Introduction
Glioblastoma (GBM) is the most common and aggressive
form of human primary brain malignancy, with ~17,000 diagnoses/year
(1). GBM is associated with poor
prognosis and a lack of effective therapeutic options, due to the
impracticality of extensive tumor resection and poor drug delivery
in the brain (2). Recently,
combined chemoradiation, using concomitant and adjuvant
temozolomide and radiotherapy, has been used with a modest degree
of efficacy (3). High-throughput
genomic data derived from patients with GBM are readily-available
for analyses aiming to identify biomarkers for measuring
therapeutic efficacy (4).
Identification of a target gene in tumors is often
complicated by broad genomic copy number aberrations, multiple
mechanisms of activating and inactivating genetic and epigenetic
alterations, and the complexity of pathway regulation (5). Target-gene identification may be
confounded by contaminating non-tumor cells or the molecular
heterogeneity of the tumor (6).
Genes do not act in isolation; it is necessary to elucidate and
annotate all functional interactions among genes in the cell to
understand cellular functions (7).
Therefore, identifying pathways or modules may be a useful and
reliable approach, and is a good choice to elucidate the
pathological mechanism of GBM. As genes in the same pathway tend to
exhibit correlated expression, analyzing the co-expression network
is an effective strategy. In the present study, multiple
differential co-expression networks (DCN) were applied to
investigate multiple differential modules of GBM in radiotherapy
and chemoradiation conditions.
Pathway dynamics may be attributed to alterations in
gene expression and connectivity among genes (i.e. pathway
rewiring). The latter has previously been demonstrated to serve a
role in disease progression and treatment responses (8,9).
However, few studies have focused on pathway dynamics in the
treatment of GBM. Therefore, the present study aimed to detect
dynamic differential modules across radiotherapy and chemoradiation
conditions in GBM. Dynamic alterations and connectivity were
evaluated using the module connectivity dynamic score (MCDS).
Dynamic modules were identified, which provided insights into the
molecular mechanisms of treatment for GBM.
Materials and methods
Gene expression profile
In the present study, the gene expression profile
E-GEOD-7696 (4,10) for GBM and normal samples was
obtained from the ArrayExpress database (ebi.ac.uk/arrayexpress). E-GEOD-7696 was composed of
four healthy samples, 28 GBM samples treated with radiotherapy, and
52 GBM samples treated with chemoradiation. In order to control the
quality of the data, standard data preprocessing was performed,
consisting of background correction using the Robust Multi-array
Average algorithm (11),
normalization based on the quantiles method (12) and probe match using the Micro Array
Suite algorithm (13). Each probe
ID was mapped to the gene symbol using the annotate package
(14) and screened using the
feature filter method to discard duplicated genes (15). A total of 20,545 genes were
identified for subsequent analysis.
Construction of multiple DCNs
For each condition, DCN construction consisted of
two steps: Constructing a binary co-expression network (BCN), and
assigning a weight to each edge in the BCN based on differential
gene expression between each treatment condition and normal
condition.
BCN
A protein-protein interaction (PPI) network was
extracted from the human STRING database (16), which was composed of 787,896
interactions among 16,730 genes. By taking intersections with the
gene expression data, a sub-PPI network was generated, including
15,130 genes and 725,216 interactions. In order to evaluate the
co-expression probability of gene pairs, the Pearson correlation
coefficient (PCC) (17) was
implemented to assess the edge scores in the PPI network, and the
absolute value of PCC for an interaction was denoted to be Δ. Genes
with interactions of Δ≥0.8 were selected to build the BCN. In order
to facilitate the analysis and comparison among different
conditions, the intersected interactions with Δ≥0.8 in the
different conditions were selected. For the BCNs in different
conditions, the genes were the same although the number of edges
and their scores were different.
Weight assignments
A weight value was assigned to each edge in the BCN
based on the differential gene expression between each treatment
and the normal condition. Prior to weight assignment, the q
value of differential gene expression between each treatment
condition (radiotherapy and chemoradiation) and the normal
condition was calculated using a one-sided t-test (18). A weight was subsequently assigned
to each edge based on the q value. The weight
Wi,j on edge (i, j) in the
co-expression network was defined as follows:
Wi,j={(logqi+logqj)1/2(2*maxl∈s|logql|)1/2ifPCC(i,j)≥Δ0,ifPCC(i,j)<Δ
Where qi and qj
stood for the q value for genes i and j,
respectively; S was the node set of the co-expression
network; and PCC (i, j) represented the
absolute value of the PCC between genes i and j based
on their expression profiles.
