Introduction
It is well established that joint function is
seriously impaired by cartilage defection, in fact even very small
defects in cartilage are difficult to repair due to the lack of
self-repair ability (1).
Mesenchymal stem cells (MSCs) are thought to be an ideal target for
repairing defective cartilage on account of their self-repair
ability, multidirectional differentiation potential and low
immunogenicity (2). One type of
MSC line derived from mouse embryos known as the C3H10T1/2 cell
line has been widely applied in research that focused on the
differentiation and regulation of stem cells (3). Bone morphogenic protein 4 (BMP4)
treatment may induce C3H10T1/2 stem cells to commit to the
adipocyte lineage (4). In
addition, curculactones, A or B, increase the expression of
osteogenic marker genes and alkaline phosphatase activity, leading
to the differentiation of the mesenchymal cell line C3H10T1/2
(5). Our previous research
demonstrated that transforming growth factor β3 (TGF-β3) induced
C3H10T1/2 MSC line differentiation (6). Therefore, an in vitro
chondrogenic differentiation model for MSCs would be of value in
the promotion of MSC chondrogenic differentiation prior to
transplantation. However, the underlying mechanism requires further
clarification.
microRNA (miRNA or miR), a type of non-coding small
RNA ~22 nucleotides in length, is found extensively in eukaryons.
The biological functions of miRNA are diverse; for example, one
miRNA may modulate a number of different target genes and one
target gene may be coregulated by several miRNAs (1). miRNA has become an important tool in
genetic engineering as it is small, specific and endogenous. An
increasing body of evidence has demonstrated that miRNA can alter
stem cell fates and modulate the underlying epigenetics in
biological behavior (7–10). It has been reported that miR-29a is
essential for the differentiation of MSCs by regulating forkhead
box O3 expression (11).
Upregulation of 573–3p by SRY-Box 9 may inhibit retinoid X
receptor-a expression and chondrogenic differentiation of MSCs
(12). In addition, miR-144-3p
acts as a negative regulator of osteogenic differentiation and
proliferation in C3H10T1/2 cells and mothers against
decapentaplegic homolog 4 serves as its specific target gene
(13). miR-125b serves as a key
regulatory factor of osteoblastic differentiation by directly
targeting core-binding factor b in C3H10T1/2 cells (14). These results indicated that miRNAs
may take part in the process of C3H10T1/2 cell differentiation or
proliferation. A previous report revealed that microRNA-140
(miR-140) served an important role in BMP4-induced C3H10T1/2
pluripotent stem cells in achieving commitment to the adipocyte
lineage (15). miR-140
overexpression promotes this process, whereas knockdown miR-140
generates the opposite result by targeting the osteopetrosis
associated transmembrane protein 1 gene (15). BMP4 and TGF-β3 are also involved in
the process of C3H10T1/2 cell differentiation (6), however, whether miR-140 could serve
as a direct downstream component of TGF-β3 during C3H10T1/2 cell
differentiation requires further study. In the present study, the
effect of miR-140 in TGF-β3-induced C3H10T1/2 cell differentiation
is analyzed.
C-X-C motif chemokine 12 (CXCL12) is encoded by the
CXCL12 gene and belongs to the chemokine family. It can
active leukocytes and is often induced by proinflammatory stimuli
including lipopolysaccharides, tumor necrosis factors or
interleukin-1. CXCL12 is a widely expressed gene and has an
extensive role in muscle, neural, liver and tissue repair (16,17).
A previous study revealed that CXCL12 is a regulator of osteogenic
differentiation induced by BMP2 (18). In addition, CXCL12/CXCR4
significantly affected early and mid-osteogenic marker expression
in C3H10T1/2 cells (19). These
findings indicated that the CXCL12/CXCR4 signal axis effects the
BMP9-induced osteogenic differentiation of MSCs. Therefore, CXCL12
may serve an important role in osteogenic differentiation.
Currently, the number of studies investigating the miRNA regulation
of CXCL12 in C3H10T1/2 is relatively low. Thus, the present study
evaluated the underlying mechanism of how miR-140 regulates
expression.
The precise molecular mechanism underlying
chondrogenic differentiation in stem cells remains elusive. The aim
of the present study was to provide a theoretical foundation for
the role of potential miRNAs in C3H10T1/2 MSC.
Materials and methods
Cell culture and preparation of cell
pellets
C3H10T1/2 MSCs were purchased from American Type
Culture Collection (Manassas, VA, USA), cultured in low-glucose
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) and were then incubated at a
37°C and 95% humidity. Once 90% confluence was achieved, C3H10T1/2
MSCs were generated in a 1:6 dilution. The passage 7 generation of
C3H10T1/2 MSCs was digested with 0.25% trypsin (Invitrogen; Thermo
Fisher Scientific, Inc.) and divided into 2 groups: The non-induced
group (culture medium contained 500 ml high-glucose DMEM and 10 ml
FBS) and the TGF-β3-induced group (culture medium contained 500 ml
high-glucose DMEM, 5 µg TGF-β3, 0.15 g vitamin C, 50 µl
dexamethasone, 0.5 ml Insulin/Transferrin/Selenium solution and 10
ml FBS). Cells were suspended and reseeded in 2 ml DMEM at a
density of 2.5×105/ml. DMEM (1 ml) was replaced every 2 to 3 days
for ~14 days to generate in vitro pellets.
