Introduction
Voltage-gated sodium channel (Nav) 1.5 is important
in the generation and propagation of action potentials, in working
myocardium and cardiac tissue conduction cells (1), therefore, it is considered the
primary cardiac Na channel. However, the present study, in
accordance with data from previous studies, demonstrated that
Nav1.5 was expressed in various mammalian tissues including the
brain, neuronal cell lines (2–14),
dorsal root ganglia (DRG) (15–17),
gastrointestinal tract (18), and
various tumor tissues and cell lines (19–32),
in addition to its established presence in cardiac tissue.
Currently, a total of nine distinct Nav1.5 channel isoforms,
Nav1.5a-f and the truncated variants Nav1.5 E28B-D, have been
identified. It has been demonstrated that four of these variants,
including Nav1.5a and Nav1.5c-e, may act as functional channels
generating an Na current (1,33,34).
Therefore, it is important to verify the distinct Nav1.5 isoforms
expressed in different tissues, in order to completely elucidate
the specific functional contribution to each tissue type.
The authors previously demonstrated the expression
of neonatal Nav1.5 (Nav1.5e), Nav1.5a and Nav1.5f in the rat brain
(9,10,13).
However, the presence of further Nav1.5 isoforms expressed in the
rat brain and their exact localization remains to be elucidated.
The function of Nav isoforms in these tissues also remains to be
elucidated. Therefore, the present study systematically
investigated the expression of distinct Nav1.5 isoforms in the
frontal lobe of Sprague Dawley (SD) rat brains via the reverse
transcription-polymerase chain reaction (RT-PCR), DNA sequencing,
restriction enzyme digestion and immunochemistry. Following this,
the functional properties of Nav1.5 in the frontal lobe brain
slices were analyzed using whole-cell patch clamping. In addition,
the expression patterns of various Nav1.5 isoforms in the rat brain
were detected and compared with those present in the rat heart.
Materials and methods
Materials
The investigation was approved by the Ethics
Committee and the Committee of Animal Experimentation of China
Medical University (Shenyang, China) and Capital Medical University
(Beijing, China). Healthy male Sprague-Dawley rats at postnatal day
(P)0-90 [total n=54; age groups: P0 (n=6); P3 (n=6); P6 (n=6); P9
(n=6); P12 (n=6); P15 (n=6); P21 (n=6); P30 (n=6); P90 (n=6)] were
provided by the Animal Experimentation Center of Capital Medical
University (Beijing, China). They were housed with 2 of each
group/cage and maintained on a 12-h light/dark cycle with food and
water available ad libitum under a constant temperature
(23±2°C). Rats were anesthetized with sodium pentobarbital (30
mg/kg body weight) and were sacrificed by cervical dislocation.
Tissues used for RT-PCR and immunochemistry were carefully excised.
The rat brain used for the patch clamp experiment (P21-30; weight
200–250 g) was dissected and placed in ice-cold, oxygenated (95%
O2 and 5% CO2) artificial cerebrospinal fluid
(ACSF; pH 7.4; 126 mM NaCl, 2.5 mM KCl, 26 mM NaHCO3,
0.5 mM CaCl2, 5 mM MgCl2, 1.25 mM
NaH2PO4 and 25 mM dextrose).
RNA isolation and RT-PCR
Total RNA was extracted from the cortical layers of
the frontal lobe of the SD rat brain tissue at differing
developmental stages. The RNeasy lipid tissue Mini kit (Qiagen,
Inc., Valencia, CA, USA) was used to extract the total RNA
according to the manufacturer's protocol. The cDNAs were
synthesized using the SuperScript® VILO™ cDNA synthesis
kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). For the
detection and isolation of different Nav1.5 isoforms from the
frontal lobe of the rat brain, the primer pairs 1–3 (Table I) were used to amplify different
fragments of Nav1.5 cDNA via PCR, according to the manufacturer's
protocol (New England Biolabs, Inc., Ipswich, MA, USA). PCR using
Platinum™ Taq DNA polymerase (Thermo Fisher Scientific, Inc.) was
performed separately according to the specific re-annealing
temperatures of the different primers. The PCR reaction conditions
were as follows: 95°C for 5 min followed by 36 cycles of 95°C for
30 sec, 60–66°C for 30 sec and 72°C for 30 sec, then final
elongation at 72°C for 5 min. PCR products were analyzed by gel
electrophoreses (1–2% agarose). The signal of each band was
determined using Quantity One 4.6 software (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The gene expression ratios of Nav1.5
splicing variants were presented as amplicon densities of Nav1.5 to
its variants. All PCRs for detecting the relative amount of Nav1.5
and its splicing variants were repeated at least three times.
 | Table I.Primer sequences used for the
isolation of Nav1.5 variants in the rat brain. |
Table I.
Primer sequences used for the
isolation of Nav1.5 variants in the rat brain.
|
| Sequence (5′- to
3′-) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Primer pair | Forward | Reverse | Targeting | Locationa | Length (bp) |
|---|
| 1 |
5′-TTCTGCCTGCATGCATTCACCTT-3′ |
5′-GCAGAAGACAGTGAGGACCA-3′ |
Exon6/6A(Nav1.5&Nav1.5e) | 724–963 | 240 |
| 2 |
5′-GTGCCCCCAGCCCGCAAGGAAA-3′ |
5′-TGCTGCCCTCGGAGTAACTGT-3′ | Nav1.5a,
Nav1.5c | 3101–3467 | 367 |
| 3 |
5′-TTCAGCGCAGACAACCTCACA-3′ |
5′-TGTTCTCTTCATCCTCTTCCT-3′ | Nav1.5d | 2870–3242 | 373 |
DNA sequencing
All PCR products were separated by electrophoresis
on a 2% agarose gel. The different fragments of expected size (240,
367 and 373 bp) were extracted and purified using a gel extraction
kit (Qiagen, Inc.) and then sequenced directly using a 3730xl DNA
sequencer (Applied BioSystems; Thermo Fisher Scientific, Inc.).
Restriction enzyme digestion
In order to distinguish the neonatal splice variant
of Nav1.5 from the total Nav1.5 cDNA present, restriction enzyme
SacI was used to digest the total PCR products. The reaction system
was 30 µl in total, containing 4 µl PCR products, 0.5 µl SacI
enzyme, 3 µl loading buffer and 22.5 µl super-purified water. The
electrophoresis was performed on 2% agarose gel to detect the
digestion results following a 1 h incubation period at 38°C. The
expression ratio of Nav1.5 variants vs. total Nav1.5 was detected
from the signal quantification of pre- and post-digestion using a
gel imaging analysis device (ChemiDoc MP; Bio-Rad Laboratories,
Inc.). The signal of each band was determined using Quantity One
v4.6 software (Bio-Rad Laboratories, Inc.).
