Introduction
Apolipoprotein M (ApoM), which was first discovered
and isolated by Xu and Dahlbäck (1) in 1999, belongs to the lipocalin
superfamily. ApoM is predominantly associated with high-density
lipoprotein (HDL) particles in plasma, accounts for ~5% of HDL and
an even smaller proportion (0.2~1%) of low-density lipoprotein
(LDL) particles. ApoM is exclusively expressed in the liver and
kidney and serves various functions. Wolfrum et al (2) demonstrated that ApoM enables
pre-β-HDL to be transformed into mature HDL particles, which are
involved in cholesterol efflux and may form the basis of the
antiatherogenic effect. The anti-inflammatory effects of ApoM may
also be associated with its anti-atherosclerotic effects (3). Arkensteijn et al (4) reported that S1P-ApoM can activate the
S1P-1 receptor and affect the functions of endothelial cells. A
previous study demonstrated that ApoM was expressed in human
intestinal tissues and may be associated with lymph node metastasis
of colorectal cancer (5). Thus,
ApoM functions in the cellular cholesterol efflux capacity,
anti-inflammatory activity, protection of the endothelium and
tumorigenesis.
The vitamin D receptor (VDR) is a karyophilic
protein that belongs to the steroid-thyroid receptor superfamily.
The bioactive, hormonal ligand for VDR is 1,25-dihydroxyvitamin D3
[1,25-(OH)2D3] (6). VDR is widely
distributed in various tissues and organs, and is highly expressed
in the colon, kidney, skeleton and lymphocytes. VDR functions as a
ligand-induced nuclear transcription factor to regulate various
biological processes (7). Li et
al (8) reported that increases
of circulating cholesterol in vitamin D deficiency is associated
with the reduction of transcriptional activity of VDR. Preclinical
studies have established that VDR additionally has multifarious
antitumor effects (particularly against colorectal cancer),
including anti-proliferation, pro-differentiation, pro-apoptosis,
anti-angiogenesis and anti-inflammatory effects (9,10).
These results suggest that ApoM and VDR may have similar functions
in cholesterol metabolism, immune and tumor regulation.
In the present study, whether ApoM affected the
expression of VDR in colorectal cancer cells was investigated. A
standardized method for determining ApoM and VDR mRNA levels is
required. In the present study, a single-tube duplex RT-qPCR system
was used to simultaneously detect VDR and GAPDH expression, which
could efficiently shorten time and reduce errors, and provide a
powerful tool in studying the functions and mechanisms of target
genes.
Materials and methods
Cell line and cell culture
Human colorectal cancer cell line HT-29 was
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). HT-29 cells were cultured in McCoy's
5A (modified) medium containing 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA),
Penicillin-Streptomycin (100 U/ml penicillin and 100 µg/ml
streptomycin; Gibco; Thermo Fisher Scientific, Inc.), and were
incubated at 37°C with 5% CO2.
Lentiviral infection and efficiency
detection
Lentiviral particles containing the GV365 expression
vector encoding ApoM and scrambled sequence (as a negative control)
were synthesized by GeneChem Co., Ltd. (Shanghai, China) and were
transduced into HT-29 cells following the manufacturer's
instructions. Cells infected with lentiviral vector were seeded at
a density of 1×105 cells/well in 6-well plates. The successfully
virus-infected cells, being green fluorescent protein
(GFP)-positive, were observed with fluorescence microscopy after 72
h, and the infection efficiency was detected by RT-qPCR.
Primers and probes design
The base sequences of ApoM (NM_019101, NM_001256169
and NR_045828), VDR (NM_000376, NM_001017535 and NM_001017536) and
GAPDH (NM_002046, NM_001256799, NM_001289745 and NM_001289746) were
obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Primers and
TaqMan probes were designed by Primer Premier 5.0 and synthesized
by Sangon Biotech Co., Ltd. (Shanghai, China). The TaqMan probe of
VDR was labeled with a BHQ1 quencher dye at 3′-end, and with FAM
reporter dye at 5′-end. The TaqMan probe of GAPDH was labeled with
a BHQ2 quencher dye at 3′-end, and with CY5 reporter dye at 5′-end.
Sequence data are presented in Table
I.
 | Table I.Sequences of primers and probes. |
Table I.
