Introduction
Hepatocellular carcinoma (HCC) is the fifth most
prevalent malignancy worldwide and the second most common cause of
cancer-associated mortality in eastern and western countries
(1,2). Each year, >700,000 patientcare
diagnosed with HCC and morbidity rates of HCC continue to increase,
possibly due to deterioration of environment, unhealthy dietary
habits and other associated factors (3). The traditional therapeutic strategy
for HCC is surgical resection, however, this is only able to cure
patients at an early stage of the disease (4). For patients in the late stage, the
introduction of sorafenib can improve survival rates, but remains
limited (5). Despite substantial
progression in therapeutics in previous decades, the prevention and
treatment of HCC remains challenging. Thus, the identification of
novel molecular markers to diagnose HCC at an early stage is of
high priority.
Long non-coding RNAs (LncRNAs) are well-defined as a
class of RNAs with a length of >200 nucleotides and <100,000
nucleotides, which are unable to translate proteins (6). LncRNAs can be classified into five
categories by their locations at nearby genes: Sense, antisense,
bidirectional, intronic and intergenic (7). LncRNAs have been demonstrated to be
involved in various biological processes, including chromatin
modification, transcription, translation, posttranscriptional
processing and posttranslational processing (8,9). Of
note, studies have shown that certain LncRNAs can function as
oncogenes or tumor suppressors, and that aberrant expression of
LncRNAs may contribute to tumorigenesis (10,11).
Emerging evidence has also linked LncRNAs with
multiple physiological and pathological processes of HCC, including
cell proliferation, metastasis, apoptosis and metabolism (12). For example, LncRNA AFAP1-AS1
functions to promote cell proliferation and metastasis, and
upregulates the RhoA/Rac2 signing pathway in HCC (13). LncRNA DILC has been shown to
repress the self-renewal of liver cancer stem cells by inhibiting
the autocrine interleukin-6/signal transducer and activator of
transcription 3 axis (14). Thus,
LncRNAs have been recognized as potential markers in the
progression of HCC.
In the present study, the role of LncRNA AB019562 in
cell proliferation and metastasis was examined in HCC. A specific
small interfering (si)RNA against AB019562 was used to decrease the
expression of AB019562 in HCC cell lines. The effects of AB019562
depletion on cell proliferation, metastasis and apoptosis were
examined using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay, transwell assay and apoptosis detection kit, respectively.
The results indicated the pro-oncogenic property of AB019562 in HCC
and may provide novel clues for the early clinical diagnosis and
treatment of HCC.
Materials and methods
Human samples
Clinical HCC tissue samples and samples of the
adjacent non-cancerous tissues were obtained from 50 patients who
had undergone surgical resection of HCC in the Department of
Geriatrics, Tianjin Medical University General Hospital (Tianjin,
China) between July 2012 and June 2015, in a non-identifiable
manner and without additional clinical information. Prior to
surgery, no chemotherapy or radiotherapy treatment was administered
to the patients. All patients confirmed their full intention to be
involved in the study and written informed consent was obtained.
The present study was approved by the Research Ethics Committee of
Tianjin Medical University General Hospital. All collected samples
were immediately frozen in liquid nitrogen and stored at −70°C
until RNA isolation.
Cell culture
The SMMC-7721, PLC/PRF/5 and C3AHCC cell lines, and
the normal THLE-3 liver cell line were all commercially obtained
from American Type Culture Collection (Manassas, VA, USA). The
HepG2 HCC cell line was purchased from Shanghai Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). All cells were
cultured in Eagle's minimum essential medium (EMEM; American Type
Culture Collection) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). During
the experiments, the cells were maintained in an incubator
containing 5% CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
RNAs from the human samples and cultured cells were
extracted using TRIzol reagent (Takara Biotechnology, Co., Ltd.,
Dalian, China) according to the manufacturer's protocol. The
isolated RNAs were eluted with ribonuclease-free water and
quantified using a Nanodrop 2000 spectrophotometer (Invitrogen;
Thermo Fisher Scientific, Inc.) by detecting the optical density
(OD)260 and OD280. Complementary DNA (cDNA)
was reverse transcribed from a total of 1ug RNAs using PrimeScript
Reverse Transcriptase (Takara Biotechnology, Co., Ltd.) with a
volume of 20 µl, as per the manufacturer's instructions. qPCR was
performed using SYBR Green reagent (Takara Biotechnology, Inc.) in
an ABI 7900 machine (Applied Biosystems, Foster City, CA, USA) and
a total of 1 µl cDNA was added into each well. The thermocycling
conditions were as follows: 95°C for 5 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 30 sec. The following primers were
used in the reaction, as described previously (15): AB019562, forward
5′-GGATGTCAGGTCTGCGAAACT-3′ and reverse
5′-GATAGTGTGGTTTATGGACTGAGGT-3′; GAPDH, forward
5′-GGGAAACTGTGGCGTGAT-3′ and reverse 5′-GAGTGGGTGTCGCTGTTGA-3′. The
RT-qPCR analysis was performed as previously described (15).
siRNA interference
The specific siRNA against LncRNA AB019562
(siAB019562) was designed and synthesized by GenePharm Co., Ltd.