Immunohistochemistry assay
Fresh specimens of rat brain tissue from P0-90 were
fixed with 4% paraformaldehyde (4°C for 24 h) immediately following
collection. The brain tissues were processed by dehydration using a
series of graded ethanol baths (70–100% ethanol), clearing (the
transparency of tissue) using xylene (100%) and wax infiltration
and were then paraffin-embedded. The streptavidin-peroxidase
immunohistochemical method was applied. Immunohistochemistry was
performed using the Histostain-SP kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Sections (4-µm) were incubated overnight with rabbit Nav1.5 primary
antibody (1:100; cat. no. ASC-013; Alomone, Jerusalem, Israel) at
4°C in a moisture chamber and were then washed three times with PBS
for 5 min. Sections were then incubated with biotinylated goat
anti-rabbit IgG secondary antibody (1:500; cat. no. #656140;
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for
30 min and washed three times with PBS for 5 min. PBS replaced the
primary antibody to serve as the negative control and the slide
with known positive Nav1.5 expression in rat atrial muscle served
as the positive control. The immunohistochemical staining results
were observed using a light microscope (Olympus CX31-LV320; Olympus
Corporation, Tokyo, Japan). The expression of brown and yellow
staining on the cells was considered to indicate positive
immunoreactions when compared with negative and positive
slices.
Western blot analysis
A total of 100 mg tissue was rinsed with pre-cooled
PBS (4°C) and mixed with 500 µl radioimmunoprecipitation assay
strong lysate buffer (Pierce radioimmunoprecipitation Buffer; cat.
no. #89900; Thermo Fisher Scientific, Inc.). A protease inhibitor
cocktail (Pierce; Thermo Fisher Scientific, Inc.) was subsequently
added at a volume ratio of 1:100 and the mixture was homogenized
and lyzed at 4°C overnight. Following centrifugation at 16,100 × g
for 30 min at 4°C, the supernatants were collected and assayed for
protein content. The protein concentration was determined using the
bicinchoninic acid method and 100 µg of protein was loaded into
each lane for a 6–10% SDS-PAGE running at a constant voltage of 80
V for 2 h. The proteins were subsequently transferred onto a
polyvinylidene fluoride membrane at a constant current of 400 mA
for 2 h. The membrane was blocked with 10% non-fat milk for 2 h and
incubated with rabbit anti-Nav1.5 polyclonal antibody overnight at
4°C (1:200; Alomone). The membrane was rinsed with TBS Tween-20
three times and incubated with horseradish peroxidase-linked
anti-rabbit IgG secondary antibody (1:2,000; cat. no. #7074; CST
Biological Reagents Company Limited, Shanghai, China) with shaking
at room temperature for 1 h, and subsequently exposed and developed
using the ECL system. Nav1.5 immunoreactive protein bands were
detected with an enhanced chemiluminescence reagent (ECL-Plus) and
densitometrically quantitated according to the manufacturer's
protocol (GE Healthcare Life Sciences, Chalfont, UK). Experiments
were repeated at least three times.
Electrophysiological recordings
The experiments were performed at room temperature.
Following anesthetization, the rat brain (P21-30) was removed and
placed in ice-cold ACSF, which was continuously bubbled with 95%
O2 and 5% CO2. Brain slices of the frontal
lobe were cut on a vibratome (250–300 µm; VT 1000 S; Leica
Microsystems, Inc., Buffalo Grove, IL, USA) in ice-cold cutting
ACSF and separated into the left and right hemispheres. The slices
were transferred into a chamber containing ACSF and incubated with
a mixture of 95% O2 and 5% CO2 for 30–45 mins
at 36°C. The slices were kept at room temperature for the
subsequent experiment. Whole-cell patch-clamp recordings were
performed on layer V pyramidal neurons of the frontal lobe with a
HEKA EPC-10 patch-clamp amplifier with associated software (PULSE
version 8.3 Software for Data Acquisition; IGOR Pro5.03 for
Graphing and Data Analysis; HEKA Electronik, Inc., Pfalz, Germany).
The pipette solution contained 145 mM CsCl, 2 mM MgCl2,
2 mM Na2ATP, 10 mM HEPES, 0.2 mM EGTA and 2 mM
tetraethylammonium (TEA). To isolate the Na+ currents,
CdCl2 (200 µM) and TEA (20 mM) were added to the bath
solution. Differing concentrations of tetrodotoxin (TTX) (10, 100
and 300 nM, and 1 µM; cat. no. A0224, CAS no. 4368-28-9; Mansite
Biotechnology Co., Ltd, Chengdu, China) were added to the bath
solution to identify TTX-sensitive (TTX-S) and TTX-resistant
(TTX-R) Na currents.
Results
Expression of Nav1.5 mRNA splice
variants in the frontal lobe of rat brain
Adult and neonatal Nav1.5 (Nav1.5e) are expressed
in the frontal lobe of rat brain and ventricular myocytes of rat
heart
Primer pair 1 targeting exon 6 and exon 6A was used
to detect the expression of adult Nav1.5 and neonatal Nav1.5
(Nav1.5e) in the frontal lobe of the rat brain at differing
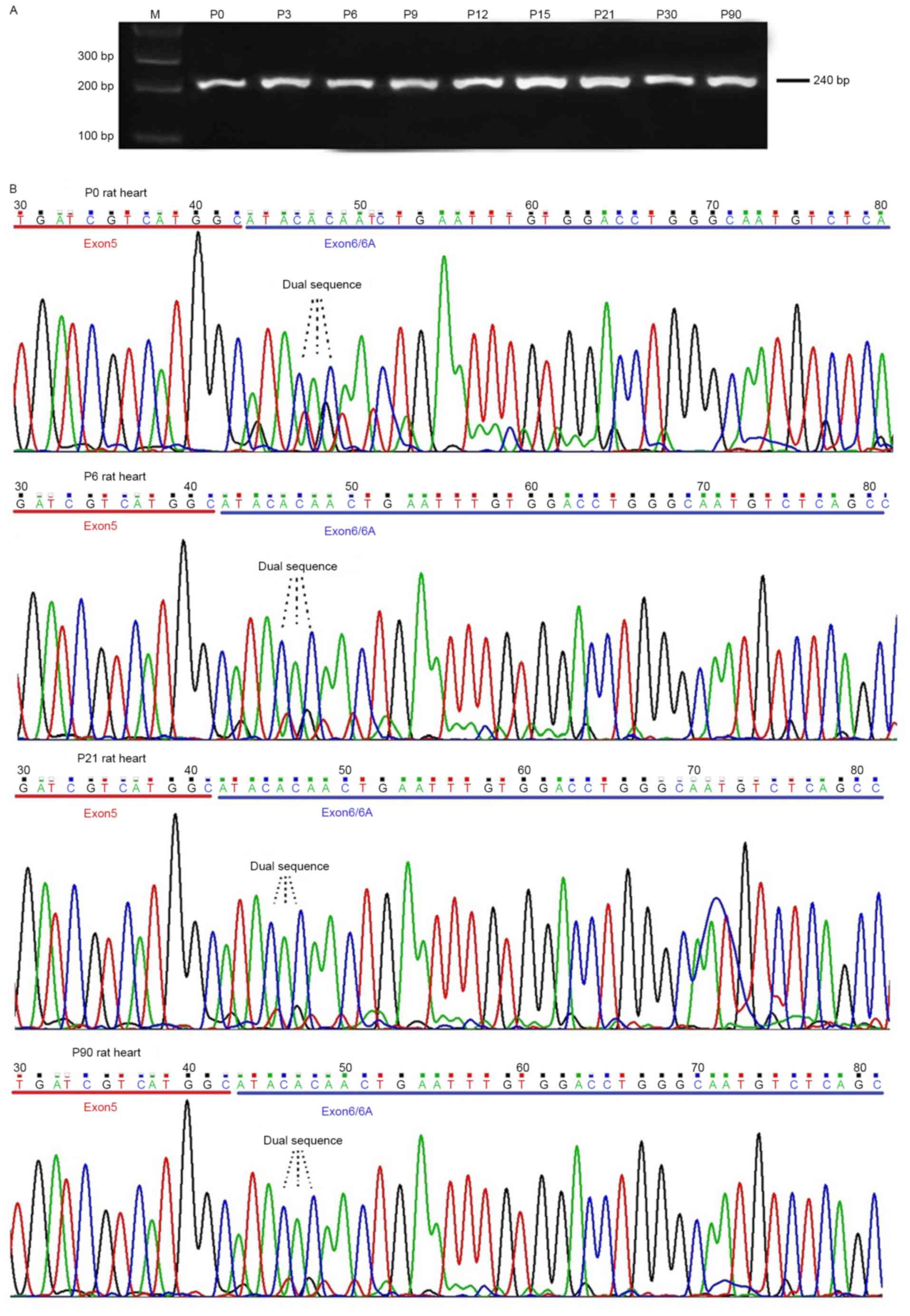
developmental ages. The results revealed that a single band with
the expected size was observed on 2% agarose gel (Fig. 1A). Direct DNA sequencing of the
purified PCR products demonstrated that a single sequence presented
in the exon 5 or 7 coding regions; however, a dual sequence
appeared in the exon 6/6A coding regions (Fig. 1B). DNA sequence analysis revealed
that exon 6 and 6A of Na voltage-gated channel α subunit 5 (SCN5A)
were inclusively expressed, indicating that adult (wild-type) and
neonatal Nav1.5 (Nav1.5e) variants were expressed in the frontal
lobe of the rat brain.
