Introduction
Human dental pulp stem cells (hDPSCs) are derived
from human teeth and have garnered attention in stem cell research
due to their biological properties, as they are highly clonogenic
and proliferative. In addition, they are characterized by
self-renewal and multi-lineage differentiation (1,2).
Therefore, it may be hypothesized that hDPSCs have potential to be
used in regenerative medicine, particularly for the development of
biologic substitutes, including artificial tissues. High rates of
proliferation and migration are critical for cells to be
successfully used in regenerative medicine applications (3).
Previous studies have demonstrated that stem cell
growth, adhesion and migration may be regulated by several
molecules, including transcriptional coactivator with PDZ-binding
motif (TAZ) (4–8). TAZ belongs to the family of 14-3-3
cytoplasmic proteins (9). Previous
studies have demonstrated its involvement in embryogenesis
(10) and the development of
several organs, including bone, lung and heart (11–13).
TAZ is one of the key downstream effectors of the Hippo signaling
pathway, which has been reported to regulate stem cell
proliferation, differentiation and survival (14). It has previously been revealed that
TAZ promoted the differentiation of mesenchymal stem cells (MSCs)
into osteoblastic lineages (15,16),
whereas cell-permeable low-molecular-weight protamine-TAZ fusion
proteins have been demonstrated to increase the odontogenic and
osteogenic differentiation of hDPSCs (17). In addition, TAZ overexpression has
been reported to promote cellular proliferation and induce
epithelial-mesenchymal transition (10). However, the effects of TAZ on the
regulation of hDPSC proliferation and migration, as well as the
molecular mechanisms underlying its actions, remain to be
elucidated.
In the present study, the effects of TAZ on the
proliferation and migration of hDPSCs were investigated. The
present results demonstrated that hDPSC proliferation and migration
were inhibited following the silencing of TAZ expression. The
molecular mechanisms underlying the effects of TAZ knockdown
appeared to be associated with the downregulation of connecting
tissue growth factor (CTGF) and cysteine-rich angiogenic inducer
(Cyr) 61 expression, and possibly implicated tumor growth factor
(TGF)-β-mediated signaling pathways.
Materials and methods
Reagents
MTT and dimethyl sulphoxide (DMSO) were obtained
from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Transwell
chambers were purchased from Corning Inc. (Corning, NY, USA).
Anti-TAZ (catalog no. 4883) primary antibodies were obtained from
Cell Signaling Technology, Inc. (Danvers, MA, USA); anti-CTGF
(catalog no. SC-365970), anti-Cyr61 (catalog no. SC-374129),
anti-mothers against decapentaplegic homolog (Smad) 3 (catalog no.
SC-101154) and anti-Smad4 (catalog no. SC-7966) primary antibodies
were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA); and anti-GAPDH primary antibody was purchased from
Sigma-Aldrich (catalog no. G8795; Sigma-Aldrich; Merck KGaA).
Cell isolation and culture
hDPSCs were isolated from healthy third molars or
premolars of healthy human subjects (male; age, 16–30 years; n=12).
Individual dental pulps were minced into small pieces (1 mm3) and
digested using type I collagenase (3.0 mg/ml) and dispase (4.0
mg/ml; Sigma-Aldrich; Merck KGaA) for 45 min at 37°C. The solution
was filtered through a 70-mm cell strainer. Single-cell suspensions
were obtained and seeded into 35 mm culture dishes at a density of
1×105/ml. Cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA), 2.0 mM/l glutamine
(Invitrogen; Thermo Fisher Scientific, Inc.), and 100 IU/ml
penicillin and streptomycin, and maintained at 37°C in a humidified
5% CO2 atmosphere. When confluent, cells were collected
by trypsinization (0.2% trypsin and 0.02% EDTA) and subcultured
with DMEM supplemented with 10% FBS. The medium was replaced every
2–3 days. Cells between 3 and 5 passages were used in
experiments.
The present study was approved by the Ethics
Committee of the Second Hospital of Hebei Medical University
(Shijiazhuang, China). Written informed consent was obtained from
all human subjects prior to enrollment in the present study.
Immunofluorescence
A total of 1×105/ml hDPSCs were seeded onto uncoated
coverslips and were fixed with 4% paraformaldehyde in PBS for 20
min at room temperature. Fixed samples were permeabilized using
0.3% Triton X-100 (Sigma-Aldrich; Merck KGaA) in PBS for 15 min at
room temperature. Following blocking with 10% normal goat serum
diluted with 1% immunoglobulin G (IgG)-free bovine serum albumin
(Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA)
and 0.3% Triton X-100 in PBS at 37°C for 2 h, the samples were
incubated with rabbit anti-TAZ polyclonal antibody (1:200)
overnight at 4°C, and with DyLight™ 488-conjugated goat anti-rabbit
IgG (catalog no. 611-141-002; 1:500; Jackson ImmunoResearch
Laboratories, Inc.) for 2 h at room temperature. A total of 5 µg/ml
Hoechst 33342 was used to stain the nuclei at 37°C for 15 min.