Verifying differentially expressed
miR-140 using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR)
RNA was reverse transcribed into cDNA using the
reverse transcriptase from the PrimeScript reverse transcription
reagent kit (Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's instructions. The qPCR reaction
mixture included SYBR Premix Ex Taq™ II (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) as well as 2 µl cDNA, 5 µl 2X master mix,
0.5 µl forward primer, 0.5 µl reverse primer and 2 µl nanopure
water to generate a final volume of 10 µl. Conditions for
amplification were as follows: Initial denaturation at 95°C for 30
sec, then 40 cycles of 10 sec at 95°C and 60 sec at 60°C, and final
extension at 72°C for 5 min. Experiments were performed with 3
replicates. The U6 gene was used as an endogenous control to
normalize differences in the amount of total RNA from each sample.
The primer sequences were as follows: Mus musculus
(mmu)-miR-140, forward 5′-GCGGCGGCAGTGGTTTTACCC-3′ and reverse
5′-ATCCAGTGCAGGGTCCGAGG-3′; and U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′CGC TTC ACG AAT TTG
CGT GTC AT-3′. The results were calculated using the
2−ΔΔCq method was performed using 30 µl Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. Cell transfection was performed
at room temperature.
Western blot analysis
To determine the expression profiles of
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and CXCL12,
C3H10T1/2 MSC extract lysates were collected and analyzed. Briefly,
samples were washed with ice-cold phosphate-buffered saline (PBS)
and homogenized in radioimmunoprecipitation lysis buffer at 4°C for
30 min which contained a cocktail of protease inhibitors and
phosphatase inhibitors (Roche Diagnostics, Shanghai, China), and
the lysates were collected following high-speed centrifugation
(12,700 × g) for 5 min at 4°C. Proteins were then separated by 10%
SDS-PAGE (Roche Diagnostics) and electrotransferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). Membranes were blocked in 5% bovine serum albumin
(Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature prior to overnight incubation at 4°C with α-tubulin
(dilution, 1:2,000; cat. no. ab7291; Abcam, Cambridge, UK) and
CXCL12 (dilution, 1:1,000; cat. no. 5712; Cell Signaling
Technology, Inc., Danvers, MA, USA) primary antibodies. Following
washing with TBST (TBS with 20% Tween-20), membranes were incubated
with the corresponding rabbit anti-mouse HRP-conjugated IgG
(dilution, 1:2,000; cat. no. A16160; Thermo Fisher Scientific,
Inc.) for 1 h at room temperature. Target proteins were visualized
using the SuperSignal® West Pico Chemiluminescent Substrate (Thermo
Fisher Scientific, Inc.) and α-tubulin was used as a loading
control.
MTT assay
C3H10T1/2 MSCs were transfected with miR-NC mimics,
miR-140 mimics, miR-NC inhibitor or miR-140 inhibitor, followed by
TGF-β3 induction. After 1 and 7 days, cell viability was measured
by MTT assay. Cells (~200 µl at 1×104/ml) were seeded into 96 well
plates. Following 24 h incubation, 20 µl of 5 mg/ml MTT solution
was added to each well and the plate was further incubated at 37°C
for 4 h. The medium was then aspirated, the wells were washed with
PBS and allowed to dry for ~4 h, and then 150 µl DMSO was added to
each well. The microtitre plate was placed on a shaker in order to
dissolve the dye. Absorbance was read at 490 nm using a Bio-Rad
iMark plate reader (Bio-Rad Laboratories, Inc.).
Alcian blue staining
Cell pellets were fixed with 4% paraformaldehyde in
0.1 M PBS for 20 min at 37°C and then rinsed with PBS three times.
The pellets were dehydrated using a gradient of ethanol (30, 50,
70, 85, 95 and 100%), placed in dimethylbenzene for 5 min, embedded
with paraffin and sectioned into 4 µm thick slices. These sections
were stained with 1% alcian blue dissolved in 3% acetic acid (ph
2.5) for 10 min at 37°C to quantify glycosaminoglycan (GAG)
synthesis. Sections were then washed with water three times and
sealed with dry neutral resins. Images were captured on a light
microscope (Nikon Eclipse Ci; Nikon Corporation, Tokyo, Japan), and
analyzed using Photoshop software CS6 (Adobe CS5; Adobe Systems,
Inc., San Jose, CA, USA).