In order to quantify the expression of the two
splice variants of Nav1.5, the restriction enzyme SacI was used to
digest the PCR products, as a specific restriction enzyme site of
SacI was observed to be present in exon 6A and not in exon 6. As
indicated in Fig. 2A, three bands
appeared on the agarose gel following enzyme digestion. The results
indicated that PCR products including exon 6A were digested into
two fragments whereas those containing exon 6 were preserved.
Therefore, the expression ratio of neonatal Nav1.5 (Nav1.5e) vs.
total Nav1.5 was detected from the signal quantification of pre-
and post-digestion by autoradiography. In order to investigate the
expression pattern of neonatal Nav1.5 in the rat brain with age
development, frontal lobes of SD rats from P0-90 were excised. The
results suggested that the expression ratio of neonatal Nav1.5 vs.
adult Nav1.5 in the rat frontal lobe decreased from 1:1 to 1:3 with
age development from P0-90 (Fig.
2B).
For comparison, the present study additionally
detected the expression of neonatal Nav1.5 in ventricular myocytes
of the rat heart from P0-90 using the aforementioned procedure.
Notably, the DNA sequencing and restriction enzyme digestion
results demonstrated the expression of neonatal and adult Nav1.5 in
the ventricular myocytes of rat heart; however, the expression
ratio of neonatal Nav1.5 vs. adult Nav1.5 mRNA decreased from a
ratio of 1:4 to 1:19 in the ventricular myocytes of rat heart from
p0-90, which was markedly different from that in the developing rat
brain (Figs. 3 and 4). The results indicated that neonatal
Nav1.5 mRNA expression was decreased in the ventricular myocytes of
the rat heart during development.
Nav1.5a and Nav1.5c are expressed in the frontal
lobe of rat brain and ventricular myocytes of rat heart
Primer pair 2, targeting partial exon 17, full exon
18 and partial exon 19, were used to detect the expression of
Nav1.5a and Nav1.5c in the frontal lobe of the rat brain. As
indicated in Fig. 5, two bands
with different sizes appeared on the agarose gel. Fragments of the
two bands were extracted and sequenced. It was suggested that the
longer fragment represented the wild-type Nav1.5 isoform and the
shorter band was the Nav1.5a splice variant, in which exon 18 was
alternatively spliced. Signal quantification via autoradiography
(Fig. 5.), revealed that the
expression ratio of wild-type Nav1.5 compared with Nav1.5a was
appropriately 1:4.5 and 1:1 in the neonatal (P0) and adult (P90)
rat brain cortex of the frontal lobe, respectively. The results
indicated that the expression ratio of wild-type Nav1.5 compared
with Nav1.5a in the frontal lobe of the rat brain increased with
age.
For comparison, the expression levels of Nav1.5a and
wild-type Nav1.5 in the ventricular myocytes of rat heart from
P0-90 were additionally detected (Fig.
6). The expression pattern of wild-type Nav1.5 compared with
Nav1.5a was similar to that observed in the frontal lobe of rat
brain with age development, however the expression ratio of the two
splice variants was different in the two distinct tissue types. In
the adult rat brain, wild-type Nav1.5 and Nav1.5a were observed to
be present at a similar abundance. However, the expression ratio of
wild-type Nav1.5 compared with Nav1.5a was appropriately 1:1 and
3.5:1 in the neonatal (P0) and adult (P90) rat heart, respectively,
indicating that wild-type Nav1.5 was the primary isoform expressed
in the adult rat ventricular myocytes. The results demonstrated
that although the expression of Nav1.5a mRNA reduced in the rat
brain and heart, its expression quantification differed in the two
tissue types.
DNA sequence analysis revealed that Nav1.5c
(additional CAG codon, Q1077) was not detected in the frontal lobe
of the rat brain and cardiac muscle in this experiment, indicating
Nav1.5c may not be expressed, or is expressed at a very low level
in these two tissue types.