Fluorescently-labeled cells were observed under an upright
fluorescence microscope (magnification, ×20; Olympus Corporation,
Tokyo, Japan).
Small interfering (si)RNA
transfection
siRNA duplex oligonucleotides targeting TAZ mRNA
(siTAZ) and non-targeting duplex oligonucleotides used as negative
controls (siCON) were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). Sequences of siTAZ and siCON were as follows:
siTAZ sense, 5′-GGCCAGAGAUAUUUCCUUATT-3′; anti-sense,
5′-UAAGGAAAUAUCUCUGGCCTT−3′; and siCON sense,
5′-UUCACCGAACGUGUCACGUTT-3′; anti-sense:
5′-ACGUGACACGUUCGGAGAATT-3′;. Following incubation for 24 h, hDPSCs
were transfected with 1 µg/ml of siRNAs, using RNAi-Mate
transfection reagent (Shanghai GenePharma Co., Ltd., Shanghai,
China), according to the manufacturer's protocol, and cultured in
DMEM. Cells were subjected to a second round of transfection prior
to subsequent experiments (18).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Following treatment, cells in different groups were
harvested and total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Total RNA was reverse transcribed into
cDNA at 42°C for 1 h using Moloney murine leukemia virus reverse
transcriptase (GeneCopoeia, Inc., Rockville, MD, USA). qPCR was
performed on cDNA using the All-in-One™ qPCR mix (GeneCopoeia,
Inc.) and an ABI7300 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following primers were
obtained from Invitrogen (Thermo Fisher Scientific, Inc.): TAZ:
sense, 5′-ATGTTGACCTCCGGACTTTGG−3′; anti-sense:
5′-GAGGAAGGGCTCGCTTTTGT-3′; Cyr61: sense,
5′-GGCAGACCTTGTGAATATA-3′; anti-sense: 5′-GTATTAGGCTTTATTTACCA-3′;
CTGF: sense, 5′-CCCAGACCCAAATATGATT-3′; anti-sense:
5′-CAATGTACGATAGTGCAGT-3′; and GAPDH sense:
5′-AGTCCAACGGCACAGTCAAGG-3′; anti-sense: 5′-AGCACCAGCATCACCCAT-3′;.
Amplification conditions were as follows: Initial denaturation at
95°C for 1 min; followed by 40 cycles of 95°C for 30 sec, 60°C for
20 sec and 72°C for 20 sec. The relative expression levels of each
gene were normalized to GAPDH expression with 2-ΔΔCt
method (19).
Western blot analysis
Total protein was extracted from hDPSCs using RIPA
lysis buffer (C1053; Applygen Technologies, Inc., Beijing, China),
according to the manufacturer's protocol. Protein concentrations
were determined using a bicinchonic acid protein assay kit (Thermo
Fisher Scientific, Inc.). A total of 50 µg extracted protein
samples were separated by 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes. Membranes were blocked with 5%
non-fat milk with TBS containing Tween-20 for 2 h at 37°C and then
incubated with anti-TAZ (1:1,000), anti-CTGF (1:200), anti-Cyr61
(1:200), anti-Smad3 (1:200), anti-Smad4B (1:200) and anti-GAPDH
(1:3,000) primary antibodies at 4°C overnight. Membranes were
subsequently incubated with IRDye800®-conjugated
secondary antibody (catalog no. 608-445-002; 1:20,000; Rockland
Immunochemicals, Inc., Limerick, PA, USA) for 1 h at 37°C, followed
by scanning with the Odyssey® Infrared Imaging System
(LI-COR Biosciences, Lincoln, NE, USA). Data were normalized to
GAPDH levels and analyzed with Image-Pro Plus software, version 7.0
(Media Cybernetics, Inc., Rockville, MD, USA).
Cellular proliferation assay
Cellular proliferation was evaluated using the MTT
assay. hDPSCs were seeded in 96-well plates, at a density of
3×103 cells/well, and incubated at 37°C for 24 h.
Subsequently, 20 µl MTT solution (5 mg/ml) was added and cells were
incubated at 37°C for 4 h. The culture medium was replaced with
DMSO (150 µl/well) and the plates were agitated at room temperature
for 10 min to dissolve the crystals. The optical density at 570 nm
for each well was measured using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Cellular proliferation was also assessed via the
detection of 5-bromo-2′-deoxyuridine (BrdU)-labeled DNA using the
Cell Proliferation ELISA, BrdU kit (Roche Applied Science,
Penzberg, Germany). Briefly, hDPSCs were seeded in 96-well plates
(3×104 cells/well) and were incubated for 24 h at 37°C
in the presence of siTAZ or siCON. Subsequently, cells were labeled
with BrdU at 37°C for 1 h. BrdU-labeled DNA was quantified using
ELISA.
Cellular migration assay
Cellular migration was assessed using a wound
healing assay. hDPSCs were seeded in 6-well plates
(~5×104 cells/well) and cultured to form a monolayer.