Bioinformatic and statistical
analysis
Prediction of miRNA target sites was performed using
TargetScan (http://www.targetscan.org) and
microRNA.org (http://www.microrna.org). Differences among groups
were analyzed by performing a two-way analysis of variance,
followed by Bonferroni post hoc tests using the statistical
software IBM SPSS version 21 (IBM Corp., Armonk, NY, USA). Data are
presented as the mean ± standard error mean of 3 repeated
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
Verification of miR-140 expression
levels using RT-qPCR
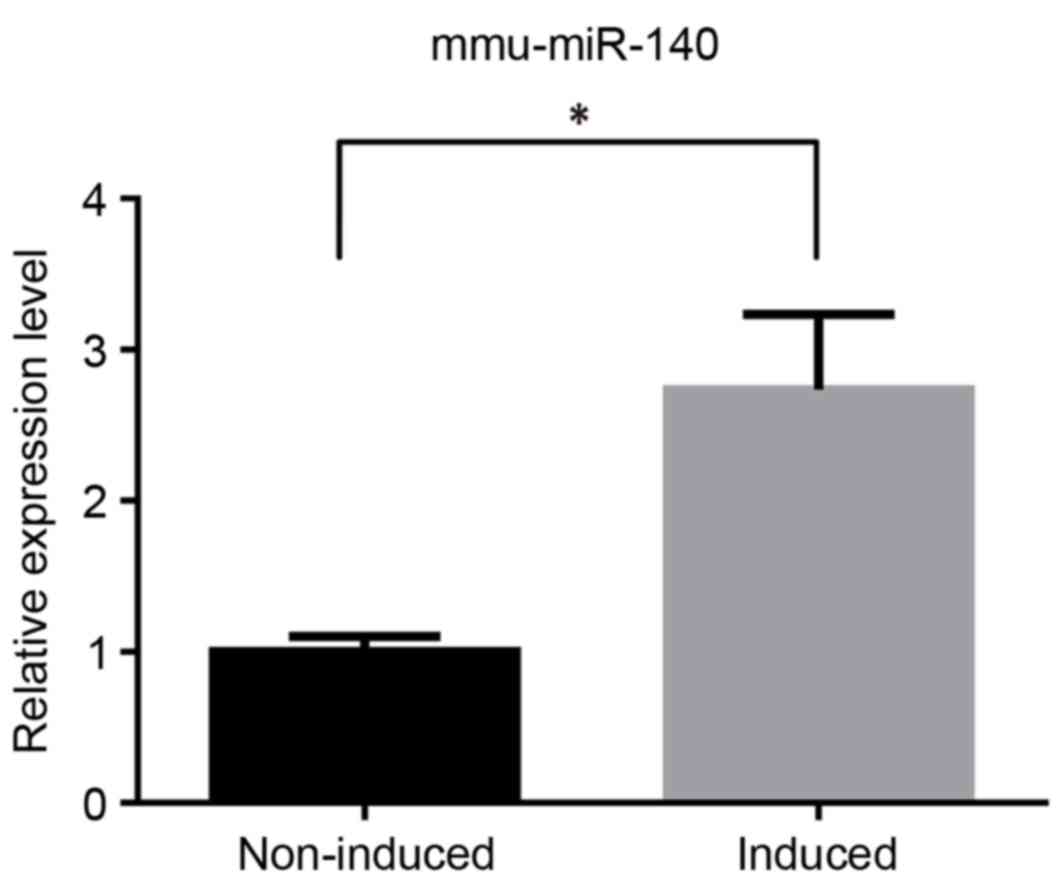
mmu-miR-140 was analyzed using RT-qPCR to determine
its different expression levels. The results of this analysis were
consistent with those observed in our microarray analysis performed
previously (data not published). In the TGF-β3-induced group,
mmu-miR-140 was significantly upregulated compared with the
non-induced group (Fig. 1).
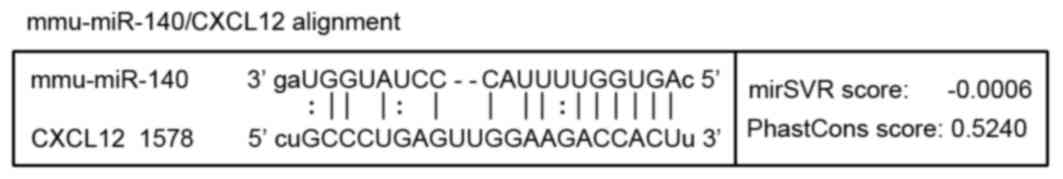
Target action site of miR-140 on
CXCL12 prediction via bioinformatics
TargetScan (http://www.targetscan.org) and microRNA.org (http://www.microrna.org) databases were used to
predict target miRNAs and cartilage-associated target genes. Genes
that exhibited relatively higher grades in the two databases were
selected as key genes. In our previous study, miR-140 was
identified as one of the miRNAs with an altered expression in the
TGF-β3-induced chondrogenic group (data not published). Thus, in
combination with the results of the microarray analysis, miR-140
was chosen as the target of the present study. It was predicted
that miR-140 may recognize and interact with the 3′ untranslated
region (3′UTR) of CXCL12 as presented in Fig. 2.