Nav1.5d is not expressed in the frontal lobe of
the rat brain
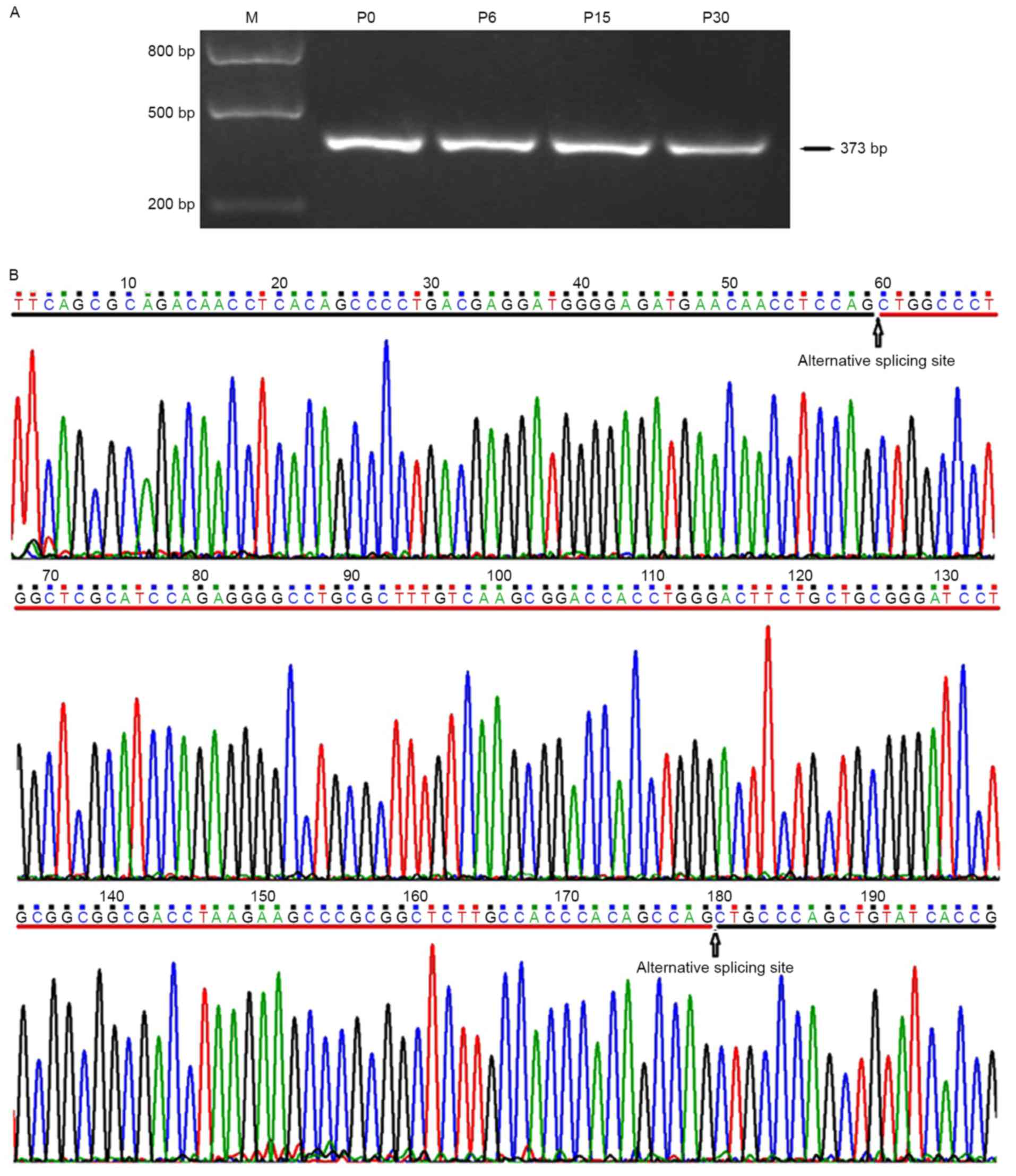
In order to detect if Nav1.5d, the splice variant
with partial splicing of exon 17 (120 bp deletion in the
intermediate region) was expressed in the frontal lobe of rat
brain, primer P3 targeting the full length of exon 17 was used in
PCR. The results demonstrated that only a single band with the
expected size of 373 bp and not 253 bp was observed on the agarose
gel (Fig. 7). Direct DNA
sequencing further confirmed the inclusive expression of the full
length exon 17 of SCN5A. This result indicated that Nav1.5d may not
be expressed or is expressed at a very low level in the frontal
lobe of the rat brain.
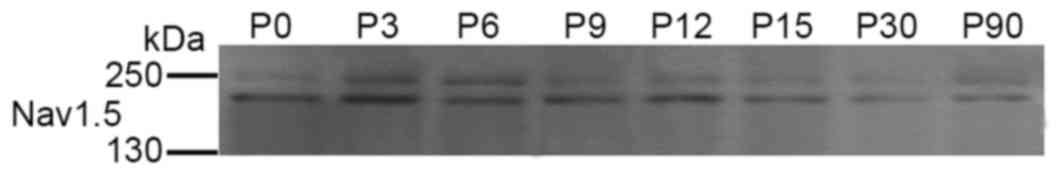
Total Nav1.5 protein is expressed in the frontal
lobe of the rat brain
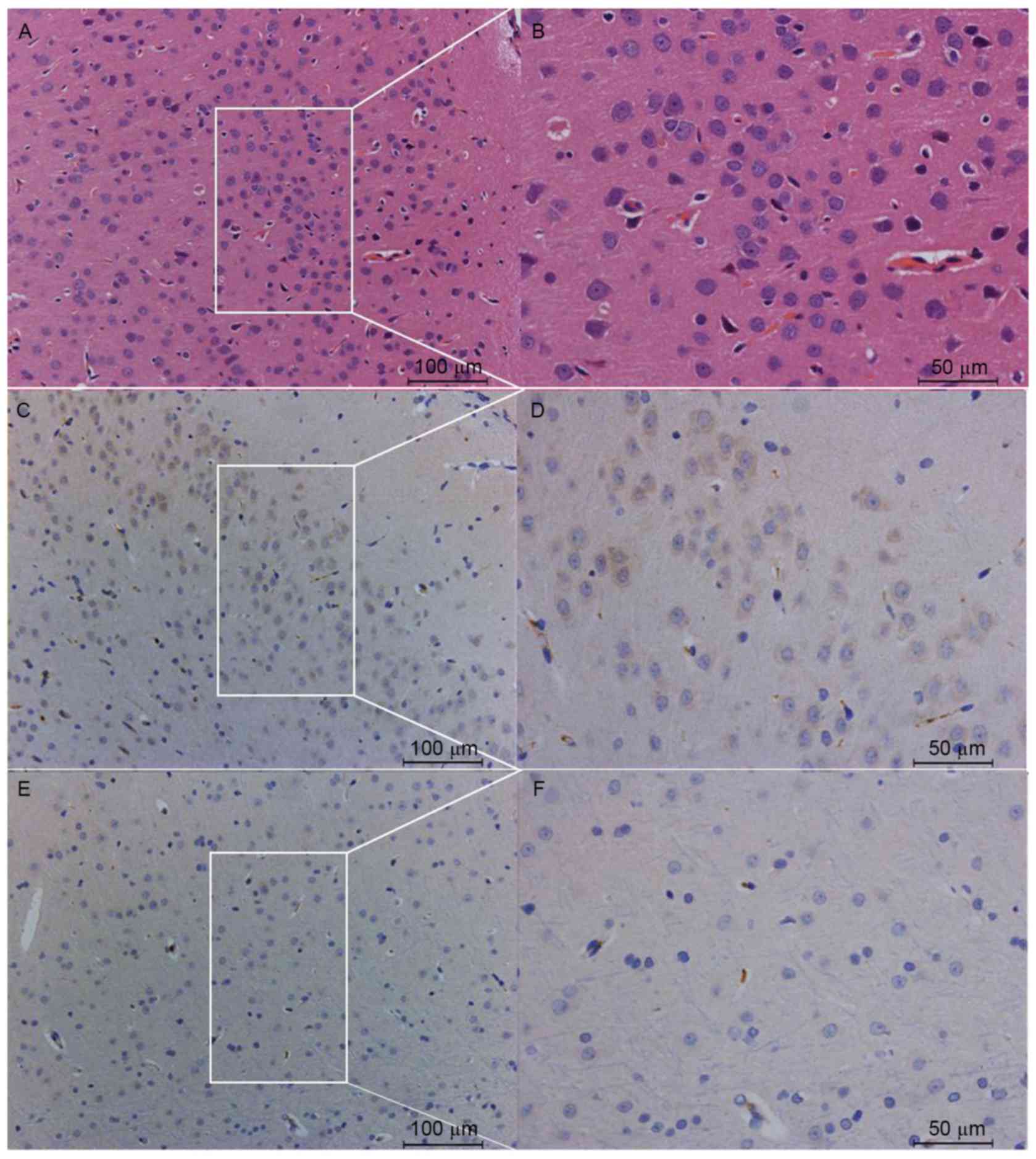
In order to investigate if the Nav1.5 protein was
expressed in the frontal lobe of rat brain, immunohistochemical and
western blotting analyses were used to detect the expression and
distribution of total Nav1.5 protein, in the neurons and glia cells
of the frontal lobe of SD rat brain. The immunohistochemical
results demonstrated that Nav1.5 protein was detected in the cortex
of the frontal lobe, with the immunoreactivity predominantly
observed in the neuronal cell bodies and processes, including axons
and dendrites, whereas little or no immunoreactivity was detected
in the glial components (Fig. 8).