The cell monolayer was scratched with a 200-µl pipette tip. Cells
were cultured for 24 h and wound healing was observed at 0 and 24 h
post-wound infliction under a phase-contrast microscope (Olympus
Corporation). Healing was assessed using Scion Image Software,
version 4.03 (Scion Corporation, Frederick, Maryland, USA).
Cellular invasion was also assessed using Transwell inserts coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Following
treatment, hDPSCs were seeded into the upper chambers at a density
of 5×104 cells in 200 µl serum-free DMEM; culture medium
supplemented with 10% FBS was added to the lower chambers.
Following incubation for 24 h at 37°C, the cells on the upper
surface of the membrane were removed. The invaded cells on the
lower membrane were fixed for 10 min with 4% paraformaldehyde at
4°C, stained at 37°C for 10 min with 5 µg/ml Hoechst 33342 and
counted under an upright fluorescence microscope (Olympus
Corporation).
Treatment with recombinant human
thrombospondin 1 (r-hTHBS1)
hDPSCs were seeded into 6-well plates at a density
of 2×105 cells/well and cultured for 24 h. Subsequently,
cells were incubated at 37°C for 10 min with r-hTHBS1, a TGF-β
activator (200 ng/ml; R&D Systems, Inc., Minneapolis, MN, USA).
Cells were harvested following treatment and used for further
analysis.
Statistical analysis
The statistical significance of the difference
between groups was assessed by the Student's t-test for pair-wise
comparisons or a one-way analysis of variance, followed by a post
hoc Student-Newman-Keuls test for multiple comparisons. Data are
expressed as the mean ± standard deviation. The experiment was
repeated three times. P<0.05 was considered to indicate a
statistically significant difference. The analysis was performed
using SPSS software (version 13.0; SPSS, Inc., Chicago, IL,
USA).
Results
TAZ is expressed in hDPSCs
hDPSCs were cultured for 48 h, fixed and processed
for immunofluorescence. TAZ was revealed to be expressed in hDPSCs
(Fig. 1).
siTAZ transfection decreases TAZ
expression in hDPSCs
Successful transfection with siTAZ in hDPSCs was
confirmed using RT-qPCR and western blot analysis. The present
results suggested that TAZ mRNA and protein expression levels were
significantly decreased following transfection with siTAZ compared
with control cells and cells transfected with siCON (Fig. 2).
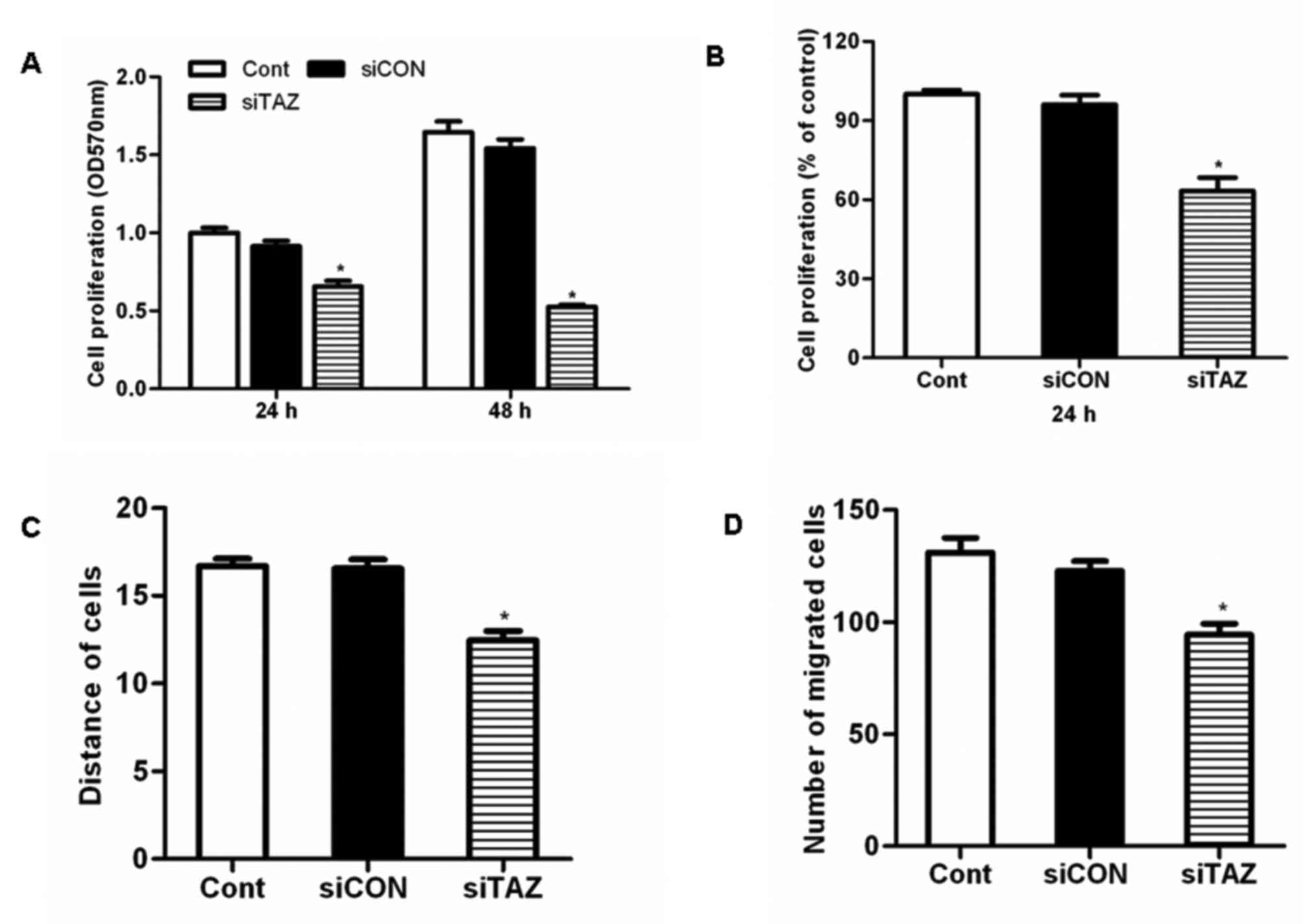
siTAZ inhibits hDPSC proliferation in
vitro
hDPSCs were transfected with siTAZ to silence TAZ
expression. Results of the MTT assay demonstrated that TAZ
silencing significantly inhibited hDPSC proliferation compared with
control cells and cells transfected with siCON (Fig. 3A). Similarly, the results of the
BrdU assay revealed that transfection with siTAZ decreased hDPSC
proliferation in vitro (Fig.
