–D122. 2010. View Article : Google Scholar
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2. 0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database issue): D92–D97. 2014. View Article : Google Scholar
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar
|
|
20
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar :
|
|
21
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar
|
|
22
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar
|
|
23
|
Ritchie W, Flamant S and Rasko JE:
Predicting microRNA targets and functions: Traps for the unwary.
Nat Methods. 6:397–398. 2009. View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar :
|
|
25
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar :
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar :
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar :
|
|
28
|
Elisei R, Schlumberger MJ, Müller SP,
Schöffski P, Brose MS, Shah MH, Licitra L, Jarzab B, Medvedev V,
Kreissl MC, et al: Cabozantinib in progressive medullary thyroid
cancer. J Clin Oncol. 31:3639–3646. 2013. View Article : Google Scholar :
|
|
29
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar
|
|
30
|
Dhillon A, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290.
2007. View Article : Google Scholar
|
|
31
|
Bradham C and McClay DR: p38 MAPK in
development and cancer. Cell Cycle. 5:824–828. 2006. View Article : Google Scholar
|
|
32
|
Zatelli MC, Piccin D, Tagliati F, Bottoni
A, Luchin A and Uberti EC degli: SRC homology-2-containing protein
tyrosine phosphatase-1 restrains cell proliferation in human
medullary thyroid carcinoma. Endocrinology. 146:2692–2698. 2005.
View Article : Google Scholar
|
|
33
|
Sumimoto H, Imabayashi F, Iwata T and
Kawakami Y: The BRAF-MAPK signaling pathway is essential for
cancer-immune evasion in human melanoma cells. J Exp Med.
203:1651–1656. 2006. View Article : Google Scholar :
|
|
34
|
Dranoff G: Cytokines in cancer
pathogenesis and cancer therapy. Nat Rev Cancer. 4:11–22. 2004.
View Article : Google Scholar
|
|
35
|
Fiore L, Pollina LE, Fontanini G, Casalone
R, Berlingieri MT, Giannini R, Pacini F, Miccoli P, Toniolo A,
Fusco A and Basolo F: Cytokine production by a new undifferentiated
human thyroid carcinoma cell line, FB-1. J Clin Endocrinol Metab.
82:4094–4100. 1997. View Article : Google Scholar
|
|
36
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-kB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar
|
|
37
|
Caamano J and Hunter CA: NF-kappaB family
of transcription factors: Central regulators of innate and adaptive
immune functions. Clin Microbiol Rev. 15:414–429. 2002. View Article : Google Scholar :
|
|
38
|
Lewander A, Butchi AK, Gao J, He LJ,
Lindblom A, Arbman G, Carstensen J, Zhang ZY and Sun XF; Swedish
Low-Risk Colorectal Cancer Study Group, : Polymorphism in the
promoter region of the NFKB1 gene increases the risk of sporadic
colorectal cancer in Swedish but not in Chinese populations. Scand
J Gastroenterol. 42:1332–1338. 2007. View Article : Google Scholar
|
|
39
|
Curran JE, Weinstein SR and Griffiths LR:
Polymorphic variants of NFKB1 and its inhibitory protein NFKBIA and
their involvement in sporadic breast cancer. Cancer Lett.
188:103–107. 2002. View Article : Google Scholar
|
|
40
|
Riemann K, Becker L, Struwe H, Rübben H,
Eisenhardt A and Siffert W: Insertion/deletion polymorphism in the
promoter of NFKB1 as a potential molecular marker for the risk of
recurrence in superficial bladder cancer. Int J Clin Pharmacol
Ther. 45:423–430. 2007. View
Article : Google Scholar
|
|
41
|
Zhang P, Wei Q, Li X, Wang K, Zeng H, Bu H
and Li H: A functional insertion/deletion polymorphism in the
promoter region of the NFKB1 gene increases susceptibility for
prostate cancer. Cancer Genet Cytogenet. 191:73–77. 2009.
View Article : Google Scholar
|
|
42
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar :
|
|
43
|
Zeki K, Morimoto I, Arao T, Eto S and
Yamashita U: Interleukin-1alpha regulates G1 cell cycle progression
and arrest in thyroid carcinoma cell lines NIM1 and NPA. J
Endocrinol. 160:67–73. 1999. View Article : Google Scholar
|
|
44
|
Hoffman B and Liebermann D: Apoptotic
signaling by c-MYC. Oncogene. 27:6462–6472. 2008. View Article : Google Scholar
|
|
45
|
Ellwood-Yen K, Graeber TG, Wongvipat J,
Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV and Sawyers CL:
Myc-driven murine prostate cancer shares molecular features with
human prostate tumors. Cancer Cell. 4:223–238. 2003. View Article : Google Scholar
|
|
46
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.
|
|
47
|
Khosla S, Oursler M, Schroeder M and
Eberhardt N: Transforming growth factor-beta 1 induces growth
inhibition of a human medullary thyroid carcinoma cell line despite
an increase in steady state c-myc messenger ribonucleic acid
levels. Endocrinology. 135:1887–1893. 1994. View Article : Google Scholar
|
|
48
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar
|
|
49
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar
|
|
50
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar
|