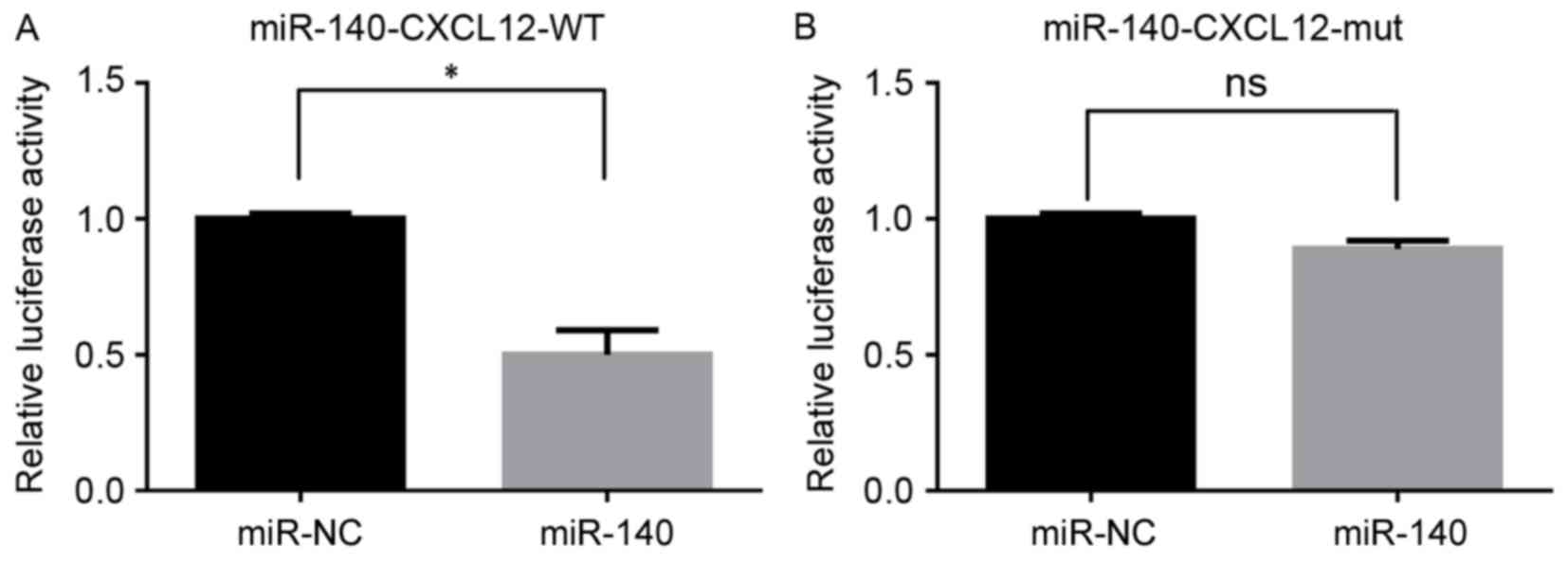
Interaction between miR-140 and the
predicted region of CXCL12
The dual luciferase reporter assay demonstrated that
miR-140 mimics significantly inhibited the fluorescence of the
CXCL12-WT group when compared with the miR-NC group (P<0.05;
Fig. 3A). However, there was no
significant difference in fluorescence between the CXCL12-mutant
(mut) group and miR-NC group (P>0.05; Fig. 3B).
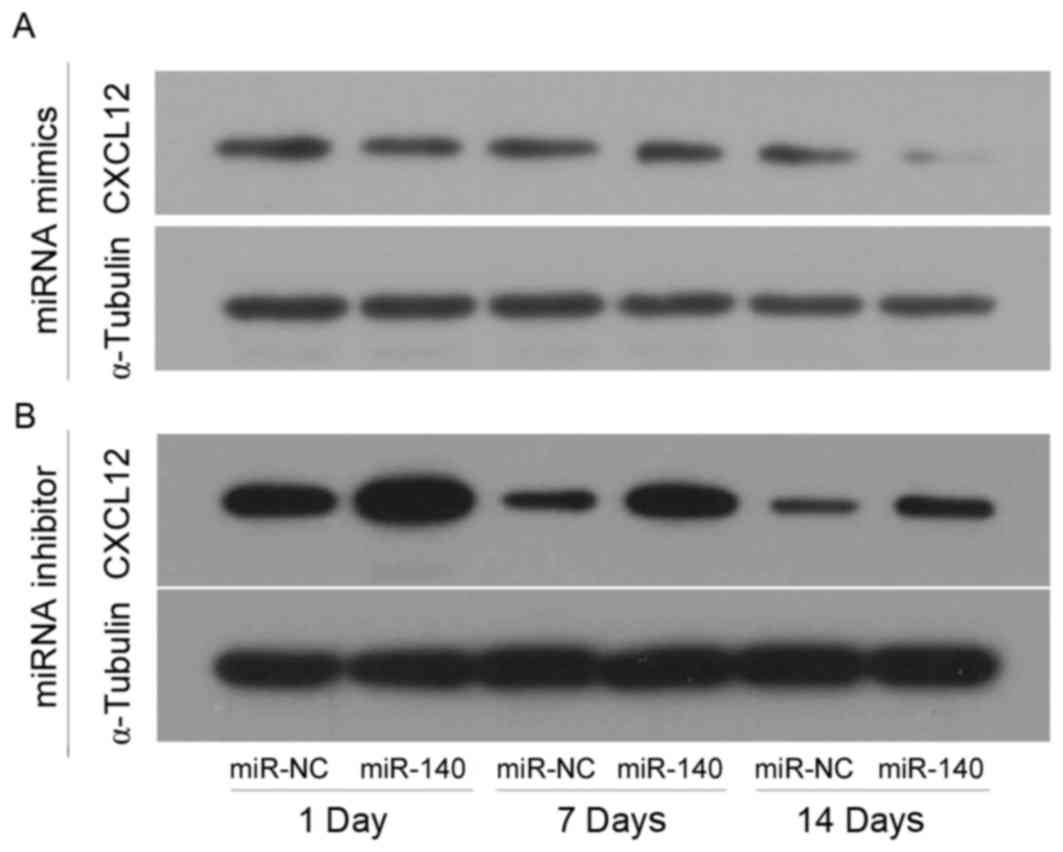
CXCL12 protein levels in the different
groups analyzed by western blotting
C3H10T1/2 MSCs were transfected with miR-140 mimics
or inhibitor in the experimental groups, and transfected with
miR-NC mimics or inhibitor in control group, and chondrogenic
differentiation was induced by TGF-β3. CXCL12 protein levels were
assessed by western blotting at 1, 7 and 14 days following TGF-β3