3B).
siTAZ inhibits hDPSC migration in
vitro
A wound-healing assay demonstrated that the
migration distance of hDPSCs transfected with siTAZ following a
scratch wound was significantly reduced compared with control cells
and cells transfected with siCON (Fig.
3C). These results suggested that TAZ silencing significantly
impaired the migratory capabilities of hDPSCs. Similarly, the
results of the Transwell invasion assay revealed that transfection
with siTAZ suppressed hDPSC invasion in vitro (Fig. 3D).
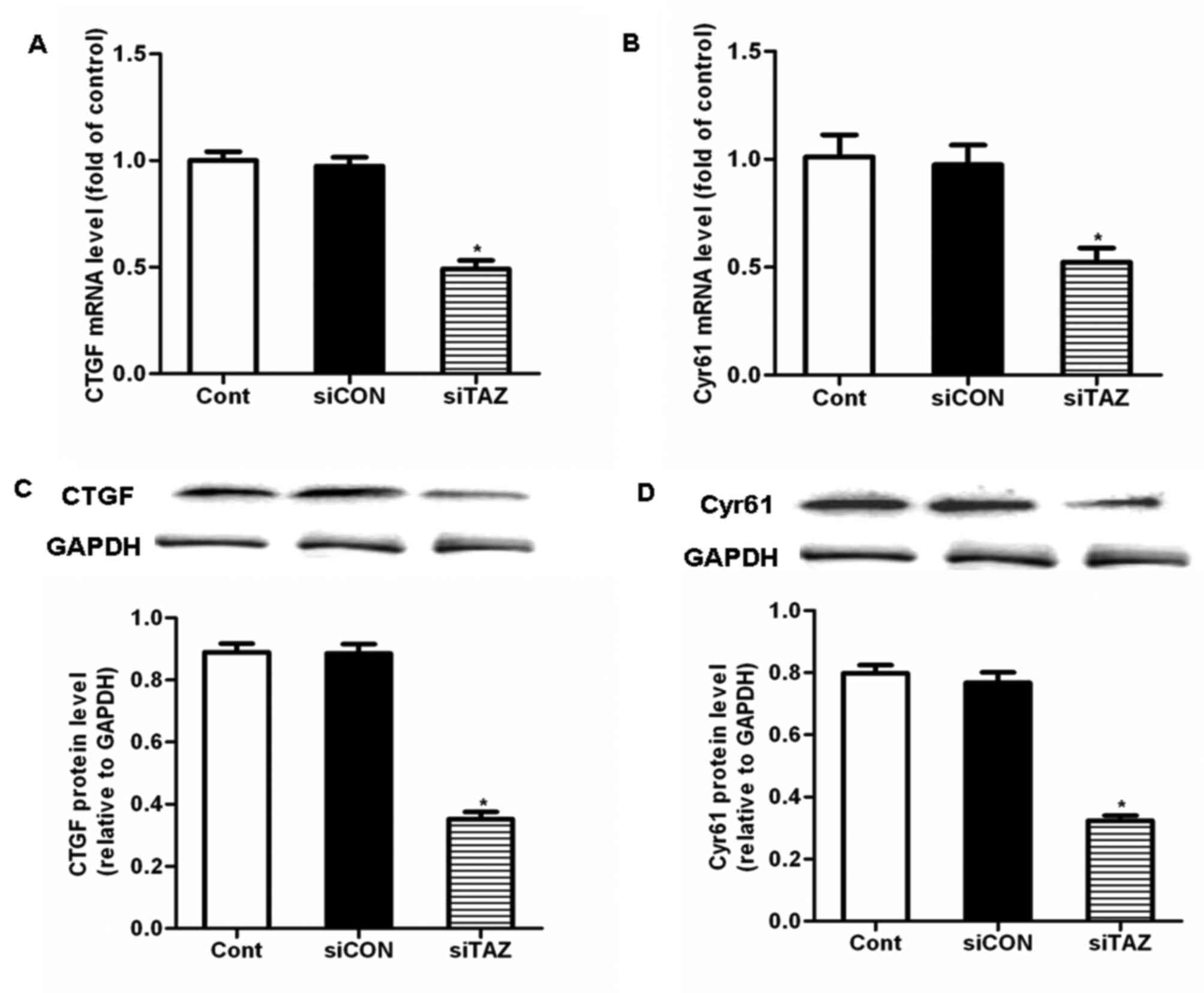
siTAZ inhibits hDPSC proliferation and
migration through the downregulation of CTGF and Cyr61
expression
To explore the molecular mechanism underlying the
inhibitory effects of TAZ silencing on hDPSC proliferation and
migration, putative downstream targets of TAZ were investigated.