(Shanghai, China). The siRNAs were dissolved in ddH2O at
a concentration of 20 µM. The negative control siRNA and the
specific siAB019562 were transfected into cells using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Briefly, the cells were washed with
pre-warmed PBS and replaced with serum-free medium. The mixture of
Lipofectamine 2000 and siRNAs was incubated for 20 min and then
added to each well containing cells at a confluence of 60%. Cells
were incubated at 37°C for 72 h post-transfection, and then used
for subsequent analyses.
Cell viability assay
An MTT assay (Promega Corporation, Madison, WI, USA)
was used to measure cell proliferative abilities, according to the
manufacturer's protocol. Briefly, the HepG2 and SMMC-7721 cells, at
an initial concentration of 2×103/well, were seeded into
96-well plates in EMEM supplemented with 10% FBS in a 37°C
humidified incubator with 5% CO2. Each experimental
group of cells was divided into six wells and the cultural medium
was refreshed every other day. At each time point (days 1, 2, 3, 4
and 5 post-transfection), the cell numbers were determined by
detecting the absorbance at 450 nm on a Tecan microplate reader
(Tecan Group, Ltd., Männedorf, Switzerland). Proliferation rate was
plotted as a ratio relative to day 1.
Cell cycle analysis
The cells were seeded into 96-well plates and
incubated for 24 h, following which the siRNAs were transfected
into the corresponding group. At 72 h post-treatment, the HepG2 and
SMMC-7721 cells were harvested and co-incubated with pre-cooled
ethanol overnight. On the subsequent day, the cells
(3×104 cells) were treated with 50 µg/ml of RNase in a
37°C incubator for 1 h, following which 20 µg/ml of propidium
iodide (PI) was added to the cells. The mixture was maintained at
4°C for an additional 30 min in the absence of light. The DNA
content was detected using flow cytometry.
Transwell assay
The HepG2 and SMMC-7721 cells were cultured in
24-well plates and transfected with siRNAs. At 48 h
post-transfection, the cells were harvested with serum-free EMEM,
following which 150 µl of cell suspension (3×104 cells)
was added to the upper chamber (Corning Incorporated, New York, NY,
USA), and the lower chamber was filled with 600 µl culture medium
containing 10% FBS. For the invasion assay, the membrane was
pre-coated with Matrigel (Corning Incorporated) for 6 h at 37°C
prior to the experiments. Following incubation for 12 h at 37°C,
the cells in each group were fixed with ice-cold methanol for 10
min and stained with crystal violet for 5 min. Images were captured
under an inverted light microscope (TS100; Nikon Corporation,
Tokyo, Japan).
Detection of apoptosis
Flow cytometry was used to determine the apoptotic
rates of cells following siAB019562 treatment. Briefly, the HepG2
and SMMC-7721 cells were harvested with low-speed centrifugation
(850 g, 4°C, 5 min) and washed three times with PBS. The cells were
then suspended in 100 µl staining buffer with 5 µl of Annexin V-APC
(20 µg/ml) and 5 µl PI (50 µg/ml; BD Pharmingen, San Diego, CA,
USA). The cells found to be Annexin V-APC-positive and PI-negative
were identified as early apoptotic cells, and those positive for
Annexin V-APC and PI were identified as late apoptotic cells. Each
experiment was repeated at least 3 times in triplicate.
Determination of relative caspase
activity
The activities of caspase-3 and caspase-8 were
determined using a Caspase-3 activity kit and a Caspase-8 activity
kit, respectively, according to the manufacturer's protocol
(Beyotime Institute of Biotechnology, Nantong, China). Briefly,
following siRNA transfection, 2×106 cells in 6-well
plates were lysed using lysis buffer (as provided in the kit) at
4°C for 15 min, centrifuged at 600 × g at 4°C for 15 min, and the
resulting cell lysates were analyzed for protein concentration by
bicinchoninic assay (Beyotime Institute of Biotechnology). An
aliquot of 10 µl proteins from the cell lysates were added to
96-well plates and mixed with 80 µl reaction buffer containing
caspase substrate (2 mM). Following incubation for 4 h at 37°C,
caspase activities were determined using a Tecan microplate reader
at an absorbance of 405 nm.