Following the BCN and weight assignment steps,
higher weight values were assigned to the co-expressed and
significantly differentially-expressed genes. A total of two
specific DCNs were identified in GBM, one for the radiotherapy
condition and one for the chemoradiation condition. Mathematically,
for the two DCNs, the node sets were the same, although the edge
sets were different due to the different weights.
Extraction of multiple differential
modules from multiple DCNs
The multiple differential module algorithm was based
on seed gene expansion and significance analysis, consistent with a
previous study identifying gene modules across multiple
co-expression networks (19). The
schema consisted of four steps: Seed selection; module search by
seed expansion; refinement of candidate modules; and significance
analysis for candidate modules.
Seed selection
The seed selection step ranked genes in two specific
DCNs according to the topological feature of the gene in the
network. For each DCN, G=(S,T), the adjacency matrix was
expressed as A=(aij). A function, g, was used
and the importance of gene i in the corresponding network
was assessed, g(i) (20).
g(i)=(1–Aij′)–1
And
Aij′=D–1/2AijD1/2
Where Aij stood for the degree
normalized weighted adjacency matrix; D was a diagonal
matrix with element Dij=∑jAij,
j belonging to N(i), which was the set of
neighbors of i in G. The equation indicated that the
importance of a node depended on the number of neighbors, the
strength of the connections and the importance of the neighbors.
Subsequent to calculating the ranks of a node in two individual
networks, a z-score for each rank was computed (21). The gene rank was obtained by
averaging the z-scores across two DCNs and the top 10% of
genes were selected as seed genes.
Module search by seed expansion
A graph entropy-based objective function (Δ
E) was used to assess the scale of a module search starting
with a seed gene (22). The seed
gene (x) was taken as a module O={ x}, and the
new candidate module as O'= O ∪{ y}. For each
vertex y in its neighborhood in all networks, N(x) =U
iNi(x), y є N(x)
was defined, in which N i (x) was the neighbor set in
Gi as the candidate for M. The entropy
decrease between O and O' was evaluated as
follows:
ΔE(O′,O)=E(O)–E(O′)
of which
E(O)=∑k=1MEk(O)|O|
Where Ek(O)=∑i∈OE(Oj) was the sum of all the
vertices in O and network k; and Oi
(1≤ i≤τ) was the group of modules being sought where τ was
the number of modules. E(O') was calculated
similarly. Δ E(O', O)>0 indicated that the
addition of vertex y improved the connectivity of the former
candidate modules. The vertex y whose addition maximized Δ
E was added to O. The searching process was not
terminated until there was no decrease subsequent to adding genes
iteratively, leading to the maximum decrease in Δ E.
Following this process, candidate modules were identified in two
DCNs.
Refinement of candidate modules
During this step, candidate modules with a gene size
of <5 were removed due to poor connectivity. In addition, the
large number of candidate modules may have led to overlap between
them; the Jaccard index (23) was
used to refine the overlap between candidate modules. In the
present study, a Jaccard index of 0.5 was utilized.
Significance analysis for candidate
modules
The statistical significance of candidate modules
from two specific DCNs was computed on the basis of the null score
distribution of the candidate modules, generated using randomized
networks. Each randomized network was composed of edges captured
from the specific PPI network, and the number of edges was the same
in the randomized network and the DCN. Using degree-preserved edge
shuffling, each network was completely randomized 100 times. A
module search was performed for the randomized networks in order to
identify the module scores. Subsequently, the empirical P-value of
a module was defined as the probability of the module exhibiting
the observed score or less by chance. P-values were corrected using
the Benjamini-Hochberg method (24), and candidate modules with P<0.05
were considered to be differential modules between each of the
treatment and normal conditions.
Connectivity dynamics of shared
differential modules
Due to condition-specific differential modules
exhibiting differential activities and mediating differential
biological processes, the module connectivity dynamic score (MCDS)
was implemented to explore dynamic alterations among them (25). The overall MCDS of a differential
module was defined as the average MCDS of all pairwise comparisons.