CTGF and Cyr61 have been suggested to be downstream targets of TAZ
(20), which may be implicated in
the regulation of cellular proliferation, migration and apoptosis.
Treatment of hDPSCs with siTAZ for 24 h significantly decreased the
mRNA and protein expression levels of CTGF and Cyr61, compared with
control cells and cells transfected with siCON (Fig. 4). These results indicated that
siTAZ inhibited hDPSC proliferation and migration through the
modulation of CTGF and Cyr61 expression.
siTAZ inhibits CTGF and Cyr61
expression via a TGF-β-mediated pathway
To further explore the molecular mechanisms
underlying the implication of TAZ in the regulation of CTGF and
Cyr61 expression, the TGF-β pathway was investigated. Following
treatment with r-hTHBS1, the protein expression levels of Smad3,
Smad4, CTGF and Cyr61 in hDPSCs were significantly upregulated
compared with the control (Fig.
5A-D). These results suggested that TAZ may be involved in the
regulation of CTGF and Cyr61 expression via a TGF-β-mediated
pathway. Therefore, the expression of Smad3/4 in hDPSCs was
investigated following silencing of TAZ expression. Western blot
analysis demonstrated that Smad3/4 protein expression levels were
significantly downregu lated in TAZ-depleted cells compared with in
control cells and cells transfected with siCON (Fig. 5E and F). These results suggested
that TAZ may be implicated in TGF-β-dependent pathways that may
modulate the expression of CTGF and Cyr61.
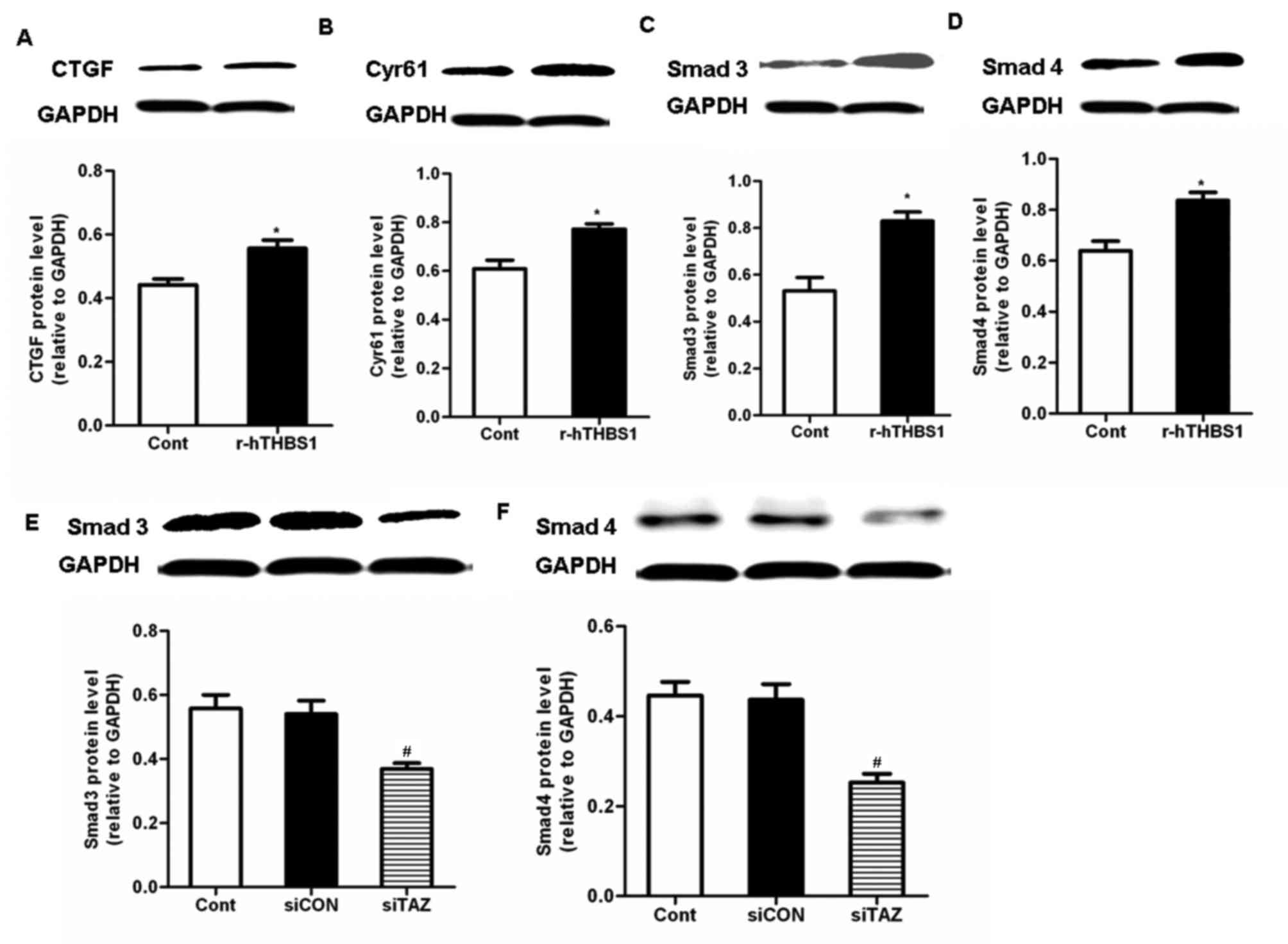 | Figure 5.TAZ modulated CTGF and Cyr61
expression via the activation of transforming growth
factor-β-mediated pathways. Western blot analysis was used to
assess the protein expression levels of (A) CTGF, (B) Cyr61, (C)
Smad3 and (D) Smad4 in hDPSCs following treatment with r-hTHBS1.
Data are expressed as the mean ± standard deviation of 3
independent experiments. *P<0.05 vs. the Cont group. Protein
expression levels of (E) Smad3 and (F) Smad4 in hDPSCs following
transfection with siRNA targeting TAZ. Data are expressed as the
mean ± standard deviation of 3 independent experiments.
#P<0.05 vs. the siCON group. TAZ, transcriptional
coactivator with PDZ-binding motif; CTGF, connecting tissue growth
factor; Cyr61, cysteine-rich angiogenic inducer 61; hDPSC, human
dental pulp stem cell; r-hTHBS, recombinant human thrombospondin;
si, small interfering; Cont, control cells received no treatment;
siCON, negative control cells transfected with non-targeting siRNA;
siTAZ, cells transfected with siRNA targeting TAZ. |
Discussion
The potential applications of human stem cells