Statistical analysis
All data are presented as the mean ± standard
deviation. Each experiment was repeated at least 3 times in
triplicate unless otherwise stated. Statistical differences between
two groups were evaluated by Student's t-test using GraphPad Prism
version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
LncRNA AB019652 is upregulated in HCC
tissues and cultured HCC cells
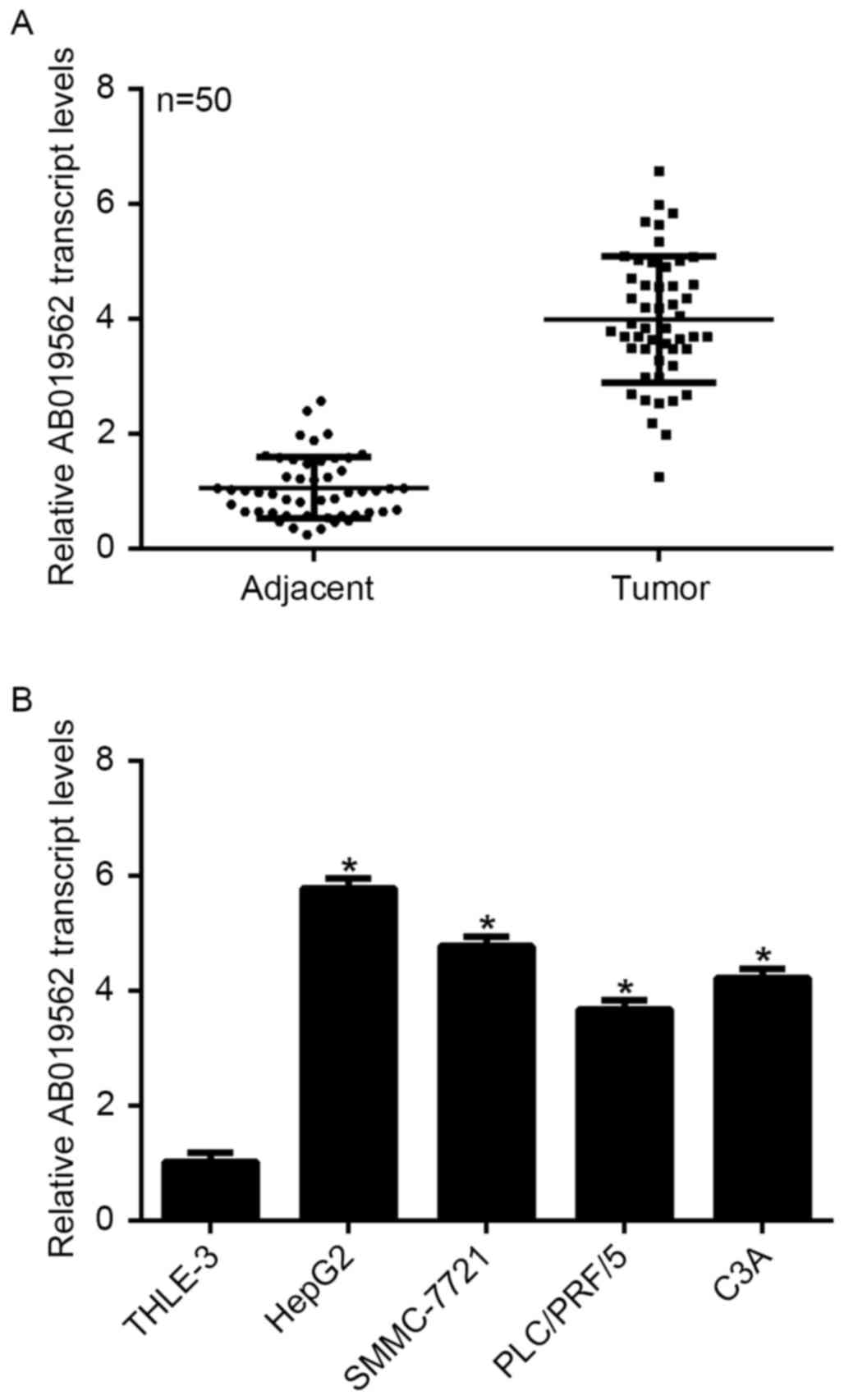
To examine the expression of LncRNA AB019562 in HCC,
tissue samples from 50 patients with HCC, including cancerous
tissues and adjacent non-cancerous tissues, were obtained. As shown
in Fig. 1A, the transcription
level of AB019562 was 4-fold higher in the HCC samples, compared
with that in the adjacent the non-cancerous samples (P<0.05).
The four HCC cell lines and a normal liver cell line were examined
to determine the transcription of AB019562 in vitro. The
results showed that, compared with the THLE-3 normal liver cells,
HepG2 cells exhibited the highest level of AB019562 (6-fold
increase), followed by the SMMC-7721 cells (5-fold increase). The
transcription of AB019562 was also significantly increased in the
PLC/PRF/5 and C3A cells (Fig. 1B).