Sequences of primers and probes.
| Gene | Primer/probe | Sequences
(5′→3′) | Product length (base
pairs) |
|---|
| ApoM | Forward primer |
CTGACAACTCTGGGCGTGGAT | 118 |
|
| Reverse primer |
TGTCCACAGGGTCAAAAGTTGC |
|
|
| Probe |
FAM-AGTTCCCAGAGGTCCACTTGGGCCA-BHQ1 |
|
| VDR | Forward primer |
GCTAAGATGATACCAGGATTCAGAGAC | 104 |
|
| Reverse primer |
AAGGACTCATTGGAGCGCAAC |
|
|
| Probe |
FAM-ACCTCTGAGGACCAGATCGTACTGCTGA-BHQ1 |
|
| GAPDH | Forward primer |
CAGGGCTGCTTTTAACTCTGGT | 104 |
|
| Reverse primer |
CATGGGTGGAATCATATTGGAAC |
|
|
| Probe |
CY5-TGGATATTGTTGCCATCAATGACCCCT-BHQ2 |
|
RNA extraction and reverse
transcription
Total RNA was extracted using the Total RNA
Purification kit (Shenergy Biocolor Bioscience and Technology,
Shanghai, China) according to the manufacturer's instructions. The
concentration and purity of total RNA were spectrophotometrically
assessed, measuring absorbance at 260/280 nm by BioPhotometer
(Eppendorf, Hamburg, Germany). RNA samples with a 260/280 nm ratio
in the range 1.8~2.0 were considered as high quality and selected
for further analysis. A total of 2 µl cDNA synthesized by the
RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.) was used for PCR.
Duplex fluorescence RT-qPCR
RT-qPCR was performed using Immolase™ DNA Polymerase
(Bioline USA, Taunton, MA, USA) and analyzed by LightCycler 480 II
PCR system (Roche Diagnostics GmbH, Mannheim, Germany).
Amplification reactions contained 2.5 µl 10X ImmoBuffer, 1 µl 10 mM
dNTPs, 2 µl 50 mM MgCl2, 0.04 µl of each 100 µM primer
and probe (Table I, synthesized by
Sangon Biotech Co., Ltd.), 0.5 µl DNA polymerase, 2 µl template
(replaced by water in no template controls) and nuclease-free water
in a final volume of 25 µl. The optimum conditions for PCR were as
follows: 95°C denaturation for 10 min, and 40 cycles of
amplification for 5 sec at 95°C and 15 sec at 60°C. PCR products
were cloned and sequenced by Sangon Biotech Co., Ltd., and the
gained recombinant plasmids were used as the standards for absolute
quantitation of DNA copy number (11).
Preparation of VDR/GAPDH plasmid
standard mixture