induction (Fig. 4). CXCL12 was
markedly decreased when miR-140 was overexpressed compared with NC
(Fig. 4A), however, it was
markedly increased when miR-140 expression was inhibited compared
with NC (Fig. 4B). In addition,
CXCL12 expression levels declined as the length of the experiment
increased in the control and experimental groups (Fig. 4).
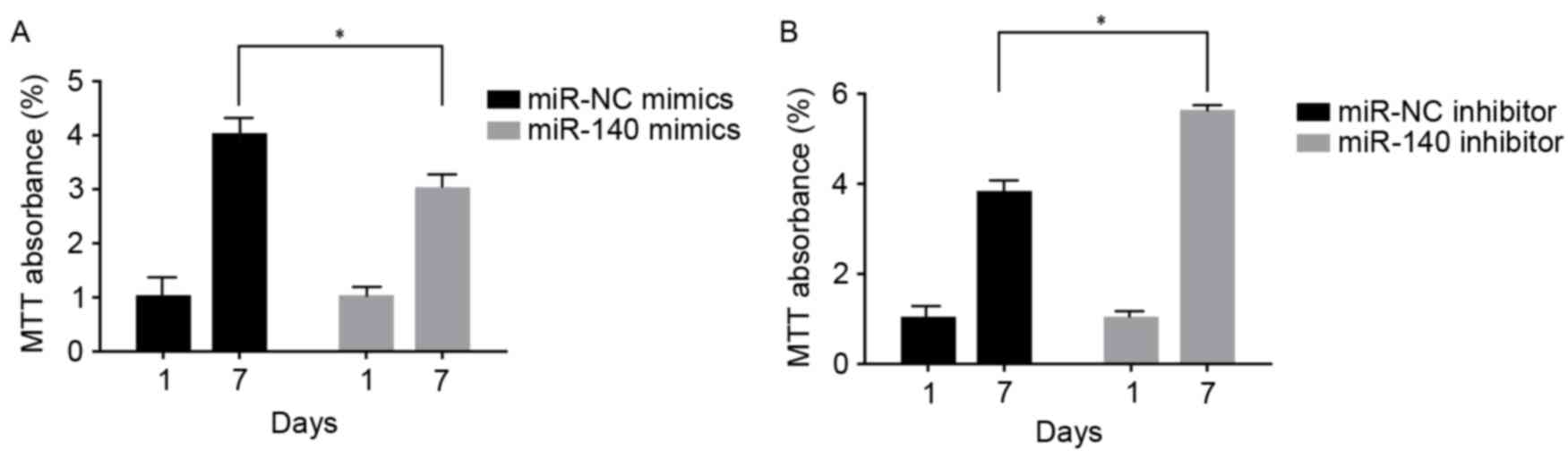
C3H10T1/2 MSC cell viability following
up-/downregulation of miR-140
At 1 and 7 days following transfection and TGF-β3
induction, C3H10T1/2 MSCs transfected with miR-140 mimics exhibited
significant inhibition of cell viability compared with NC
(P<0.05; Fig. 5A). However,
those transfected with the miR-140 inhibitor exhibited a
significant increase in cell viability compared with the control
group (P<0.05; Fig. 5B).