The HepG2 and SMMC-7721 cells were selected for subsequent
analyses. These data suggested that AB019562 was significantly
overexpressed in HCC in vivo and in vitro.
Knockdown of AB019562 inhibits cell
proliferation in HCC cells
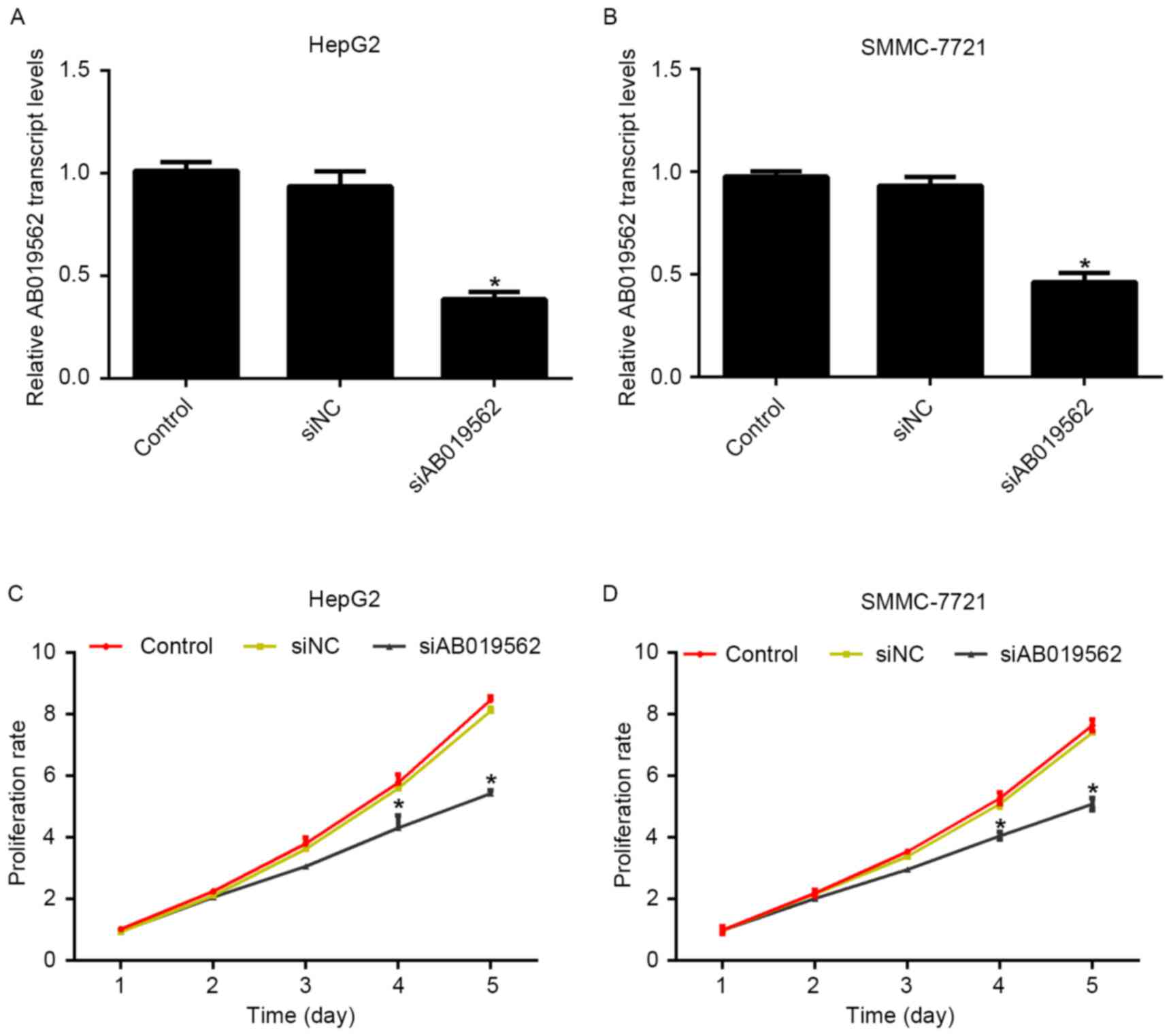
To evaluate the role of AB019562 in HCC cell
proliferation, specific siRNA against AB019562 (siAB019562) was
designed and transfected into the HepG2 and SMMC-7721 cells. The
transcription levels of AB019562 were decreased by 58% in HepG2
cells (Fig. 2A) and 51% in
SMMC-7721 cells (Fig. 2B)
following transfection with siAB019562, which suggested the high
efficiency of the synthesized siRNA. With this effective siRNA,
cell proliferative rates were examined following siRNA transfection
using MTT assays. For the first 3 days post-siRNA treatment, no
significant changes in the viability of the two cell lines were
observed, compared with the control cells. However, on day 4
post-AB019562 knockdown, the proliferation rates were decreased by
28.6% in the HepG2 cells and 24%in the SMMC-7721. This inhibitory
effect was more pronounced on day 5 following AB019562 knockdown in
the HCC cells (Fig. 2C and D).
These results suggested that AB019562 promoted cell proliferation