The concentrations of recombinant plasmids were
determined using a spectrophotometer. VDR and GAPDH plasmid were
mixed with equal volumes and copies (4×106 copies/µl) and used as
the templates for PCR amplification. Subsequently, 10-fold serial
dilutions (from 4×105~4×101 copies/µl) of extracted plasmid DNA
were used to generate a standard curve by plotting the cycle
threshold vs. the log initial copy number of input plasmid DNA.
Sensitivity and repeatability of
duplex RT-qPCR method
The sensitivity of our assay was assessed using a
10-fold dilution series (from 4×105-4×101 copies/µl) of VDR/GAPDH
plasmid standard mixture. The limit of detection was based on the
final dilution at which the signal of the TaqMan probes was
exponentially amplified. Each sample was amplified five times in
five parallel reactions to detect the intra- and
inter-repeatability of the assay.
Statistical analysis
Statistical testing was conducted with the
assistance of GraphPad Prism software, version 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). All data are presented as the
mean ± standard error or the mean ± standard deviation. Student's
t-test (two-tailed) was used to compare two groups. One-way
analysis of variance was used to analyze multiple groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
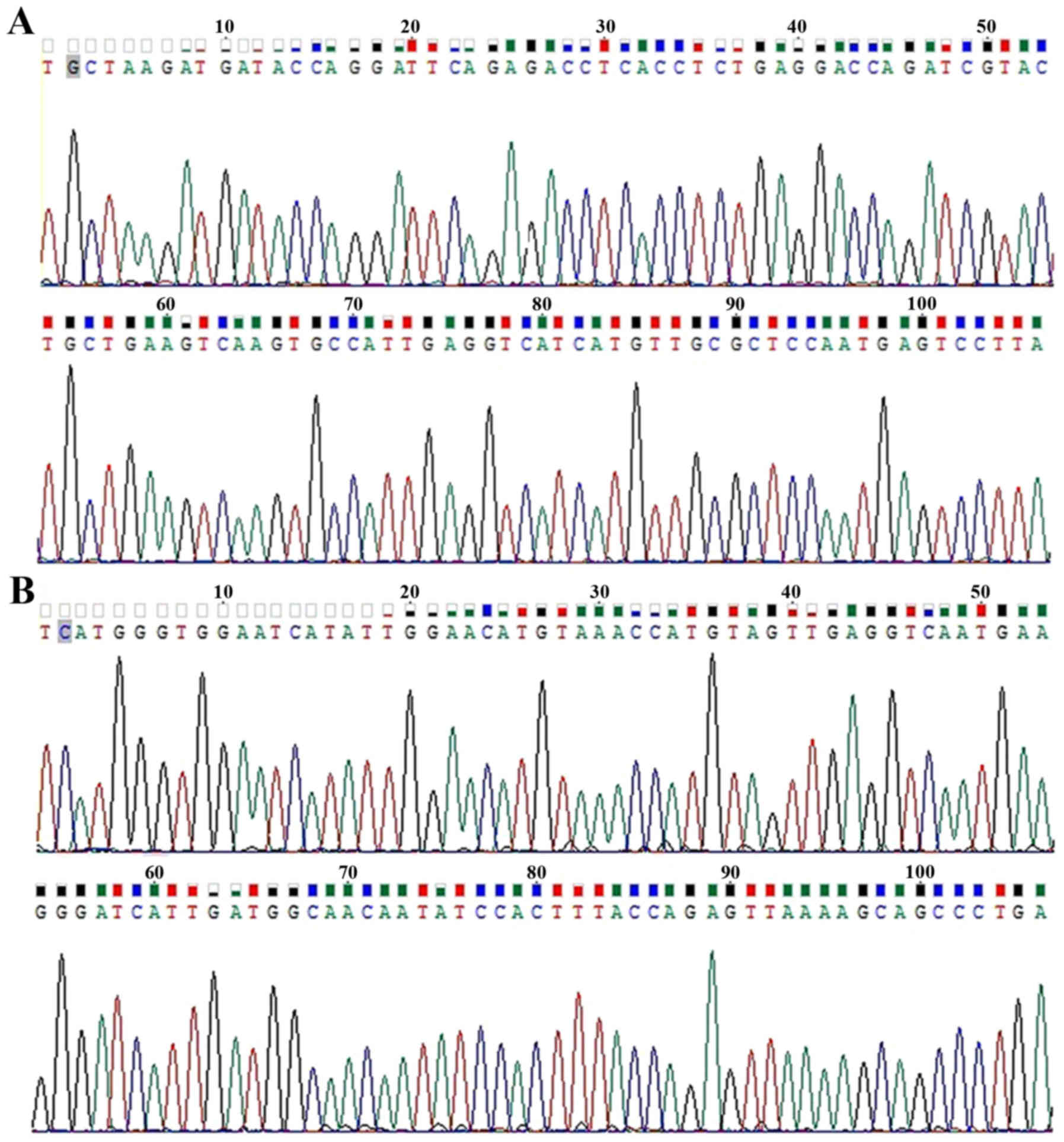
Sequence analysis of recombinant
plasmid
The sequence alignment indicated that the sequence
of VDR (Fig. 1A) and GAPDH
(Fig. 1B) recombinant plasmids
completely matched with their respective gene sequences, which
confirmed that the amplification products were the specific
fragments of VDR and GAPDH, respectively.
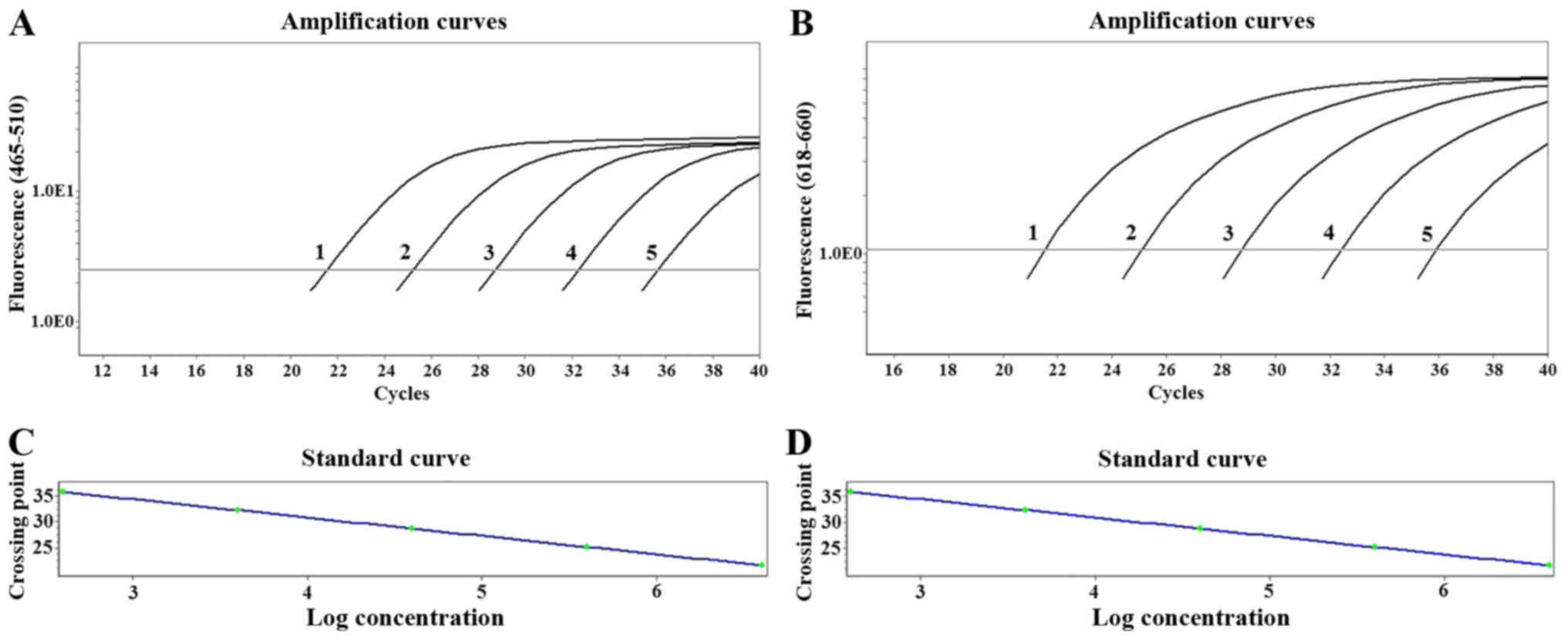
Amplification curve of VDR/GAPDH and
sensitivity analysis
The results indicated that a typical amplification
curve of VDR gene was present in the FAM (465–510 nm) channels
(Fig. 2A), and that of GAPDH gene
was produced in the CY5 (618–660 nm) channels (Fig. 2B). Amplification of 10-fold
serially diluted VDR/GAPDH plasmid mixture indicated that the
sensitivity of this method was 4×101 copies/µl.