The statistical significance of MCDS for a differential module was
computed in a similar way as that for candidate modules. If the
MCDS for a differential module had P<0.05, it was denoted to be
a dynamic module with increased dynamic alterations.
Results
Analysis of pathway dynamics is a
novel approach in GBM
An important innovation in the present method was
the ability to identify shared differential modules from two
specific DCNs, each of which represented a different perturbation
condition. There were two conditions of GBM (radiotherapy and
chemoradiation) in the present study. Therefore, two DCNs and two
group differential modules were obtained for the two conditions,
compared with the normal condition.
Construction of DCNs
For each treatment condition, the gene co-expression
for the PPI (including 15,130 genes and 725,216 interactions) was
calculated based on the PCC, and gene pairs with Δ≥0.8 were
considered significantly co-expressed and used to build the BCN. A
weight value was assigned to each edge in the BCN based on
differential gene expression between radiotherapy/chemoradiation
and the normal condition. A total of two condition-specific DCNs
with 287 nodes and 1,052 edges were constructed in GBM, one for the
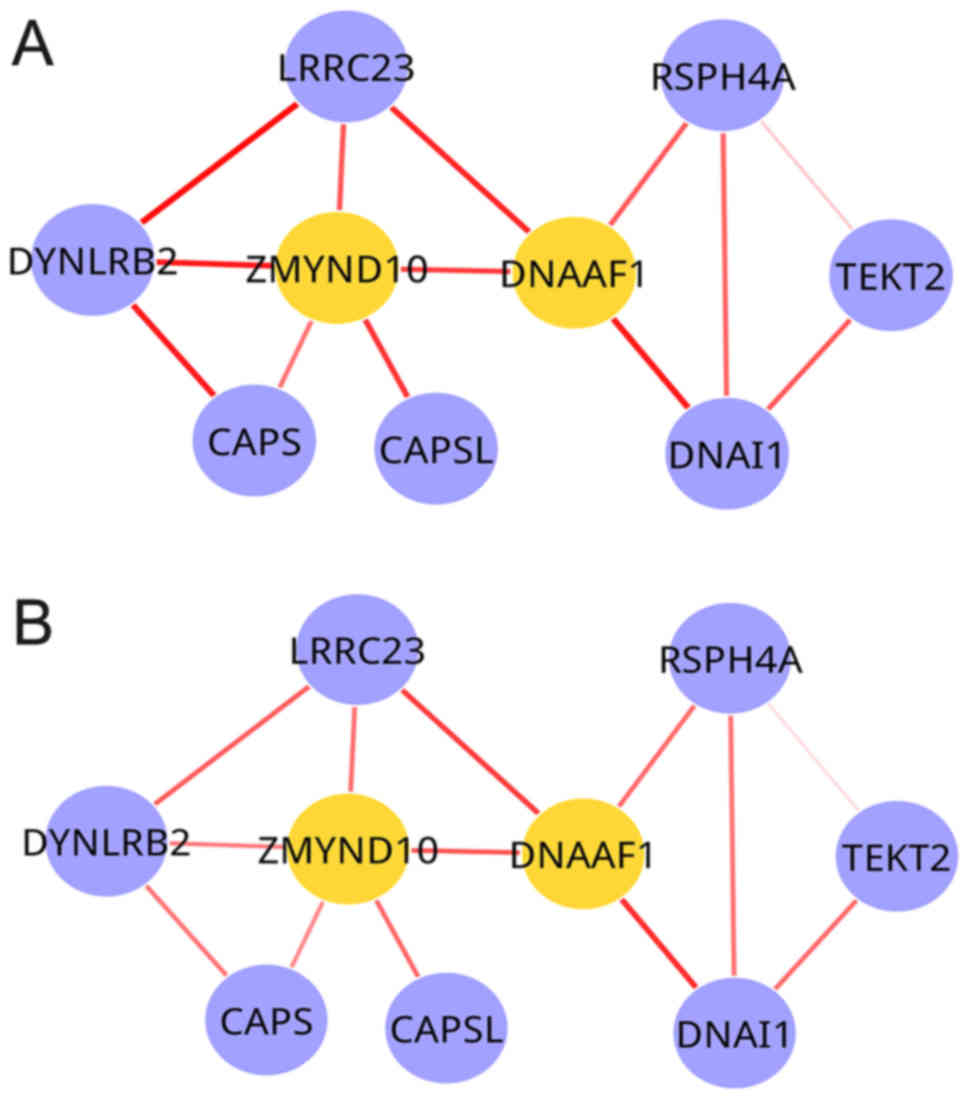
radiotherapy condition (Fig. 1A)
and one for the chemoradiation condition (Fig. 1B). These DCNs exhibited the same
node sets, although the weights of edges were different. The
frequency distribution of edge weights between the two conditions
is presented in Fig. 2. In the
chemoradiation-specific DCN, the average weight (0.0801) was
decreased compared with the radiotherapy condition (0.0997). Out of
1,052 edges, 89 exhibited increased weight values and 956 exhibited
decreased weights in the chemoradiation condition compared with the
radiotherapy condition, and the weights of seven edges were the
same in the two conditions.