in the HCC cells in vitro.
Knockdown of AB019562 arrests cell
cycle at the G0/G1 phase in HCC cells
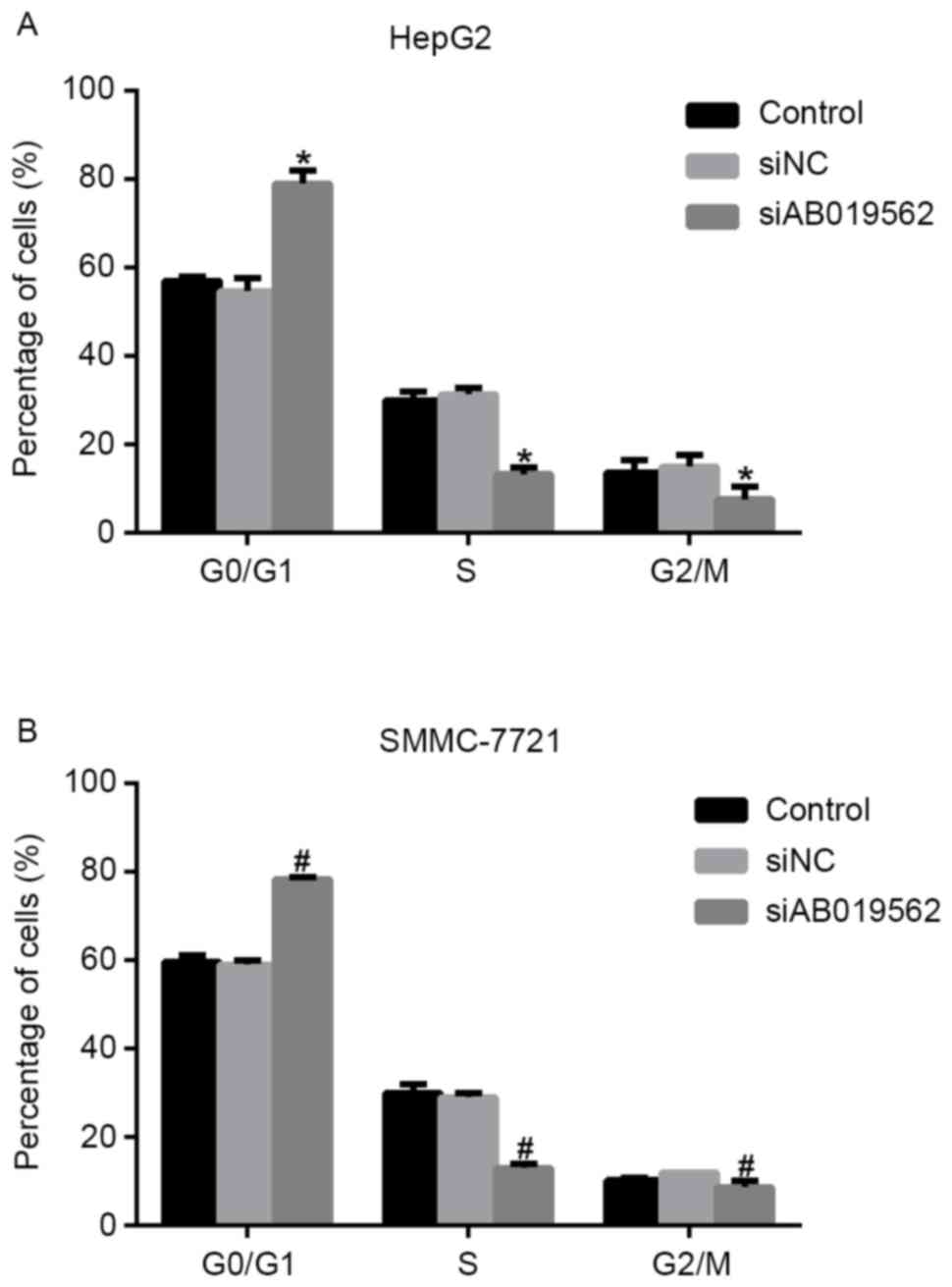
The present study subsequently analyzed cell cycle
progression. As shown in Fig. 3A,
when AB019562 was knocked down in the HepG2 cells, the proportion
of cells in the G0/G1 phase increased by 22%, whereas the
proportions of cells in the S phase and G2/M phase decreased by 14
and 8%, respectively. Similarly, in the siAB019562-transfected
SMMC-7721 cells, the percentage of cells in the G0/G1 phase was
elevated by 18%, whereas the percentages of cells in the S phase
and G2/M phase decreased 12 and 9%, respectively (Fig. 3B). These data suggested that the
knockdown of LncRNA AB019562 arrested cell cycle in the G0/G1 phase
in HCC cell lines. The cell cycle arrest following AB019562
knockdown provided further support that AB019562 promoted the
proliferation of HCC cells.
Knockdown of AB019562 suppresses the
metastasis of HCC cells
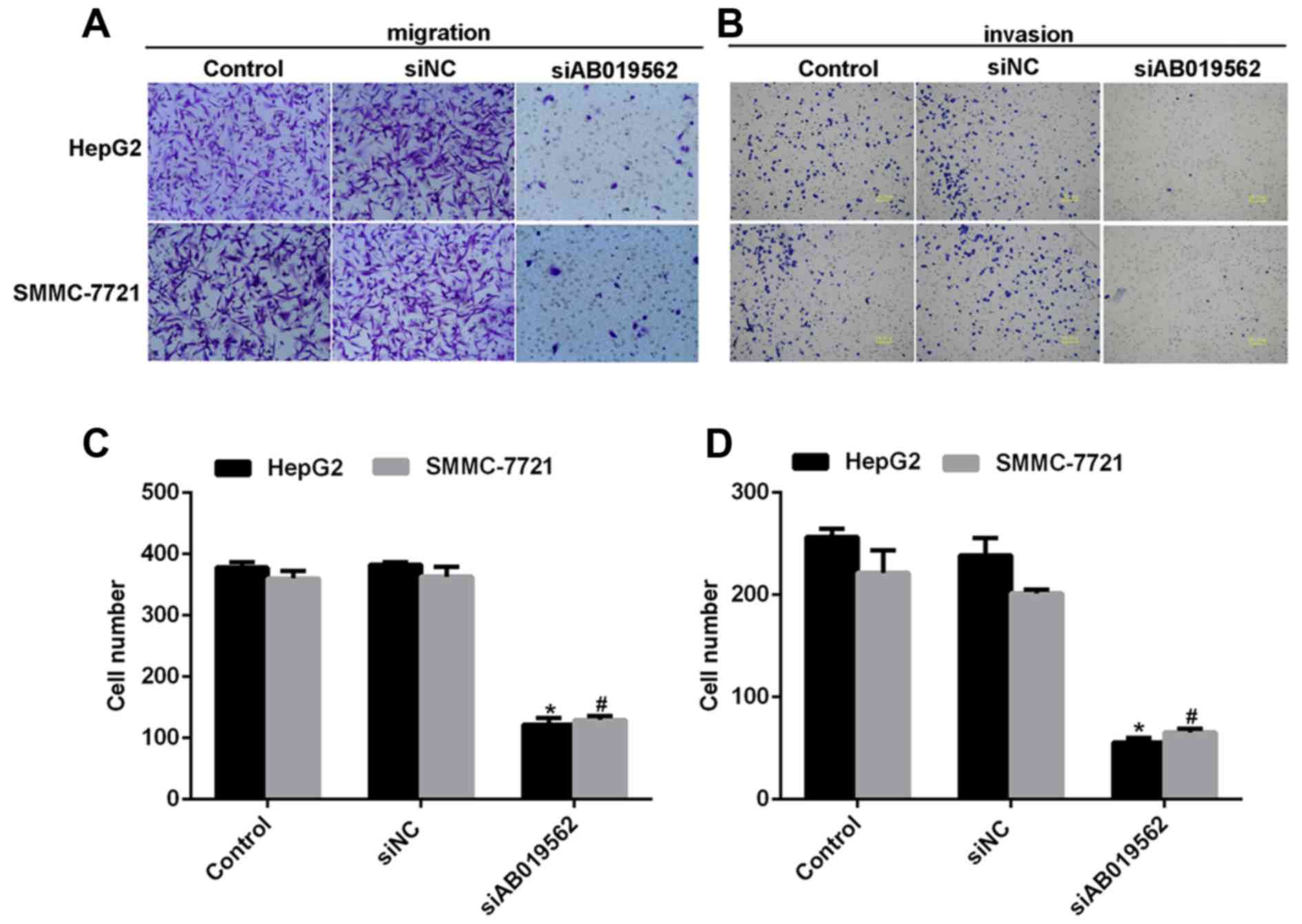
Subsequently, a transwell assay was performed to
examine the effects of AB019562 knockdown on cell migration
(Fig. 4A) and invasive abilities
(Fig. 4B) in the HCC cell lines.