Establishment of standard curve and
linearity range
Standard curves were generated from serial dilutions
of plasmid cDNA standards for VDR and GAPDH. The standard curve
equation of VDR and GAPDH amplification were Y=−3.518X+41.18
(Fig. 2C), and Y=−3.369X+41.50
(Fig. 2D), respectively, where Y
represented Cq and X stood for the logarithm of initial copy
number. The linearity range of the standard curve was between 4×101
and 4×105 copies/µl. Standard curves for each plasmid cDNA standard
demonstrated linearity over the complete range of dilution series
with correlation coefficients of 0.999. The amplification
efficiency of VDR and GAPDH were 92.42% and 98.07%,
respectively.
Repeatability of duplex RT-qPCR
The amplification of VDR and GAPDH had a precise
intra- and inter-assay repeatability. The intra-assay coefficient
of variation (CV) [intra-assay CV] respectively were 0.09–0.34% and
0.19–0.43% (Table II).
Inter-assay CV respectively were 0.32–0.65% and 0.40–0.75%
(Table III). Data are presented
as the mean ± standard error.
 | Table II.Intra-batch repeatability of VDR/GAPDH
standards (mean ± SD). |
Table II.
Intra-batch repeatability of VDR/GAPDH
standards (mean ± SD).
|
| VDR | GAPDH |
|---|
|
|
|
|
|---|
| Standards | Mean ± SD | CV (%) | Mean ± SD | CV (%) |
|---|
|
4.0×105 | 21.84±0.04 | 0.18 | 21.92±0.07 | 0.32 |
|
4.0×104 | 25.20±0.03 | 0.12 | 25.32±0.11 | 0.43 |
|
4.0×103 | 28.67±0.04 | 0.14 | 28.82±0.04 | 0.14 |
|
4.0×102 | 32.21±0.03 | 0.09 | 32.20±0.06 | 0.19 |
|
4.0×101 | 35.47±0.12 | 0.34 | 35.61±0.10 | 0.28 |
 | Table III.Inter-batch repeatability of
VDR/GAPDH standards (mean ± SD). |
Table III.
Inter-batch repeatability of
VDR/GAPDH standards (mean ± SD).