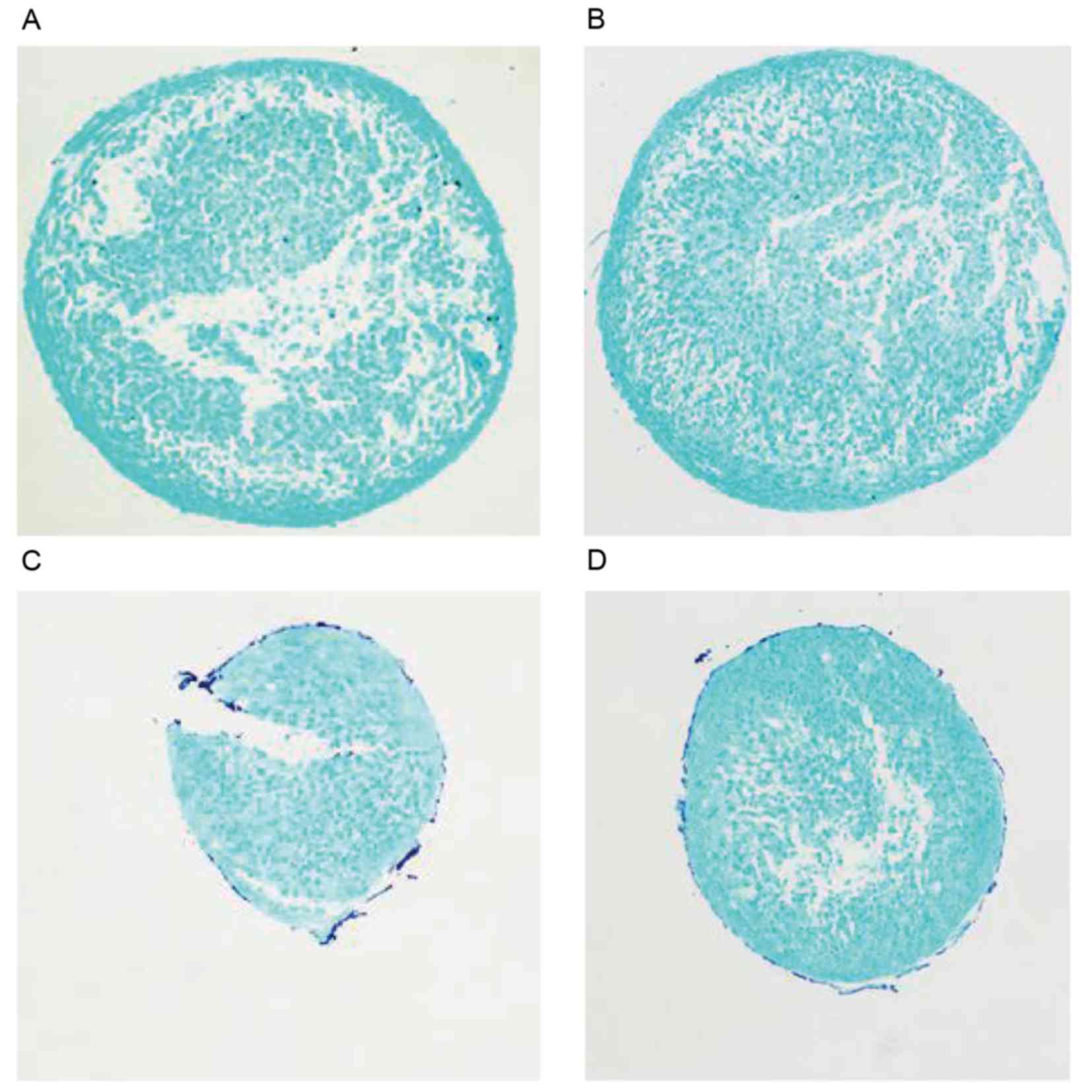
Analysis of GAG synthesis using alcian
blue staining
In cell pellets generated by chondrogenic
differentiation following the induction of C3H10T1/2 for 7 days,
there was no visible difference in alcian blue staining between the
miR-140 up-/downregulation and control groups (Fig. 6).
Discussion
Microarray analysis demonstrated that, in the
TGF-β3-induced group, miR-140 expression levels were significantly
higher than those observed in the control group. Further analysis
using bioinformatics prediction and a dual luciferase assay
validated that CXCL12 was the target gene of miR-140. C3H10T1/2
MSCs were transfected with miR-140 mimics or miR-NC mimics,
followed by TGF-β3 induction. Western blotting after 1, 7 and 14
days revealed that CXCL12 was markedly downregulated as a result of
miR-140 overexpression. By contrast, CXCL12 was significantly
upregulated when miR-140 was inhibited. In addition, the expression
of CXCL12 decreased as the length of the experiment increased.
Following TGF-β3 induction, at 1 and 7 days after transfection,
C3H10T1/2 MSCs transfected with miR-140 mimics exhibited reduced
cell viability, whereas, those transfected with the miR-140
inhibitor displayed increased cell viability when compared with the
control groups. In cell pellets generated by the induction of
C3H10T1/2 MSCs for 7 days, no visible difference in GAG synthesis
was observed between the miR-140 up-/downregulation and control
groups.