The results showed that transmigrated cells were visually decreased
when the cells were transfected with siAB019562 (Fig. 4A and B). Quantification of the
transmigrated cells further supported that migration abilities were
reduced by siAB019562 transfection, with decreases by up to 67% in
HepG2 cells and 63% in SMMC-7721 cells (Fig. 4C). Similarly, it was also shown
that 75% of HepG2 cells and 60% of SMMC-7721 cells were inhibited
from invading through the Matrigel upon siAB019562 treatment in the
invasion assay (Fig. 4D). These
data suggested that AB019562 promoted cell migration and invasion
in the HCC cells in vitro.
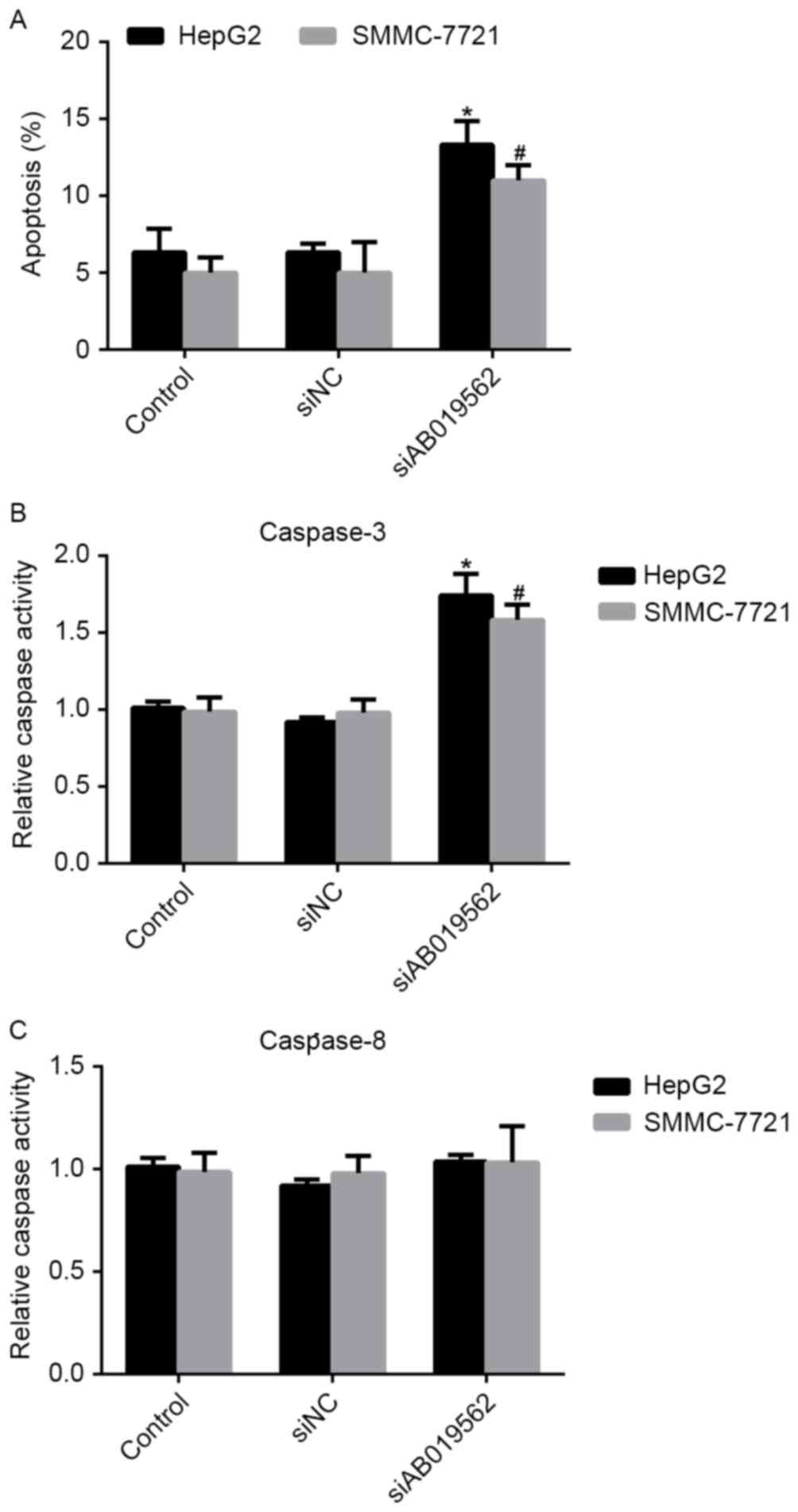
Knockdown of AB019562 induces cell
apoptosis and affects the activity of caspase-3 in HCC
Cell apoptosis has been demonstrated to be important
in the tumorigenesis of HCC (16).