Notably, pyramid cells in layer V of the gray matter of the frontal
lobe cortex demonstrated a greater level of immunoreactivity to the
Nav1.5 antibody, compared with neurons in other layers (Fig. 8). Western blot analyses confirmed
the expression of total Nav1.5 protein in the frontal lobe of SD
rats (Fig. 9). These results
confirmed the expression and distribution of Nav1.5 in the frontal
lobe cortex of rat brain.
Electrophysiological properties of Nav1.5 in the
rat brain: TTX-R current isolated from layer V pyramidal neurons of
the frontal lobe
Whole-cell patch-clamp recordings were conducted on
layer V pyramidal neurons of the frontal lobe to confirm if Nav1.5
was functionally expressed in the neurons. In order to isolate the
Na current from the total current, CdCl2 (200 µM) and
TEA (20 mM) were added to the bath solution to block
Ca2+ and K+ currents, respectively. To
isolate the TTX-R Na current from the total Na current, a protocol
specific to obtaining the total and TTX-R Na currents was used
(35). TTX was applied at
differing concentrations (10, 100 and 300 nM, and 1 µM) to record
the Na current via whole-cell patch clamping. As indicated in
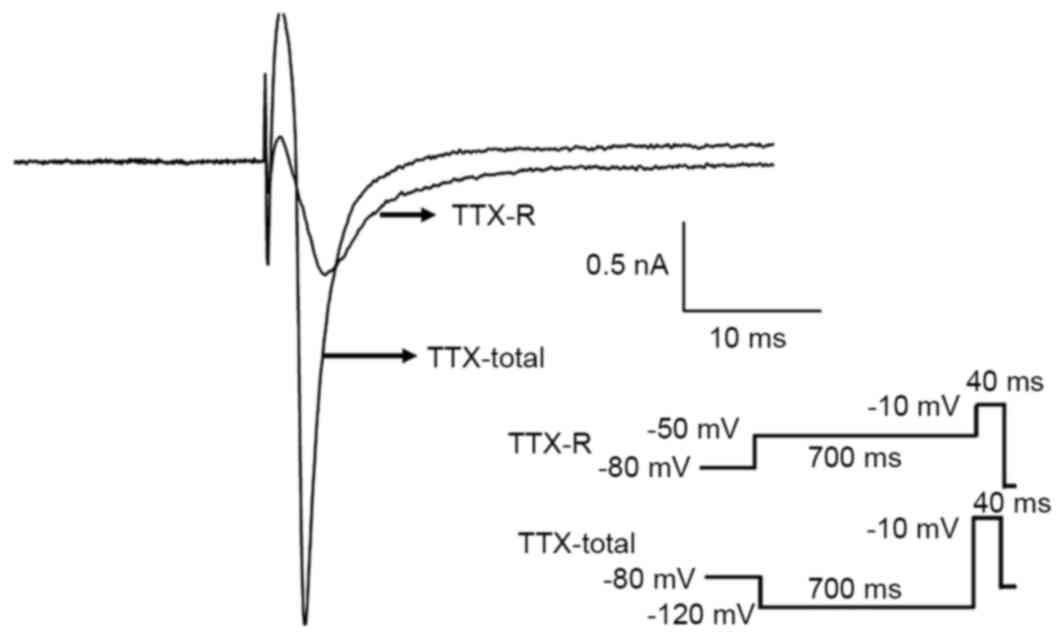
Fig. 10, the TTX-R current was
isolated and recorded in accordance with the aforementioned
protocol. The TTX-R Na current was activated at −40 mV and reached
the maximum amplitude at 0 mV. To further confirm the results, TTX
was added to the bath solution. As presented in Fig. 11, the Na current was recorded at
300 nM TTX, a concentration at which the TTX-S Na current was
completely blocked. When the TTX concentration was increased to 1
µM, the Na current was not detected.
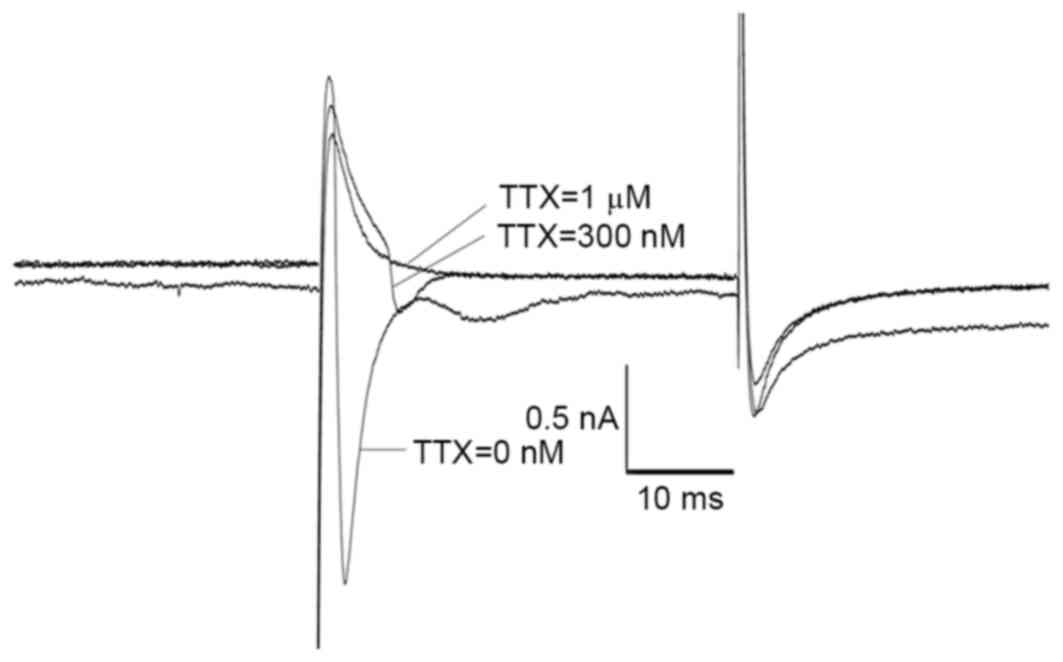 | Figure 11.Isolation of TTX-R current recorded
in the layer V pyramidal neurons of the frontal lobe via TTX. In
order to isolate the TTX-R Na current from the total Na current,
differing concentrations of TTX (10 nM, 100 nM, 300 nM and 1 µM)
were added to the solutions. CdCl2 (200 µM) and
tetraethylammonium (20 mM) were added to the bath solution to block
Ca2+ and K+ currents, respectively. As
indicated, the Na current was recorded when the TTX concentration
was 300 nM and TTX-S Na current was blocked completely. However,
when the TTX concentration was elevated to 1 µM, the Na current
disappeared completely. This value of TTX-sensitivity was
consistent with those reported for other Nav1.5 clones (0.3–10 µM)
and lower than those reported for Nav1.8 and Nav1.9 (>40 µM)
channels, indicating that the TTX-R Na current recorded in the
neurons of frontal lobe of rat brain was generated by Nav1.5. R,
resistant; TTX, tetrodotoxin; S, sensitive; Nav, voltage gated
sodium channel; tetraethylammonium. |
Discussion
Alternative splicing in Nav genes may generate
structurally and functionally distinct Na channels (33,34,36,37).