Identification of shared differential
modules
In the present study, a systemic differential module
algorithm was employed to identify the shared differential modules
across radiotherapy and chemoradiation conditions in GBM. According
to the topological properties of the importance of genes and the
rank of genes across two DCNs (calculated by averaging the
z-scores), a total of 28 seed genes were obtained and are presented
in Table I. The top rank seed gene
was solute carrier family 17 member 7 (z-score=21.797). A module
search by seed expansion was carried out based on the entropy
decrease Δ E (O, O) between O and
O, and a total of seven shared candidate modules were
detected following refinement with the Jaccard index ≥0.5 in the
two conditions.
 | Table I.Seed genes and their average
z-scores in differential co-expression networks. |
Table I.
Seed genes and their average
z-scores in differential co-expression networks.
| No. | Seed gene | Average
z-score |
|---|
| 1 | SLC17A7 | 21.797 |
| 2 | GAD2 | 21.693 |
| 3 | CPLX2 | 18.762 |
| 4 | CCK | 17.799 |
| 5 | SLC12A5 | 14.862 |
| 6 | PACSIN1 | 11.924 |
| 7 | SYN1 | 11.897 |
| 8 | CHGA | 11.799 |
| 9 | GABRA1 | 10.869 |
| 10 | CORO1A | 6.718 |
| 11 | CD53 | 6.685 |
| 12 | TYROBP | 6.253 |
| 13 | UNC13C | 5.989 |
| 14 | PDYN | 5.701 |
| 15 | STX1A | 4.964 |
| 16 | CACNG3 | 3.985 |
| 17 | LAPTM5 | 3.973 |
| 18 | LY86 | 3.935 |
| 19 | ARHGAP9 | 3.593 |
| 20 | C3AR1 | 3.448 |
| 21 | GMFG | 3.164 |
| 22 | HCK | 3.091 |
| 23 | CYBA | 3.036 |
| 24 | DPT | 3.026 |
| 25 | DNAAF1 | 2.959 |
| 26 | HCLS1 | 2.771 |
| 27 | PSMD10 | 2.750 |
| 28 | ZMYND10 | 2.737 |
In order to calculate the statistical significance
of the seven shared candidate modules across the two conditions,
the null score distribution analysis was performed. A randomized
network with 1,052 edges was constructed in order to examine the
modules, and a total of 1,846 modules were obtained. Candidate
modules which met the threshold of P<0.05 were regarded as
differential modules, and five shared differential modules were
identified (Fig. 3). It was
observed that the weights of the five shared differential modules
in the two conditions were different. The properties of the five
differential modules are presented in Table II.
 | Table II.Properties of the five shared
differential modules. |
Table II.
Properties of the five shared
differential modules.
| Module | Nodes | Edges | No. of seed
genes | Start seed
gene | P-value |
|---|
| 1 | 29 | 88 | 11 | CORO1A | 0.0376 |
| 2 | 44 | 125 | 11 | TYROBP | 0.0451 |
| 3 | 22 | 66 | 9 | LY86 | 0.0259 |
| 4 | 9 | 13 | 2 | DNAAF1 | 0.0436 |
| 5 | 11 | 15 | 1 | PSMD10 | 0.0038 |
Connectivity dynamics of shared
differential modules
As component modules share the same set of genes
between two DCNs, although they may differ in their connectivity,
MCDS was used to capture dynamic alterations in the shared
differential modules and the significance of the MCDS was
calculated. Among the five shared differential modules, only one
exhibited a significant dynamic alteration between the radiotherapy
and chemoradiation conditions (P=0.0436; Fig. 4). Consequently, this module was
denoted to be a dynamic module.