Therefore, the present study examined the effects of AB019562
knockdown on the apoptosis of HCC cells. It was shown that,
compared with the control cells, the percentages of apoptotic HepG2
and SMMC-7721 cells were increased by 8 and 7%, respectively,
following AB019562 knockdown (Fig.
5A). The activities of caspase-3 and caspase-8 in the HCC cells
were also examined. As shown in Fig.
5B, AB019562 knockdown increased the activity of caspase-3 by
60% in the HepG2 cells and 50% in the SMMC-7721 cells. However, the
activity of caspase-8 remained unchanged, even when cells were
transfected with siAB019562 (Fig.
5C). Taken together, these results confirmed that the knockdown
of AB019562 induced cell apoptosis and promoted the activity of
caspase-3, but not caspase-8.
Discussion
HCC is frequently one of the top five causes of
liver cirrhosis. However, the early diagnosis of HCC remains
challenging (17,18). The primary treatment for HCC is
surgery, which accounts for a five-year survival rate of 30–70%
(19,20). However, <20% of patients with
HCC are eligible for surgery following confirmation of diagnosis
(21), indicating the necessity of
diagnosing HCC at an early stage. The most refractory limitation in
the early detection and prevention of HCC is the absence of
symptoms (22). Therefore, an
increasing number of studies have focused on identifying novel
biomarkers, which can be used to diagnose and prevent HCC in the
early stage (23,24).
LncRNAs are distinct from housekeeping RNAs,
including tRNAs, rRNAs and siRNAs (25). They are important in various
aspects of cell biology. The aberrant expression of LncRNAs is a
basic feature of several types of cancer, and has been shown to
affect cell proliferation, metastasis and apoptosis in human cancer
(26,27). The LncRNA AB019562 was first
identified by Zhou et al using gene microarray analysis
(28). In this pioneer study,
AB019562 was shown to be upregulated in human hypopharyngeal
squamous cell carcinoma. However, the role of AB019562 in HCC and
the detailed mechanisms underlying how AB019562 regulates the
tumorigenesis of HCC remain to be fully elucidated.
In the present study, the transcription levels of
AB019562 were determined in HCC tissues and in a series of HCC cell
lines. It was shown that the expression of AB019562 was markedly
upregulated in HCC. Furthermore, it was observed that the knockdown
of AB019562 significantly reduced the rate of cell proliferation
and arrested cell cycle at the G0/G1 phase, suggesting the
promotion of proliferation by AB019562. The induction of cell
apoptosis by AB019562 knockdown further confirmed that AB019562
functioned to promote cell proliferation in HCC, as the induction
of apoptosis is a sound basis for the inhibition of proliferation
(16). In addition, the knockdown
of AB019562 impaired cell migration and invasion abilities in the
HCC cell lines. These data demonstrated that AB019562 promoted cell
proliferation and metastasis in HCC.
However, whether the intrinsic or extrinsic
apoptotic signal pathway predominantly contributes to the
AB019562-mediated biological changes remains to be elucidated. The
induction of apoptosis usually has two signaling pathways, the
intrinsic and extrinsic pathways (29). The initiation of the intrinsic
pathway is associated with the pro-apoptotic factors, B-cell
lymphoma 2 (Bcl-2)-associated X protein and Bcl-2-associated death
promoter, which leads to increased permeability of the mitochondria
membrane, loss of membrane potential and the release of cytochrome
C into the cytosol. The intrinsic pathway is associated with
activated caspase-3, whereas the extrinsic pathway is associated
with the activation of caspase-8 (30). As shown in Fig. 5C, the activities of caspase-8 were
stable upon siAB019562 administration, which indicated that the
extrinsic pathway may not be critically involved. Instead, the
relative activities of caspase-3 were markedly increased following
AB019562 knockdown in HepG2 and SMMC-7721 cells. This observation
indicated that the intrinsic pathway maybe involved in the
induction of apoptosis by siAB019562 transfection. However, further
investigations are required to systemically reveal the detailed
mechanisms.
In conclusion, the present study examined the role
of LncRNA AB019562 in human HCC in vivo and in vitro.
AB019562 was expressed at high levels in patients with HCC and
cultured HCC cells. The knockdown of AB019562 caused cell cycle
arrest at the G0/G1 phase, leading to eventual cell apoptosis and
thereby inhibiting the proliferation of HCC cells. Furthermore, the
knockdown of AB019562 impaired cell migration and invasion of the
HepG2 and SMMC-7721 cells. These data suggested that AB019562 may
promote cell proliferation and metastasis in HCC, and provided
evidence that AB019562 may serve as a novel biomarker and
therapeutic target for the diagnosis and clinical treatment of
HCC.