To the best of our knowledge, these results
demonstrate for the first time that during the process of
TGF-β3-induced chondrogenic differentiation, miR-140 inhibits
C3H10T1/2 MSC viability by targeting CXCL12, however, there it has
no significant influence on C3H10T1/2 MSC differentiation.
As a member of non-coding RNA family, which are a
type of endogenous regulatory RNA, miRNA consists of ~21 to 23
nucleotides. An increasing body of evidence has indicated that
miRNA may participate in a number of physiological and pathological
processes, including development, cell differentiation, metabolism
and cancer (9,10,20,21).
miR-140 is expressed in cartilage tissue and serves an important
role in proliferation and differentiation (22,23).
In the present study, microarray analysis and RT-qPCR demonstrated
that miR-140 is upregulated in TGF-β3-induced C3H10T1/2 MSCs.
Following this result, the aim was to further clarify the
biological role and target gene of miR-140.
Firstly, online software was used to predict the
target gene of miR-140 and the interaction site between miR-140 and
its target gene. A dual luciferase reporter assay was performed to
validate that miR-140 could recognize and interact with
CXCL12-3′UTR. As the target gene of miR-140, CXCL12 is also known
as stromal cell-derived factor-1 (SDF-1) (24). CXCL12 has been reported to serve a
number of biological functions, including bone-marrow myelopoiesis
(25), organogenesis and
tumorigenesis (26). Kanbe et
al (27) indicated that SDF-1
may participate in the destruction of cartilage in osteoarthritis
and rheumatoid arthritis. The present study further investigated
whether the level of CXCL12 expression was affected by miR-140
activation/inhibition in TGF-β3-induced C3H10T1/2 MSCs.
Based on our previous study of cell pellets
generated from TGF-β3-induced chondrogenic differentiation in
C3H10T1/2 MSCs (6), it was
revealed that CXCL12 was markedly decreased when miR-140 is
overexpressed, however, it increased when miR-140 expression was
inhibited. In addition, CXCL12 expression levels declined as the
length of experiment increased in the control and experimental
groups. The results of the present study combined with those of our
previous study (6), suggest that
miR-140 may take part in the formation of cell pellets by
regulating CXCL12. However, it is unknown which process miR-140
regulates, C3H10T1/2 MSC proliferation or differentiation.
In the present study C3H10T1/2 MSC viability was
detected, and the results revealed that following transfection with
miR-140 mimics, it was significantly reduced when compared with the
miR-NC group. However, C3H10T1/2 MSC proliferation was
significantly promoted following miR-140 mimics transfection.
Alcian blue staining identifies the characteristic extracellular
matrix GAGs secreted by chondrocytes (28). In the present study, GAG synthesis
was also measured by alcian blue staining and no visible difference
in GAG synthesis was observed between the groups with
overexpression or inhibition of miR-140 at 7 days following
TGF-β3-induced chondrogenic differentiation.
The present study investigated the biological
effects of miRNA-140 on CXCL12 and C3H10T1/2 MSC
proliferation/differentiation based on our previous study (6). Collectively, the results indicate
that prior to progression in TGF-β3-induced chondrogenesis,
C3H10T1/2 MSC proliferation may be effectively upregulated by
promoting CXCL12, which could be induced by inhibiting miRNA-140
expression. The results highlight the potential role of miRNA-140
in future clinical therapies to treat cartilage defection via
CXCL12 targeting. Future studies will further investigate the
function and precise regulatory mechanism of miRNA-140.
Acknowledgements
The present study was financially supported by the
National Basic Research Program of China (973 Program; grant no.
2012CB619105).
References
|
1
|
Redman SN, Oldfield SF and Archer CW:
Current strategies for articular cartilage repair. Eur Cell Mater.
9:23–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xian CJ and Foster BK: Repair of injured
articular and growth plate cartilage using mesenchymal stem cells
and chondrogenic gene therapy. Curr Stem Cell Res Ther. 1:213–229.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ji YH, Ji JL, Sun FY, Zeng YY, He XH, Zhao
JX, Yu Y, Yu SH and Wu W: Quantitative proteomics analysis of
chondrogenic differentiation of C3H10T1/2 mesenchymal stem cells by
iTRAQ labeling coupled with on-line two-dimensional LC/MS/MS. Mol
Cell Proteomics. 9:550–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang QQ, Otto TC and Lane MD: Commitment
of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc
Natl Acad Sci USA. 101:9607–9611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Son HE, Kim TH and Jang WG: Curculactones
A and B induced the differentiation of C3H10T1/2 and MC3T3-E1 cells
to osteoblasts. Bioorg Med Chem Lett. 27:1301–1303. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wa Q, Gao M, Dai X, Yu T, Zhou Z, Xu D and
Zou X: Induction of chondrogenic differentiation of mouse embryonic
mesenchymal stem cells through an in vitro pellet model. Cell Biol
Int. 39:657–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Megosh HB, Cox DN, Campbell C and Lin H:
The role of PIWI and the miRNA machinery in Drosophila germline
determination. Curr Biol. 16:1884–1894. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park JK, Liu X, Strauss TJ, McKearin DM
and Liu Q: The miRNA pathway intrinsically controls self-renewal of
Drosophila germline stem cells. Curr Biol. 17:533–538. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Medvid R, Melton C, Jaenisch R and
Blelloch R: DGCR8 is essential for microRNA biogenesis and
silencing of embryonic stem cell self-renewal. Nat Genet.