The authors previously demonstrated that Nav1.5a, Nav1.5e and
Nav1.5f splicing variants were expressed in the rat brain (9,10,13).
The present study systematically investigated the expression of
further Nav1.5 splice variants in the frontal lobe of rat brain.
The expression pattern of Nav1.5 splice variants with age
development was additionally detected in this experiment. RT-PCR
and DNA sequencing confirmed the expression of neonatal and adult
Nav1.5 isoforms in the frontal lobe of the rat brain at different
developmental stages. However, the expression level of neonatal
Nav1.5 decreased with age development. Specifically, the expression
of neonatal Nav1.5 mRNA compared with adult Nav1.5 mRNA decreased
from postnatal day 0 (1:1) to 9 (1:3) and appeared to be constant
from P12 to 90 in the frontal lobe of the rat brain, which
indicated a reducing expression pattern of neonatal Nav1.5 with age
development. The expression pattern of neonatal Nav1.5 splice
variants with age development is similar to that of various other
Navs, including Nav1.3 (38).
However, as the ‘neonatal’ isoform of Nav1.5 may be detected in the
adult rat brain cortex and the ‘adult’ Nav1.5 isoform is expressed
in the neonatal rat brain cortex, the terms ‘neonatal’ and ‘adult’
are commonly used and do not necessarily denote the strict
expression of the neonatal isoform to neonates and the adult
isoform to adults. Therefore, the present study, in accordance with
previous studies, selected the terminology ‘Nav1.5e’ rather than
‘neonatal Nav1.5’ to describe this splicing variant in normal
tissues. Conversely, numerous authors decide to use the terminology
‘neonatal Nav1.5’ when investigating the expression of Nav1.5 in
tumor cell lines (8,23,27,29,32),
as ‘neonatal’ is there used to represent the re-expression of an
embryonic gene or oncogene and its expression is associated with
the occurrence and development of tumors.
The Nav1.5a splicing variant, with the alternative
splicing of exon 18 of SCN5A gene, was additionally detected in
this investigation. The expression ratio of Nav1.5a compared with
wild-type Nav1.5 altered with age development. In the neonatal rat
brain cortex (P0), the expression ratio of these two variants was
4.5:1; however, in the adult rat brain cortex, the expression ratio
was 1:1. The results indicated that wild-type Nav1.5 and Nav1.5a
were expressed in the adult rat brain cortex and presented with a
similar abundance, which was consistent with previous studies
(7,13). Similar expression patterns of
Nav1.5a were observed in the human neuroblastoma cell line NB-1
(22). However, Nav1.5a was not
detected in the human brain cortex, indicating that the Nav1.5a
transcript may be specific only for small rodents or certain tumor
cell lines (10). The
electrophysiological properties of wild-type Nav1.5 and Nav1.5a
were similar to those indicted in the author's previous study of
Nav1.5a cloned from the human neuroblastoma cell line NB-1
(22). However, they were
different in the kinetics of steady-state inactivation and
activation, indicating the alternative splicing of exon 18 did
alter the electrophysiological properties of Nav1.5.
The splicing variant Nav1.5c, characterized by an
additional CAG trinucleotide (encoding an additional glutamine at
position 1077) at the starting site of exon 18, was not detected in
the frontal lobe of the rat brain in the present study, which was
concordant with the author's previous study (13). Notably, Nav1.5c has been detected
in the human brain cortex (10),
with the expression ratio of Nav1.5c vs. wild-type Nav1.5 at 1:5,
indicating the differing expression patterns of Nav.5 splice
variants in different species. The electrophysiological properties
of the Nav1.5c variant were indistinguishable to wild-type Nav1.5
under normal physiological conditions, however the Nav1.5 kinetics
alter significantly when expressing various mutations in the
wild-type Nav1.5 (39–41).
Overall, with the exception of Nav1.5c and Nav1.5d,
adult Nav1.5 and various Nav1.5 splice variants, including Nav1.5a,
Nav1.5e and Nav1.5f, were expressed in the frontal lobe of the rat
brain.
Nav1.5 is the primary cardiac Na channel as it
demonstrates the greatest expression in the heart and is important
in the generation and propagation of action potentials in the
electrophysiological activities of cardiac tissues (1,33).
Previous studies suggest that neonatal Nav1.5 is not detected in
the adult mouse, rat and human hearts (8,42,43).
The present study systematically investigated the expression of
adult and neonatal Nav1.5 in the rat ventricular myocytes with age
development via RT-PCR, DNA sequencing and restriction enzyme
digestion methods. DNA sequencing of the PCR products indicated
that neonatal and adult Nav1.5 variants were expressed in the rat
ventricular myocytes of P0 to 90. However, the expression level of
neonatal Nav1.5 was low in adult rat ventricular myocytes compared
with that in the neonatal rat heart. Direct DNA sequencing of the
PCR products from the rat ventricular myocytes revealed the dual
sequences in the exon 6/6A coding region of SCN5A gene in neonatal
and adult rat heart, however the additional band generated by the
enzyme digestion was only observed in the neonatal rat ventricular
myocytes and not in the adult, which further confirmed the low
expression level of neonatal Nav1.5 in the adult rat ventricular
myocytes (undetectable level by electrophoresis on the agarose
gel). As indicated by electrophoresis results following enzyme
digestion, the expression level of neonatal Nav1.5 reduced with age
development from p0 to 6. The band signal on the agarose gel
suggested that the expression ratio of neonatal Nav1.5 vs. adult
Nav1.5 decreased from 1:4 to 1:19 from P0 to 90. These results
confirmed the adult Nav1.5 mRNA expressed in the adult rat heart
accounted for >95% of total Nav1.5 mRNA whereas the neonatal
Nav1.5 mRNA represented <5%. As the expression level of neonatal
Nav1.5 mRNA is low, the neonatal Nav1.5 protein has previously been
demonstrated to be undetectable in the adult heart (8).
The expression of Nav1.5a in the developing rat
ventricular myocytes was additionally observed in the present
study. The expression pattern of wild-type Nav1.5 compared with
Nav1.5a was similar to that in the rat brain with age development,
the expression ratio of the two splice variants was different in
the two distinct tissue types. In the adult rat brain cortex,
wild-type Nav1.5 and Nav1.5a were present at a similar abundance.