Discussion
Networking is able to provide significant
instructions for mining unknown connections in incomplete networks.
Although the availability of data on large-scale protein
interactions is increasing with the development of high throughput
testing technology, certain important interactions are rarely
studied, including significant pathways (26). This may be resolved to some extent
by utilizing modules of the complex network (27). However, from a systems biology
outlook, diseases are caused by perturbations of the gene network
and such perturbations dynamically alter the disease process
(25). Therefore, the present
study investigated dynamic alterations in module activity and
connectivity across the radiotherapy and chemoradiation conditions
of GBM.
The present method presents an opportunity to study
the dynamics of gene modules in ≥2 conditions, and the MCDS metric
is able to distinguish between dynamic and static shared
differential modules. The dynamic module identified in the present
study differed from the others in multiple aspects, including their
topological properties. In particular, the activity and
connectivity of the dynamic module was correlated with alterations
in the different tumor conditions, suggesting that dynamic modules
may serve a more important role during the treatment of tumor
progression. Therefore, the study of pathway dynamics may lead to
novel insights into tumor pathogenesis and therapy.
In the present study, two DCNs were constructed,
five shared differential modules identified, and one dynamic module
identified with increased dynamic alterations in activity and
connectivity, across the radiotherapy and chemoradiation conditions
in GBM. In the two DCNs, the number of nodes and edges were the
same, although the weights were different; this was additionally
observed in the shared differential modules across the radiotherapy
and chemoradiation conditions. By applying MCDS and the
significance analyses, one dynamic module with nine nodes and 13
edges was identified, of which dynein axonemal assembly factor 1
(DNAAF1) and zinc finger MYND type-containing 10
(ZMYND10) were the seed genes. DNAAF1 was the start
seed gene and ZMYND10 exhibited the highest degree. ZMYND
domain-containing proteins are a protein family whose members are
associated with transcriptional regulators and may modulate the
process of malignant transformation (28). As a member of the ZMYND family,
ZMYND10 has been demonstrated to localize to the cytoplasmic
puncta in respiratory epithelial cells and to regulate
transcription of dynein proteins (29). In addition, ZMYND10 has been
demonstrated to act as a tumor suppressor, arresting the cell cycle
at the G1 phase, downregulating cyclin D1 promoter
activity and inhibiting the clonogenic growth of nasopharyngeal
carcinoma cells (30). It has been
reported that methylation of β-catenin in lung
cancer/ZMYND10 was detected in >95% of primary glioma
tumors (28). Therefore, it is
hypothesized that ZMYND10 may be associated with the
treatment of GBM. Future studies will investigate the association
of the dynamic module identified in the present study with the
treatment of GBM.
In conclusion, the present study investigated the
dynamic alterations of module activity and connectivity, and
identified 1 dynamic module across the radiotherapy and
chemoradiation conditions in GBM, which provides insights into the
molecular mechanism of GBM treatment and potential biomarkers for
the therapy of GBM.
References
|
1
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sottoriva A, Spiteri I, Piccirillo SG,
Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C and Tavaré
S: Intratumor heterogeneity in human glioblastoma reflects cancer
evolutionary dynamics. Proc Natl Acad Sci USA. 110:4009–4014. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, Van Den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lambiv WL, Vassallo I, Delorenzi M, Shay
T, Diserens AC, Misra A, Feuerstein B, Murat A, Migliavacca E,
Hamou MF, et al: The Wnt inhibitory factor 1 (WIF1) is targeted in
glioblastoma and has a tumor suppressing function potentially by
induction of senescence. Neuro Oncol. 13:736–747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Werner T: Target gene identification from
expression array data by promoter analysis. Biomol Eng. 17:87–94.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu T, Reinhart BJ, Magnani E, Huang T,
Kerstetter R and Barton M: Of blades and branches: Understanding
and expanding the Arabidopsis Ad/Abaxial regulatory network through
target gene identification. Cold Spring Harb Symp Quant Biol.