isolated from dental pulp in tissue regeneration are promising, as
hDPSCs may be easily isolated from discarded teeth following
extraction. The procedure is non-invasive and lacking ethical
concerns, whereas isolated cells may be stored for future use
(21,22). Furthermore, harvesting stem cells
from patients' dental tissue is a reasonable and simple alternative
to MSC harvesting (17).
Therefore, hDPSCs were used in the present study for in
vitro experiments.
TAZ is a transcriptional factor that has been
implicated in the development of various types of human tissue
(11,23). It serves critical roles in cellular
proliferation and apoptosis, as well as in the regulation of organ
size (24,25). Previous studies have reported that
TAZ overexpression enhanced the proliferation, migration,
transformation and epithelial-to-mesenchymal transition of
immortalized mammary epithelial cells; conversely, silencing TAZ
impaired the proliferative and migratory capabilities of mammary
epithelial cells (26,27). Notably, hDPSCs have been
demonstrated to express TAZ (17).
However, the effects of TAZ on the proliferation and migration of
hDPSCs have yet to be elucidated. The present study investigated
the roles of TAZ in hDPSCs. Cellular proliferation assays
demonstrated that following TAZ knockdown, hDPSC proliferation was
impaired. In addition, in vitro transwell and wound healing
assays revealed that following TAZ silencing, the migratory and
invasive capabilities of hDPSCs were markedly impaired. These
results suggested that TAZ may be involved in the regulation of
hDPSC proliferation and migration.
The implication of TAZ in processes of cellular
proliferation has previously been reported (28); however, the molecular mechanisms
underlying the role of TAZ in hDPSCs remain to be elucidated. CTGF
and Cyr61 have been identified to be downstream transcriptional
targets of TAZ, and have been reported to serve diverse roles in
several cellular processes, including development, differentiation,
proliferation, adhesion and migration, as well as angiogenesis and
tumorigenesis (20,29–31).
Furthermore, TAZ, through the activation of its target genes CTGF
and Cyr61, has been demonstrated to induce cellular differentiation
(32). In the present study,
RT-qPCR and western blot analysis revealed that CTGF and Cyr61 mRNA
and protein expression levels were downregulated following TAZ
silencing in hDPSCs. These results suggested that the mechanisms
underlying the actions of TAZ on hDPSC proliferation and migration
may involve the modulation of the expression of its downstream
target genes CTGF and Cyr61.