39:380–385. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guerit D, Brondello JM, Chuchana P,
Philipot D, Toupet K, Bony C, Jorgensen C and Noël D: FOXO3A
regulation by miRNA-29a Controls chondrogenic differentiation of
mesenchymal stem cells and cartilage formation. Stem Cells Dev.
23:1195–1205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guerit D, Philipot D, Chuchana P, Toupet
K, Brondello JM, Mathieu M, Jorgensen C and Noël D: Sox9-regulated
miRNA-574-3p inhibits chondrogenic differentiation of mesenchymal
stem cells. PLoS One. 8:e625822013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang C, Geng J, Wei X, Zhang R and Jiang
S: MiR-144-3p regulates osteogenic differentiation and
proliferation of murine mesenchymal stem cells by specifically
targeting Smad4. FEBS Lett. 590:795–807. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang K, Fu J, Zhou W, Li W, Dong S, Yu S,
Hu Z, Wang H and Xie Z: MicroRNA-125b regulates osteogenic
differentiation of mesenchymal stem cells by targeting Cbfβ in
vitro. Biochimie. 102:47–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Zhang ZC, Qian SW, Zhang YY, Huang
HY, Tang Y, Guo L, Li X and Tang QQ: MicroRNA-140 promotes
adipocyte lineage commitment of C3H10T1/2 pluripotent stem cells
via targeting osteopetrosis-associated transmembrane protein 1. J
Biol Chem. 288:8222–8230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schrader AJ, Lechner O, Templin M, Dittmar
KE, Machtens S, Mengel M, Probst-Kepper M, Franzke A, Wollensak T,
Gatzlaff P, et al: CXCR4/CXCL12 expression and signalling in kidney
cancer. Br J Cancer. 86:1250–1256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Blanchet X, Langer M, Weber C, Koenen RR
and von Hundelshausen P: Touch of chemokines. Front Immunol.
3:1752012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hosogane N, Huang Z, Rawlins BA, Liu X,
Boachie-Adjei O, Boskey AL and Zhu W: Stromal derived factor-1
regulates bone morphogenetic protein 2-induced osteogenic
differentiation of primary mesenchymal stem cells. Int J Biochem
Cell Biol. 42:1132–1141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu C, Weng Y, Yuan T, Zhang H, Bai H, Li
B, Yang D, Zhang R, He F, Yan S, et al: CXCL12/CXCR4 signal axis
plays an important role in mediating bone morphogenetic protein
9-induced osteogenic differentiation of mesenchymal stem cells. Int
J Med Sci. 10:1181–1192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pfeffer S and Voinnet O: Viruses,
microRNAs and cancer. Oncogene. 25:6211–6219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miyaki S, Nakasa T, Otsuki S, Grogan SP,
Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK and Asahara H:
MicroRNA-140 is expressed in differentiated human articular
chondrocytes and modulates interleukin-1 responses. Arthritis
Rheum. 60:2723–2730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swingler TE, Wheeler G, Carmont V, Elliott
HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP,
Hajihosseini MK, Münsterberg A, et al: The expression and function
of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum.
64:1909–1919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karakus S, Bagci B, Bagci G, Sancakdar E,
Yildiz C, Akkar O and Cetin A: SDF-1/CXCL12 and CXCR4 gene
variants, and elevated serum SDF-1 levels are associated with
preeclampsia. Hypertens Pregnancy. 1-7:2016.(Epub ahead of
print).
|
|
25
|
Nagasawa T, Hirota S, Tachibana K,
Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H and
Kishimoto T: Defects of B-cell lymphopoiesis and bone-marrow
myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature.
382:635–638. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ratajczak MZ, Zuba-Surma E, Kucia M, Reca
R, Wojakowski W and Ratajczak J: The pleiotropic effects of the
SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis.
Leukemia. 20:1915–1924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanbe K, Takemura T, Takeuchi K, Chen Q,
Takagishi K and Inoue K: Synovectomy reduces stromal-cell-derived
factor-1 (SDF-1) which is involved in the destruction of cartilage
in osteoarthritis and rheumatoid arthritis. J Bone Joint Surg Br.
86:296–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Björnsson S: Simultaneous preparation and
quantitation of proteoglycans by precipitation with alcian blue.
Anal Biochem. 210:282–291. 1993. View Article : Google Scholar : PubMed/NCBI
|