|
| VDR | GAPDH |
|---|
|
|
|
|
|---|
| Standards | Mean ± SD | CV (%) | Mean ± SD | CV (%) |
|---|
|
4.0×105 | 21.65±0.14 | 0.65 | 21.78±0.11 | 0.51 |
|
4.0×104 | 25.18±0.08 | 0.32 | 25.19±0.19 | 0.75 |
|
4.0×103 | 28.62±0.17 | 0.59 | 28.75±0.18 | 0.63 |
|
4.0×102 | 32.24±0.16 | 0.50 | 32.20±0.13 | 0.40 |
|
4.0×101 | 35.65±0.21 | 0.59 | 35.47±0.16 | 0.45 |
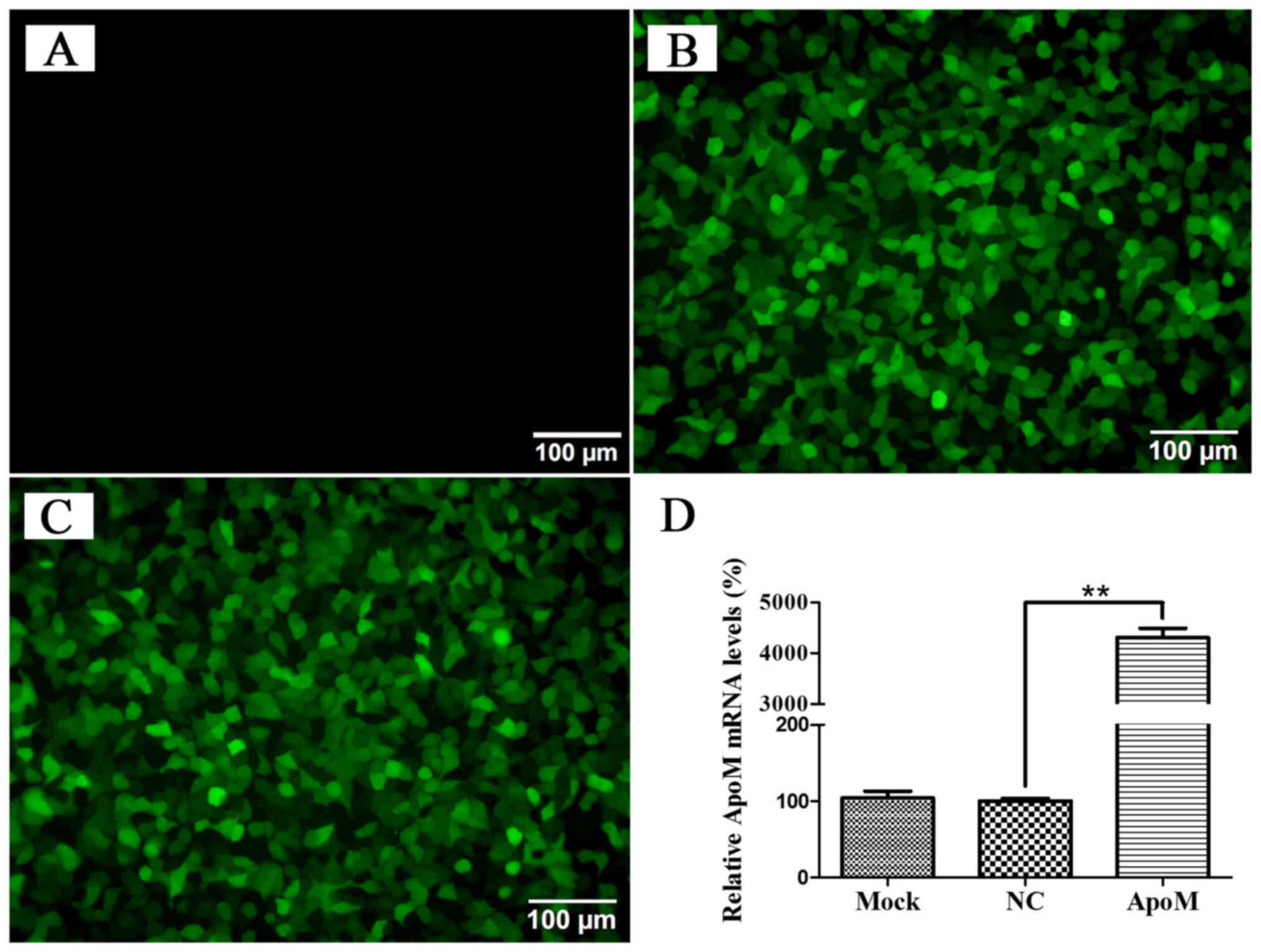
ApoM overexpression in HT-29
cells
To further study the biological role of ApoM in
colorectal cancer progression, HT-29 cells were transfected with
the GFP-labeled lentiviral vector carrying ApoM. As shown in
Fig. 3A-C, transfection efficiency
was >90% based on the percentages of GFP-positive cells after
transfection with a GFP expression vector. RT-qPCR was conducted in
order to detect the expression levels of ApoM in HT-29 cells of
each group. The PCR results indicated that the levels of ApoM mRNA
in the experimental group were significantly higher (~40 fold) than
that of the negative control group (Fig. 3D).