It has previously been reported that TGF-β-mediated
pathways are critical in the regulation of tumor formation and
invasion (33), whereas CTGF and
Cyr61 have been identified to be target genes in TGF-β signaling
(34). Smad3 and Smad4 have also
been reported to participate in TGF-β-mediated processes (28). The present results suggested that
the activity of TGF-β pathways was impaired following TAZ
depletion. In conclusion, the results of the present study
suggested that TAZ may regulate the proliferation and migration of
hDPSCs. In addition, TAZ appeared to exert its regulatory actions
on CTGF and Cyr61 expression through TGF-β-mediated pathways.
Therefore, it may be hypothesized that TAZ has potential to be a
target for tissue engineering applications.
Acknowledgements
The present study was supported by the Second
Hospital of Hebei Medical University (grant no. 2h1201504).
References
|
1
|
Karaöz E, Demircan PC, Sağlam O, Aksoy A,
Kaymaz F and Duruksu G: Human dental pulp stem cells demonstrate
better neural and epithelial stem cell properties than bone
marrow-derived mesenchymal stem cells. Histochem Cell Biol.
136:455–473. 2011. View Article : Google Scholar
|
|
2
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:pp. 13625–13630. 2000;
View Article : Google Scholar :
|
|
3
|
Liu P, Cai J, Dong D, Chen Y, Liu X, Wang
Y and Zhou Y: Effects of SOX2 on proliferation, migration and
adhesion of human dental pulp stem cells. PLoS One.
10:e01413462015. View Article : Google Scholar :
|
|
4
|
Dieterich LC, Huang H, Massena S,
Golenhofen N, Phillipson M and Dimberg A: αB-crystallin/HspB5
regulates endothelial-leukocyte interactions by enhancing
NF-κB-induced up-regulation of adhesion molecules ICAM-1, VCAM-1
and E-selectin. Angiogenesis. 16:975–983. 2013. View Article : Google Scholar :
|
|
5
|
Grudzinska MK, Kurzejamska E, Bojakowski
K, Soin J, Lehmann MH, Reinecke H, Murry CE, Soderberg-Naucler C
and Religa P: Monocyte chemoattractant protein 1-mediated migration
of mesenchymal stem cells is a source of intimal hyperplasia.
Arterioscler Thromb Vasc Biol. 33:1271–9. 2013. View Article : Google Scholar
|
|
6
|
Luo Z, Li D, Kohli MR, Yu Q, Kim S and He
WX: Effect of Biodentine™ on the proliferation, migration and
adhesion of human dental pulp stem cells. J Dent. 42:490–497. 2014.
View Article : Google Scholar
|
|
7
|
Tate MC, Garcia AJ, Keselowsky BG, Schumm
MA, Archer DR and LaPlaca MC: Specific beta1 integrins mediate
adhesion, migration and differentiation of neural progenitors
derived from the embryonic striatum. Mol Cell Neurosci. 27:22–31.
2004. View Article : Google Scholar
|
|
8
|
Yang D, Sun S, Wang Z, Zhu P, Yang Z and
Zhang B: Stromal cell-derived factor-1 receptor
CXCR4-overexpressing bone marrow mesenchymal stem cells accelerate
wound healing by migrating into skin injury areas. Cell Reprogram.
15:206–215. 2003.
|
|
9
|
Kanai F, Marignani PA, Sarbassova D, Yagi
R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC
and Yaffe MB: TAZ: A novel transcriptional co-activator regulated
by interactions with 14-3-3 and PDZ domain proteins. EMBO J.
19:6778–6791. 2000. View Article : Google Scholar :
|
|
10
|
Murakami M, Tominaga J, Makita R, Uchijima
Y, Kurihara Y, Nakagawa O, Asano T and Kurihara H: Transcriptional
activity of Pax3 is co-activated by TAZ. Biochem Biophys Res
Commun. 339:533–539. 2006. View Article : Google Scholar
|
|
11
|
Hong JH, Hwang ES, McManus MT, Amsterdam
A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp
PA, et al: TAZ, a transcriptional modulator of mesenchymal stem
cell differentiation. Science. 309:1074–1078. 2005. View Article : Google Scholar
|
|
12
|
Murakami M, Nakagawa M, Olson EN and
Nakagawa O: A WW domain protein TAZ is a critical coactivator for
TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc
Natl Acad Sci USA. 102:pp. 18034–18039. 2005; View Article : Google Scholar :
|
|
13
|
Park KS, Whitsett JA, Di Palma T, Hong JH,
Yaffe MB and Zannini M: TAZ interacts with TTF-1 and regulates
expression of surfactant protein-C. J Biol Chem. 279:17384–17390.
2004. View Article : Google Scholar
|
|
14
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar :
|
|
15
|
Hong JH and Yaffe MB: TAZ: A
beta-catenin-like molecule that regulates mesenchymal stem cell
differentiation. Cell Cycle. 5:176–179. 2006. View Article : Google Scholar
|
|
16
|
Jeong H, Bae S, An SY, Byun MR, Hwang JH,
Yaffe MB, Hong JH and Hwang ES: TAZ as a novel enhancer of
MyoD-mediated myogenic differentiation. FASEB J. 24:3310–3320.