Acknowledgements
This study was sponsored by National Natural Science
Foundation of China (grant nos. 81670086 and 81370183), Tianjin
Natural Science Foundation (grant no. 14JCYBJC27800) and
International S&T Cooperation Program of China (grant no.
2015DFA50310).
References
|
1
|
Laursen L: A preventable cancer. Nature.
516:(Suppl). S2–S3. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park YN: Update on precursor and early
lesions of hepatocellular carcinomas. Arch Pathol Lab Med.
135:704–715. 2011.PubMed/NCBI
|
|
3
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Worns MA and Galle PR: HCC
therapies-lessons learned. Nat Rev Gastroenterol Hepatol.
11:447–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang JW, Jiang R, Deng L, Zhang X, Wang K
and Sun B: Circulation long non-coding RNAs act as biomarkers for
predicting tumorigenesis and metastasis in hepatocellular
carcinoma. Oncotarget. 6:4505–4515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: A new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
10
|
Guo XQ, Xia J and Deng K: Long non-coding
RNAs: Emerging players in gastric cancer. Tumor Biol.
35:10591–10600. 2014. View Article : Google Scholar
|
|
11
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q,
Wu JY, Qin J, Jin T and Xu JM: Long noncoding RNA AFAP1-AS1
indicates a poor prognosis of hepatocellular carcinoma and promotes
cell proliferation and invasion via upregulation of the RhoA/Rac2
signaling. Int J Oncol. 48:1590–1598. 2016.PubMed/NCBI
|
|
14
|
Wang X, Sun W, Shen W, Xia M, Chen C,
Xiang D, Ning B, Cui X, Li H, Li X, et al: Long non-coding RNA DILC
represses self-renewal of liver cancer stem cells via inhibiting
autocrine IL-6/STAT3 axis. J Hepatol. 64:1283–1294. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou J, Li M, Yu W, Li W, Wang J, Xiang X,
Li G, Pan X and Lei D: AB209630, a long non-coding RNA decreased
expression in hypopharyngeal squamous cell carcinoma, influences
proliferation, invasion, metastasis, and survival. Oncotarget.
7:14628–14638. 2016.PubMed/NCBI
|
|
16
|
He R, Yang L, Lin X, Chen X, Lin X, Wei F,
Liang X, Luo Y, Wu Y, Gan T, et al: MiR-30a-5p suppresses cell
growth and enhances apoptosis of hepatocellular carcinoma cells via
targeting AEG-1. Int J Clin Exp Pathol. 8:15632–15641.
2015.PubMed/NCBI
|
|
17
|
Eggert T, McGlynn KA, Duffy A, Manns MP,
Greten TF and Altekruse SF: Epidemiology of fibrolamellar
hepatocellular carcinoma in the USA, 2000–10. Gut. 62:1667–1668.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
El-Serag HB: Epidemiology of
hepatocellular carcinoma in USA. Hepatol Res. 37:(Suppl 2).
S88–S94. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
European Association For The Study Of The
Liver; European Organisation For Research And Treatment Of Cancer:
ASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abdo AA, Hassanain M, AlJumah A, Al Olayan
A, Sanai FM, Alsuhaibani HA, Abdulkareem H, Abdallah K, AlMuaikeel
M, Al Saghier M, et al: Saudi guidelines for the diagnosis and
management of hepatocellular carcinoma: Technical review and
practice guidelines. Ann Saudi Med. 32:174–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
European Association for Study of Liver;
European Organisation for Research and Treatment of Cancer:
EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. Eur J Cancer. 48:599–641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chatterjee R and Mitra A: An overview of
effective therapies and recent advances in biomarkers for chronic
liver diseases and associated liver cancer. Int Immunopharmacol.
24:335–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma X, Hui H, Jin Y, Dong D, Liang X, Yang
X, Tan K, Dai Z, Cheng Z and Tian J: Enhanced immunotherapy of
SM5-1 in hepatocellular carcinoma by conjugating with gold
nanoparticles and its in vivo bioluminescence tomographic
evaluation. Biomaterials. 87:46–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jaganathan A, Murugan K, Panneerselvam C,
Madhiyazhagan P, Dinesh D, Vadivalagan C, Aziz AT, Chandramohan B,
Suresh U, Rajaganesh R, et al: Earthworm-mediated synthesis of
silver nanoparticles: A potent tool against hepatocellular
carcinoma, Plasmodium falciparum parasites and malaria mosquitoes.
Parasitol Int. 65:276–284. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sanchez Y and Huarte M: Long non-coding
RNAs: Challenges for diagnosis and therapies. Nucleic Acid Ther.
23:15–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou JY, Li W, Jin T, Xiang X, Li M, Wang
J, Li G, Pan X and Lei D: Gene microarray analysis of lncRNA and
mRNA expression profiles in patients with hypopharyngeal squamous
cell carcinoma. Int J Clin Exp Med. 8:4862–4882. 2015.PubMed/NCBI
|
|
29
|
Spencer SL and Sorger PK: Measuring and
modeling apoptosis in single cells. Cell. 144:926–939. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du Y, Gong J, Tian X, Yan X, Guo T, Huang
M, Zhang B, Hu X, Liu H, Wang Y, et al: Japonicone A inhibits the
growth of non-small cell lung cancer cells via
mitochondria-mediated pathways. Tumor Biol. 36:7473–7482. 2015.
View Article : Google Scholar
|