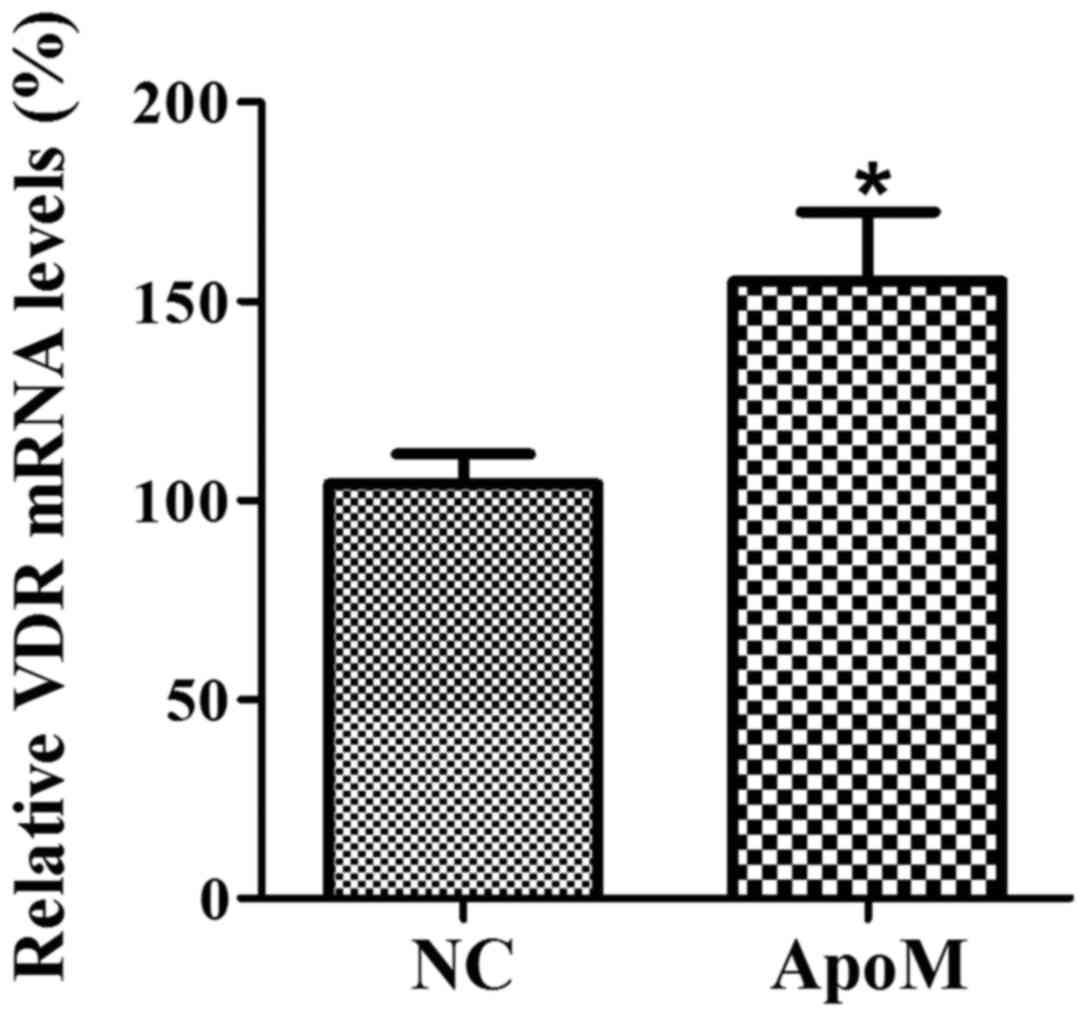
ApoM increases the expression of VDR
mRNA in HT-29 cells
In order to evaluate whether overexpression of ApoM
can affect the expression of VDR in HT-29 cells, the VDR mRNA
levels were detected by duplex RT-PCR. The results indicated that
ApoM can significantly increase the expression of VDR in HT-29
cells, compared with the negative control group (Fig. 4).
Discussion
RT-qPCR is known as the gold standard for accurate,
sensitive and fast quantification of nucleic acid sequences. It is
commonly used to analyze alterations of gene expression levels in
tumors, microbes and other disease states. However, data
normalization is a crucial step in gene quantification analysis.
Usually, the expression level of the target genes are normalized
using internal control genes known as reference genes, including
GAPDH, β-actin, 18S ribosomal RNA and β-2 microglobulin to derive
changes in gene expression levels (12). Traditionally, target and reference
genes are detected in two divided tubes, which is wasteful, time
consuming and may lead to experimental errors in the cDNA addition
step (13,14). In the developed duplex qPCR used in
the present study, the two gene expression levels could be
determined simultaneously in a single tube, which effectively
improved the efficiency of amplification and eliminated errors in
sample addition steps. The methodology used produced high
sensitivity, repeatability and amplification efficiency intra- and
inter-assay. The linearity range of the standard curve was between
4×101 and 4×105 copies/µl.
In the methods used in the current study, the design
and selection of primers were crucial to obtain the desired results
of PCR. The ideal qPCR primers and probes design should consider
the following points: i) All annotated splice variants of each gene
should be taken into account when the biological function of
individual gene splice variants remain to be understood (15). For example, VDR has three splice
variants while GAPDH has four. The primers and probes of VDR and
GAPDH were all designed in the common sequence of all splice
variants; ii) primers are required to span exons to avoid
amplification of contaminating genomic DNA; iii) a similarity
search was used to ensure the specificity of each primer (16). All the above points are important
for the method to obtain accurate and optimal results.
ApoM is a lipoprotein-associated plasma protein of
the apolipoprotein family. ApoM protects against atherosclerosis
primarily via partaking in pre-β-HDL formation and promoting
cholesterol efflux to HDL (17,18).