77:31–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in: Functional interaction networks
of proteins, globally integrated and scored. Nucleic Acids Res.
39:(Database issue). D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taylor IW, Linding R, Warde-Farley D, Liu
Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q and Wrana JL:
Dynamic modularity in protein interaction networks predicts breast
cancer outcome. Nat Biotechnol. 27:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee MJ, Ye AS, Gardino AK, Heijink AM,
Sorger PK, MacBeath G and Yaffe MB: Sequential application of
anticancer drugs enhances cell death by rewiring apoptotic
signaling networks. Cell. 149:780–794. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolstad B: Affy: Built-in Processing
Methods. 2015.http://120.52.72.36/www.bioconductor.org/c3pr90ntcsf0/packages/release/bioc/vignettes/affy/inst/doc/builtinMethods.pdf
|
|
14
|
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin
SM, Lapointe DS and Green MR: ChIPpeakAnno: A Bioconductor package
to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics.
11:2372010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gentleman R, Carey V, Huber W and Hahne F:
Genefilter: Methods for filtering genes from microarray
experiments. R package. 2011.http://www.bioconductor.org/packages/2.3/bioc/html/genefilter.html
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Benesty J, Chen J, Huang Y and Cohen I:
Pearson correlation coefficient. Noise reduction in speech
processing Berlin Heidelberg: Springer; pp. 1–4. 2009, View Article : Google Scholar
|
|
18
|
Cohen J, Cohen P, West SG and Aiken LS:
Applied multiple regression/correlation analysis for the behavioral
sciences. 3rd. Hoboken: Taylor and Francis; 2013
|
|
19
|
Ma X, Gao L and Tan K: Modeling disease
progression using dynamics of pathway connectivity. Bioinformatics.
30:2343–2350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vanunu O, Magger O, Ruppin E, Shlomi T and
Sharan R: Associating genes and protein complexes with disease via
network propagation. PLoS Comput Biol. 6:e10006412010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou D, Bousquet O, Lal TN, Weston J and
Schölkopf B: Learning with local and global consistency. Adv Neural
Inform Pro Systems. 16:321–328. 2004.
|
|
22
|
Fortunato S and Barthélemy M: Resolution
limit in community detection. Proc Natl Acad Sci USA. 104:36–41.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bouchard M, Jousselme A-L and Doré P-E: A
proof for the positive definiteness of the Jaccard index matrix.
Int J Approximate Reasoning. 54:615–626. 2013. View Article : Google Scholar
|
|
24
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Stat Soc Series B (Methodological).
57:289–300. 1995.
|
|
25
|
Ma X, Gao L, Karamanlidis G, Gao P, Lee
CF, Garcia-Menendez L, Tian R and Tan K: Revealing pathway dynamics
in heart diseases by analyzing multiple differential networks. PLoS
Comput Biol. 11:e10043322015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nibbe RK, Chowdhury SA, Koyutürk M, Ewing
R and Chance MR: Protein-protein interaction networks and
subnetworks in the biology of disease. Wiley Interdiscip Rev Syst
Biol Med. 3:357–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, Jing R, Jiang L, Jiang Y, Kuang Q,
Ye L, Yang L, Li Y and Li M: Combination use of protein-protein
interaction network topological features improves the predictive
scores of deleterious non-synonymous single-nucleotide
polymorphisms. Amino Acids. 46:2025–2035. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hesson LB, Cooper WN and Latif F:
Evaluation of the 3p21. 3 tumour-suppressor gene cluster. Oncogene.
26:7283–7301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zariwala MA, Gee HY, Kurkowiak M,
Al-Mutairi DA, Leigh MW, Hurd TW, Hjeij R, Dell SD, Chaki M,
Dougherty GW, et al: ZMYND10 is mutated in primary ciliary
dyskinesia and interacts with LRRC6. Am J Hum Genet. 93:336–345.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Liu H, Li B, Huang P, Shao J and
He Z: Tumor suppressor BLU inhibits proliferation of nasopharyngeal
carcinoma cells by regulation of cell cycle, c-Jun N-terminal
kinase and the cyclin D1 promoter. BMC Cancer. 12:2672012.
View Article : Google Scholar : PubMed/NCBI
|