2010. View Article : Google Scholar
|
|
17
|
Suh JS, Kim KS, Lee JY, Choi YJ, Chung CP
and Park YJ: A cell-permeable fusion protein for the mineralization
of human dental pulp stem cells. J Dent Res. 91:90–96. 2012.
View Article : Google Scholar
|
|
18
|
Xue P, Wu X, Zhou L, Ma H, Wang Y, Liu Y,
Ma J and Li Y: IGF1 promotes osteogenic differentiation of
mesenchymal stem cells derived from rat bone marrow by increasing
TAZ expression. Biochem Biophys Res Commun. 433:226–231. 2013.
View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Lai D, Ho KC, Hao Y and Yang X: Taxol
resistance in breast cancer cells is mediated by the hippo pathway
component TAZ and its downstream transcriptional targets Cyr61 and
CTGF. Cancer Res. 71:2728–2738. 2011. View Article : Google Scholar
|
|
21
|
Iohara K, Zheng L, Wake H, Ito M, Nabekura
J, Wakita H, Nakamura H, Into T, Matsushita K and Nakashima M: A
novel stem cell source for vasculogenesis in ischemia: Subfraction
of side population cells from dental pulp. Stem Cells.
26:N2408–N2418. 2008. View Article : Google Scholar
|
|
22
|
Nakamura S, Yamada Y, Katagiri W, Sugito
T, Ito K and Ueda M: Stem cell proliferation pathways comparison
between human exfoliated deciduous teeth and dental pulp stem cells
by gene expression profile from promising dental pulp. J Endod.
35:1536–1542. 2009. View Article : Google Scholar
|
|
23
|
Hong W and Guan KL: The YAP and TAZ
transcription co-activators: Key downstream effectors of the
mammalian Hippo pathway. Semin Cell Dev Biol. 23:785–793. 2012.
View Article : Google Scholar :
|
|
24
|
Wang K, Degerny C, Xu M and Yang XJ: YAP,
TAZ and Yorkie: A conserved family of signal-responsive
transcriptional coregulators in animal development and human
disease. Biochem Cell Biol. 87:77–91. 2009. View Article : Google Scholar
|
|
25
|
Zeng Q and Hong W: The emerging role of
the hippo pathway in cell contact inhibition, organ size control,
and cancer development in mammals. Cancer Cell. 13:188–192. 2008.
View Article : Google Scholar
|
|
26
|
Chan SW, Lim CJ, Guo K, Ng CP, Lee I,
Hunziker W, Zeng Q and Hong W: A role for TAZ in migration,
invasion, and tumorigenesis of breast cancer cells. Cancer Res.
68:2592–2598. 2008. View Article : Google Scholar
|
|
27
|
Lei QY, Zhang H, Zhao B, Zha ZY, Bai F,
Pei XH, Zhao S, Xiong Y and Guan KL: TAZ promotes cell
proliferation and epithelial-mesenchymal transition and is
inhibited by the hippo pathway. Mol Cell Biol. 28:2426–2436. 2008.
View Article : Google Scholar :
|
|
28
|
Wang Q, Xu Z, An Q, Jiang D, Wang L, Liang
B and Li Z: TAZ promotes epithelial to mesenchymal transition via
the upregulation of connective tissue growth factor expression in
neuroblastoma cells. Mol Med Rep. 11:982–988. 2015.
|
|
29
|
Dhar A and Ray A: The CCN family proteins
in carcinogenesis. Exp Oncol. 32:2–9. 2010.
|
|
30
|
Leask A and Abraham DJ: All in the CCN
family: Essential matricellular signaling modulators emerge from
the bunker. J Cell Sci. 119:4803–4810. 2006. View Article : Google Scholar
|
|
31
|
Leivonen SK and Kähäri VM: Transforming
growth factor-beta signaling in cancer invasion and metastasis. Int
J Cancer. 121:2119–2124. 2007. View Article : Google Scholar
|
|
32
|
Kim KM, Choi YJ, Hwang JH, Kim AR, Cho HJ,
Hwang ES, Park JY, Lee SH and Hong JH: Shear stress induced by an
interstitial level of slow flow increases the osteogenic
differentiation of mesenchymal stem cells through TAZ activation.
PLoS One. 9:e924272014. View Article : Google Scholar :
|
|
33
|
Labbé E, Lock L, Letamendia A, Gorska AE,
Gryfe R, Gallinger S, Moses HL and Attisano L: Transcriptional
cooperation between the transforming growth factor-beta and Wnt
pathways in mammary and intestinal tumorigenesis. Cancer Res.
67:75–84. 2007. View Article : Google Scholar
|
|
34
|
Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du
ZP, Lv Z, Gu W, Pan F, Xu XE, et al: Involvement of CYR61 and CTGF
in the fascin-mediated proliferation and invasiveness of esophageal
squamouscell cell carcinomas cells. Am J Pathol. 76:939–951. 2010.
View Article : Google Scholar
|