The process of atherosclerosis is always accompanied by an
inflammatory reaction (3). As the
human ApoM gene is located in the major histocompatibility complex
class III region on chromosome 6, with multiple immune and
inflammatory response-associated genes, it is suggested be involved
in immune response (19). The
anti-inflammatory effect of ApoM is regarded as one pathway of its
atheroprotective. Additionally, it has been previously reported
that ApoM is abnormally expressed in hepatocellular carcinoma and
the colorectal cancer tissues (5),
which indicates that ApoM may participate in tumor regulation.
However, the role and regulatory mechanism for ApoM in tumor
progression remains to be further elucidated.
VDR functions as a transcription factor that
regulates various biological processes including the regulation of
proliferation, differentiation, apoptosis, angiogenesis, immunity
and miRNAs (9). Studies have
reported an antineoplastic effect of VDR, particularly in
colorectal cancer; VDR expression is increased in
well-differentiated or moderately differentiated colorectal cancer
tissues, however is decreased in poorly differentiated tumors
(20), and high VDR expression is
correlated with an advantageous prognosis in colorectal patients
(21). In addition, studies in
different animal models and cell lines of colorectal cancer support
the antineoplastic effect of VDR via various mechanisms (22–24).
A previous study indicated that mRNA and protein
levels of ApoM are significantly reduced in the colorectal cancer
tissues when compared with their matched adjacent normal tissues.
From this result, it was hypothesized that ApoM may serve an
anti-tumor role in colorectal cancer. In the present study, it was
demonstrated that overexpression of ApoM could significantly
increase the expression of VDR mRNA in HT-29 cells. In addition,
associated studies have confirmed that VDR has antineoplastic
effect (25–27). This may suggest that ApoM serves an
antineoplastic role during the progress of the colorectal cancer by
upregulating VDR expression. As chronic inflammation is regarded as
a risk factor for the development of cancer, the suppression of
inflammation by ApoM and VDR may contribute to their antineoplastic
activity. It is suggested that the duplex RT-qPCR used in the
present study could ensure objectivity and credibility.
In conclusion, a single-tube duplex RT-qPCR to
simultaneously detect VDR and GAPDH expression in colorectal cancer
cells was successfully developed. The methodology results
demonstrated that the duplex RT-qPCR with high sensitivity and
specificity could ensure the accuracy of detection. Subsequently,
it was identified that ApoM significantly increased the expression
of VDR in HT-29 cells. In addition, it was suggested that ApoM may
be involved in antineoplastic activity via upregulation of VDR
expression, which provided novel directions for the investigation
of ApoM in cancer.
Acknowledgements
The present study was supported by the Changzhou
High-Level Medical Talents Training Project (grant no.
2016ZCLJ002).
References
|
1
|
Xu N and Dahlbäck B: A novel human
apolipoprotein (apoM). J Biol Chem. 274:31286–31290. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wolfrum C, Poy MN and Stoffel M:
Apolipoprotein M is required for prebeta-HDL formation and
cholesterol efflux to HDL and protects against atherosclerosis. Nat
Med. 11:418–422. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang XS, Zhao SP, Hu M and Luo YP:
Apolipoprotein M likely extends its anti-atherogenesis via
anti-inflammation. Med Hypotheses. 69:136–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arkensteijn BW, Berbée JF, Rensen PC,
Nielsen LB and Christoffersen C: The apolipoprotein
m-sphingosine-1-phosphate axis: Biological relevance in lipoprotein
metabolism, lipid disorders and atherosclerosis. Int J Mol Sci.
14:4419–4431. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo G, Zhang X, Mu Q, Chen L, Zheng L, Wei
J, Berggren-Söderlund M, Nilsson-Ehle P and Xu N: Expression and
localization of apolipoprotein M in human colorectal tissues.
Lipids Health Dis. 9:1022010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gonzalez-Parra E, Rojas-Rivera J, Tuñón J,
Praga M, Ortiz A and Egido J: Vitamin D receptor activation and
cardiovascular disease. Nephrol Dial Transplant. 27 Suppl
4:iv17–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Christakos S, Dhawan P, Verstuyf A,
Verlinden L and Carmeliet G: Vitamin D: Metabolism, molecular
mechanism of action, and pleiotropic effects. Physiol Rev.
96:365–408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li S, He Y, Lin S, Hao L, Ye Y, Lv L, Sun
Z, Fan H, Shi Z, Li J, et al: Increase of circulating cholesterol
in vitamin D deficiency is linked to reduced vitamin D receptor
activity via the Insig-2/SREBP-2 pathway. Mol Nutr Food Res.