However, in the adult (P30~90) rat ventricular myocytes, the
expression quantification of wild-type Nav1.5 was ~3.5 fold
compared with Nav1.5a, indicating the wild-type Nav1.5 was the
major isoform in the adult rat heart. These results were similar to
those observed in previous studies (7,44)
and demonstrated that although the expression of Nav1.5a reduced in
the rat heart and brain with age development, its expression
quantification was different in these two tissue types.
Neonatal Nav1.5, adult Nav1.5 and Nav1.5a were all
expressed in the rat brain cortex and cardiac muscle, however the
expression ratios among these splice variants differed in the
distinct tissue types. Further studies may explore the underlying
mechanisms regulating the alternative splicing of the SCN5A gene in
the same or differing tissue types with age development.
Previous studies have detected the TTX-insensitive
or resistant heart-like Na (Nav1.5) current in the striatal,
hippocampal, medial entorhinal and olfactory sensory neurons of the
rat brain (3,14,45,46).
The present study recorded the Na current from the pyramidal cells
in layer V of the frontal lobe of the SD rat brain via the
whole-cell patch clamp technique. As K+ and
Ca2+ currents may be recorded simultaneously, TEA and
CdCl2 were added to the bath solution in order to
respectively block them. Various Na channel types, including
Nav1.1, Nav1.2, Nav1.3, Nav1.5 and Nav1.6, may be co-expressed in
brain neurons, therefore, the Na currents recorded in this
experiment were compound products, including the TTX-S and TTX-R Na
currents. In order to isolate the TTX-R Na current from the total
Na current, different concentrations of TTX were added to the bath
solution. In accordance with previous results, the TTX-R Na current
was detected in the neurons of the frontal lobe of the rat brain.
However, two independent approaches were used to distinguish the
TTX-S and TTX-R Na currents in the present study. The TTX-R Na
current was recorded in 300 nM TTX, in which TTX-S Na channels were
blocked completely. The TTX-R Na current disappeared when the
concentration of TTX was increased to 1 µM. This value of
TTX-sensitivity was consistent with those reported for other Nav1.5
variants (0.3–10 µM) and was lower than those reported for Nav1.8
and Nav1.9 (>40 µM) channels, indicating that the TTX-R Na
current recorded in the neurons of the frontal lobe of the rat
brain was generated by Na channel Nav1.5 (1,3,14,37).
Further studies are necessary in order to clarify the specific
contributions of each Nav1.5 isoform to the total Nav1.5 current
and the generation of action potential in neurons.
In conclusion, the results of the present study
demonstrated that various Nav1.5 isoforms, particularly the
neonatal and adult isoforms, were expressed in the rat brain,
however their expression ratios varied. The electrophysiological
analysis conducted using the whole-cell patch clamp technique
further confirmed the functional expression of Nav1.5 in the brain
neurons.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 31100770) and the
Liaoning Provincial Natural Science Foundation of China (grant no.
2014021097).
References
|
1
|
Rook MB, Evers MM, Vos MA and Bierhuizen
MF: Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc
Res. 93:12–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yarowsky PJ, Krueger BK, Olson CE,
Clevinger EC and Koos RD: Brain and heart sodium channel subtype
mRNA expression in rat cerebral cortex. Proc Natl Acad Sci USA.
88:9453–9457. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoehn K, Watson TW and MacVicar BA: A
novel tetrodotoxin-insensitive, slow sodium current in striatal and
hippocampal neurons. Neuron. 10:543–552. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schaller KL, Krzemien DM, Yarowsky PJ,
Krueger BK and Caldwell JH: A novel, abundant sodium channel
expressed in neurons and glia. J Neurosci. 15:3231–3242.
1995.PubMed/NCBI
|
|
5
|
Hartmann HA, Colom LV, Sutherland ML and
Noebels JL: Selective localization of cardiac SCN5A sodium channels
in limbic regions of rat brain. Nat Neurosci. 2:593–595. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Donahue LM, Coates PW, Lee VH, Ippensen
DC, Arze SE and Poduslo SE: The cardiac sodium channel mRNA is
expressed in the developing and adult rat and human brain. Brain
Res. 887:335–343. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Korsgaard Gersdorff MP, Christophersen P,
Ahring PK and Olesen SP: Identification of a novel voltage-gated
Na+ channel rNa(v)1.5a in the rat hippocampal progenitor stem cell
line HiB5. Pflugers Arch. 443:18–30. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chioni AM, Fraser SP, Pani F, Foran P,
Wilkin GP, Diss JK and Djamgoz MB: A novel polyclonal antibody
specific for the Na(v)1.5 voltage-gated Na(+) channel ‘neonatal’
splice form. J Neurosci Methods. 147:88–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Ou SW, Wang YJ, Zong ZH, Lin L,
Kameyama M and Kameyama A: New variants of Nav1.5/SCN5A encode Na+
channels in the brain. J Neurogenet. 22:57–75. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Ou SW, Wang YJ, Kameyama M,
Kameyama A and Zong ZH: Analysis of four novel variants of
Nav1.5/SCN5A cloned from the brain. Neurosci Res. 64:339–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Black JA, Newcombe J and Waxman SG:
Astrocytes within multiple sclerosis lesions upregulate sodium
channel Nav1.5. Brain. 133:835–846. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu L, Nishiyama K, Hollyfield JG and Wang
Q: Localization of Nav1.5 sodium channel protein in the mouse
brain. Neuroreport. 13:2547–2551. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren CT, Li DM, Ou SW, Wang YJ, Lin Y, Zong
ZH, Kameyama M and Kameyama A: Cloning and expression of the two
new variants of Nav1.5/SCN5A in rat brain. Mol Cell Biochem.
365:139–148. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Frenz CT, Hansen A, Dupuis ND, Shultz N,
Levinson SR, Finger TE and Dionne VE: NaV1.5 sodium channel window
currents contribute to spontaneous firing in olfactory sensory
neurons. J Neurophysiol. 112:1091–1104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Renganathan M, Dib-Hajj S and Waxman SG:
Na(v)1.5 underlies the ‘third TTX-R sodium current’ in rat small
DRG neurons. Brain Res Mol Brain Res. 106:70–82. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kerr NC, Gao Z, Holmes FE, Hobson SA,
Hancox JC, Wynick D and James AF: The sodium channel Nav1.5a is the
predominant isoform expressed in adult mouse dorsal root ganglia
and exhibits distinct inactivation properties from the full-length
Nav1.5 channel. Mol Cell Neurosci. 35:283–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kerr NC, Holmes FE and Wynick D: Novel
isoforms of the sodium channels Nav1.8 and Nav1.5 are produced by a
conserved mechanism in mouse and rat. J Biol Chem. 279:24826–24833.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Osorio N, Korogod S and Delmas P:
Specialized functions of Nav1.5 and Nav1.9 channels in
electrogenesis of myenteric neurons in intact mouse ganglia. J
Neurosci. 34:5233–5244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng D, Kyle JW, Martin RL, Ambler KS and
Hanck DA: Cardiac sodium channels expressed in a peripheral
neurotumor-derived cell line, RT4-B8. Am J Physiol.
270:C1522–C1531. 1996.PubMed/NCBI
|
|
20
|
Gu XQ, Dib-Hajj S, Rizzo MA and Waxman SG:
TTX-sensitive and -resistant Na+ currents and mRNA for the
TTX-resistant rH1 channel, are expressed in B104 neuroblastoma
cells. J Neurophysiol. 77:236–246. 1997.PubMed/NCBI
|
|
21
|
Fraser SP, Diss JK, Chioni AM, Mycielska
ME, Pan H, Yamaci RF, Pani F, Siwy Z, Krasowska M, Grzywna Z, et
al: Voltage-gated sodium channel expression and potentiation of
human breast cancer metastasis. Clin Cancer Res. 11:5381–5389.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ou SW, Kameyama A, Hao LY, Horiuchi M,
Minobe E, Wang WY, Makita N and Kameyama M: Tetrodotoxin-resistant
Na+ channels in human neuroblastoma cells are encoded by new
variants of Nav1.5/SCN5A. Eur J Neurosci. 22:793–801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brackenbury WJ, Chioni AM, Diss JK and
Djamgoz MB: The neonatal splice variant of Nav1.5 potentiates in
vitro invasive behaviour of MDA-MB-231 human breast cancer cells.
Breast Cancer Res Treat. 101:149–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao R, Wang J, Shen Y, Lei M and Wang Z:
Functional expression of voltage-gated sodium channels Nav1.5 in
human breast cancer cell line MDA-MB-231. J Huazhong Univ Sci
Technolog Med Sci. 29:64–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Onkal R and Djamgoz MB: Molecular
pharmacology of voltage-gated sodium channel expression in
metastatic disease: Clinical potential of neonatal Nav1.5 in breast
cancer. Eur J Pharmacol. 625:206–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
House CD, Vaske CJ, Schwartz AM, Obias V,
Frank B, Luu T, Sarvazyan N, Irby R, Strausberg RL, Hales TG, et
al: Voltage-gated Na+ channel SCN5A is a key regulator of a gene
transcriptional network that controls colon cancer invasion. Cancer
Res. 70:6957–6967. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chioni AM, Shao D, Grose R and Djamgoz MB:
Protein kinase A and regulation of neonatal Nav1.5 expression in
human breast cancer cells: Activity-dependent positive feedback and
cellular migration. Int J Biochem Cell Biol. 42:346–358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao R, Shen Y, Cai J, Lei M and Wang Z:
Expression of voltage-gated sodium channel alpha subunit in human
ovarian cancer. Oncol Rep. 23:1293–1299. 2010.PubMed/NCBI
|
|
29
|
Brisson L, Driffort V, Benoist L, Poet M,
Counillon L, Antelmi E, Rubino R, Besson P, Labbal F, Chevalier S,
et al: NaV1.5 Na+ channels allosterically regulate the
NHE-1 exchanger and promote the activity of breast cancer cell
invadopodia. J Cell Sci. 126:4835–4842. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dulong C, Fang YJ, Gest C, Zhou MH,
Patte-Mensah C, Mensah-Nyagan AG, Vannier JP, Lu H, Soria C, Cazin
L, et al: The small GTPase RhoA regulates the expression and
function of the sodium channel Nav1.5 in breast cancer cells. Int J
Oncol. 44:539–547. 2014.PubMed/NCBI
|
|
31
|
Shan B, Dong M, Tang H, Wang N, Zhang J,
Yan C, Jiao X, Zhang H and Wang C: Voltage-gated sodium channels
were differentially expressed in human normal prostate, benign
prostatic hyperplasia and prostate cancer cells. Oncol Lett.
8:345–350. 2014.PubMed/NCBI
|
|
32
|
Xing D, Wang J, Ou S, Wang Y, Qiu B, Ding
D, Guo F and Gao Q: Expression of neonatal Nav1.5 in human brain
astrocytoma and its effect on proliferation, invasion and apoptosis
of astrocytoma cells. Oncol Rep. 31:2692–2700. 2014.PubMed/NCBI
|
|
33
|
Schroeter A, Walzik S, Blechschmidt S,
Haufe V, Benndorf K and Zimmer T: Structure and function of splice
variants of the cardiac voltage-gated sodium channel Na(v)1.5. J
Mol Cell Cardiol. 49:16–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Walzik S, Schroeter A, Benndorf K and
Zimmer T: Alternative splicing of the cardiac sodium channel
creates multiple variants of mutant T1620K channels. PLoS One.
6:e191882011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang ZJ and Song XJ: Differing
alterations of sodium currents in small dorsal root ganglion
neurons after ganglion compression and peripheral nerve injury. Mol
Pain. 4:202008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Makielski JC, Ye B, Valdivia CR, Pagel MD,
Pu J, Tester DJ and Ackerman MJ: A ubiquitous splice variant and a
common polymorphism affect heterologous expression of recombinant
human SCN5A heart sodium channels. Circ Res. 93:821–828. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Onkal R, Mattis JH, Fraser SP, Diss JK,
Shao D, Okuse K and Djamgoz MB: Alternative splicing of Nav1.5: An
electrophysiological comparison of ‘neonatal’ and ‘adult’ isoforms
and critical involvement of a lysine residue. J Cell Physiol.
216:716–726. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gazina EV, Richards KL, Mokhtar MB, Thomas
EA, Reid CA and Petrou S: Differential expression of exon 5 splice
variants of sodium channel alpha subunit mRNAs in the developing
mouse brain. Neuroscience. 166:195–200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan BH, Valdivia CR, Rok BA, Ye B, Ruwaldt
KM, Tester DJ, Ackerman MJ and Makielski JC: Common human SCN5A
polymorphisms have altered electrophysiology when expressed in
Q1077 splice variants. Heart Rhythm. 2:741–747. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tan BH, Valdivia CR, Song C and Makielski
JC: Partial expression defect for the SCN5A missense mutation
G1406R depends on splice variant background Q1077 and rescue by
mexiletine. Am J Physiol Heart Circ Physiol. 291:H1822–H1828. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang DW, Desai RR, Crotti L, Arnestad M,
Insolia R, Pedrazzini M, Ferrandi C, Vege A, Rognum T, Schwartz PJ
and George AL Jr: Cardiac sodium channel dysfunction in sudden
infant death syndrome. Circulation. 115:368–376. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gellens ME, George AL Jr, Chen LQ, Chahine
M, Horn R, Barchi RL and Kallen RG: Primary structure and
functional expression of the human cardiac tetrodotoxin-insensitive
voltage-dependent sodium channel. Proc Natl Acad Sci USA.
89:554–558. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zimmer T, Bollensdorff C, Haufe V,
Birch-Hirschfeld E and Benndorf K: Mouse heart Na+ channels:
Primary structure and function of two isoforms and alternatively
spliced variants. Am J Physiol Heart Circ Physiol. 282:H1007–H1017.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Blechschmidt S, Haufe V, Benndorf K and
Zimmer T: Voltage-gated Na+ channel transcript patterns in the
mammalian heart are species-dependent. Prog Biophys Mol Biol.
98:309–318. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
White JA, Alonso A and Kay AR: A
heart-like Na+ current in the medial entorhinal cortex. Neuron.
11:1037–1047. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deisz RA: A tetrodotoxin-insensitive
[corrected] sodium current initiates burst firing of neocortical
neurons. Neuroscience. 70:341–351. 1996. View Article : Google Scholar : PubMed/NCBI
|