60:798–809. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dou R, Ng K, Giovannucci EL, Manson JE,
Qian ZR and Ogino S: Vitamin D and colorectal cancer: Molecular,
epidemiological and clinical evidence. Br J Nutr. 115:1643–1660.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feldman D, Krishnan AV, Swami S,
Giovannucci E and Feldman BJ: The role of vitamin D in reducing
cancer risk and progression. Nat Rev Cancer. 14:342–357. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nurmi J, Ylikoski A, Soukka T, Karp M and
Lövgren T: A new label technology for the detection of specific
polymerase chain reaction products in a closed tube. Nucleic Acids
Res. 28:E282000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Angelone-Alasaad S, Min A Molinar,
Pasquetti M, Alagaili AN, D'Amelio S, Berrilli F, Obanda V, Gebely
MA, Soriguer RC and Rossi L: Universal conventional and real-time
PCR diagnosis tools for Sarcoptes scabiei. Parasit Vectors.
8:5872015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parker J, Fowler N, Walmsley ML, Schmidt
T, Scharrer J, Kowaleski J, Grimes T, Hoyos S and Chen J:
Analytical Sensitivity Comparison between Singleplex Real-Time PCR
and a Multiplex PCR Platform for Detecting Respiratory Viruses.
PLoS One. 10:e01431642015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shen Z: Effect of reaction tube on the
accuracy of polymerase chain reaction. J Prac Medi Tech.
17:142–143. 2010.
|
|
15
|
Gubelmann C, Gattiker A, Massouras A, Hens
K, David F, Decouttere F, Rougemont J and Deplancke B: GETPrime: A
gene- or transcript-specific primer database for quantitative
real-time PCR. Database (Oxford). 2011:bar0402011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Spandidos A, Wang H and Seed B:
PrimerBank: A PCR primer database for quantitative gene expression
analysis, 2012 update. Nucleic Acids Res. 40(Database issue):
D1144–D1149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang LZ, Gao JL, Pu C, Zhang PH, Wang LZ,
Feng G and Zhang Y: Apolipoprotein M: Research progress, regulation
and metabolic functions (Review). Mol Med Rep. 12:1617–1624.
2015.PubMed/NCBI
|
|
18
|
Elsøe S, Christoffersen C, Luchoomun J,
Turner S and Nielsen LB: Apolipoprotein M promotes mobilization of
cellular cholesterol in vivo. Biochim Biophys Acta. 1831:1287–1292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo G, Zhang X, Nilsson-Ehle P and Xu N:
Apolipoprotein M. Lipids Health Dis. 3:212004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matusiak D, Murillo G, Carroll RE, Mehta
RG and Benya RV: Expression of vitamin D receptor and
25-hydroxyvitamin D3-1{alpha}-hydroxylase in normal and malignant
human colon. Cancer Epidemiol Biomarkers Prev. 14:2370–2376. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Milczarek M, Filip-Psurska B, Swiętnicki
W, Kutner A and Wietrzyk J: Vitamin D analogs combined with
5-fluorouracil in human HT-29 colon cancer treatment. Oncol Rep.
32:491–504. 2014.PubMed/NCBI
|
|
22
|
Elimrani I, Koenekoop J, Dionne S, Marcil
V, Delvin E, Levy E and Seidman EG: Vitamin D Reduces Colitis- and
Inflammation-Associated Colorectal Cancer in Mice Independent of
NOD2. Nutr Cancer. 69:276–288. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vidigal VM, Silva TD, de Oliveira J,
Pimenta CAM, Felipe AV and Forones NM: Genetic polymorphisms of
vitamin D receptor (VDR), CYP27B1 and CYP24A1 genes and the risk of
colorectal cancer. Int J Biol Markers. 32:e224–e230. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takada I and Makishima M: Control of
inflammatory bowel disease and colorectal cancer by synthetic
vitamin D receptor ligands. Curr Med Chem. Dec 2–2016.(Epub ahead
of print).
|
|
25
|
Yazdani S, Poosti F, Toro L, Wedel J,
Mencke R, Mirković K, de Borst MH, Alexander JS, Navis G, van Goor
H, et al: Vitamin D inhibits lymphangiogenesis through
VDR-dependent mechanisms. Sci Rep. 7:444032017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chiang KC, Yeh TS, Huang CC, Chang YC,
Juang HH, Cheng CT, Pang JS, Hsu JT, Takano M, Chen TC, et al:
MART-10 represses cholangiocarcinoma cell growth and high vitamin D
receptor expression indicates better prognosis for
cholangiocarcinoma. Sci Rep. 7:437732017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tavera-Mendoza LE, Westerling T, Libby E,
Marusyk A, Cato L, Cassani R, Cameron LA, Ficarro SB, Marto JA,
Klawitter J and Brown M: Vitamin D receptor regulates autophagy in
the normal mammary gland and in luminal breast cancer cells. Proc
Natl Acad Sci USA. 114:E2186–E2194. 2017. View Article : Google Scholar : PubMed/NCBI
|