Introduction
RNA interference (RNAi) is an evolutionarily
conserved mechanism for silencing specific genes (1). The process of RNAi can be moderated
by either microRNA (miRNA) or small interfering RNA (siRNA). miRNA
and siRNA are processed inside the cell by the enzyme, Drosha,
Dicerand a complex called RNA-induced silencing complex (RISC)
(2–6). The Argonaute (Ago) family of proteins
are essential components of RISC, which are involved in mRNA
cleavage. Ago2 is the only enzyme conferring this activity in
mammals (7). RNAi molecules, which
target the degradation of mRNA have the ability to alter cellular
pathways and events. RNAi is a useful tool in the treatment of
diseases and functional investigations of genes. Furthermore,
targeting specific genes using RNAi molecules has been applied in
preclinical studies (8,9).
RA is a chronic inflammatory autoimmune disorder,
which is high in prevalence and characterized by persistent
synovitis and systemic inflammation, often leading to other serious
complications. RA can finally lead to joint destruction and
functional disability, and patients may succumb to mortality
(10,11). However, the pathogenesis of RA
remains to be fully elucidated and there are no satisfactory
therapeutic strategies to cure this disease (12,13).
Alterations of miRNAs in RA have been reported previously. Several
aberrantly expressed miRNAs have been found to contribute to
various aspects of the pathogenesis of RA and may have applications
in biotherapeutic approaches for the diagnosis and treatment of RA
(14). For example, miRNA
(miR)-24, miR-26a, miR-125a-5p and miR-323-3p are increased in RA,
indicating that these miRNAs may be RA biomarkers (15,16).
miR-146a is significantly upregulated in RA, and is associated with
the level of tumor necrosis factor (TNF)-α and disease activity
(17). miR-19 can regulate the
expression of Toll-like receptor 2 in rheumatoid fibroblast-like
synoviocytes (18). Of note,
therapeutic trials aimed at targeting miRNA in arthritis have been
performed in vivo models (19,20).
Thus, targeting miRNAs offers a novel advanced therapeutic strategy
for treating RA.
Alterations of miRNAs in RA have been reported,
however, the regulation of these molecules remains to be fully
elucidated. The present study investigated whether the mRNA levels
of Dicer, Ago2 and Drosha, which are components of the RNAi
mechanism, are associated with the clinical signature of RA.
Investigations were also performed to investigate the contribution
of Dicer, which was identified in our pilot study, to the
pathogenesis of RA.
Materials and methods
Subjects
A total of 50 patients with RA and 25 healthy
controls were recruited following the provision of informed consent
from February 2014 to July 2014. The procedure was approved by the
Medical Ethics Committee of Central Hospital of Zibo in January
2014. All patients with RA fulfilled the American College of
Rheumatology classification criteria for RA. An RA Disease Activity
Score (DAS28) for each patient was determined at the time of blood
sample collection. Additional clinical information is listed in
Table I.
 | Table I.Characteristics of the study
subjects. |
Table I.
Characteristics of the study
subjects.
| Characteristic | Patients with
rheumatoid arthritis (n=50) | Healthy controls (n
=25) |
|---|
| Female, n | 38 | 19 |
| Male, n | 12 | 6 |
| Age, years
(range) | 55.88 (28–85) | 56.25 (31–78) |
| Disease duration,
months | 54 | – |
| ILD, n (%) | 4 (8) | – |
| Infection, n (%) | 9
(18) | – |
| ANA, n (%) | 22 (44) | – |
| RF, n (%) | 42 (84) | – |
| Anti-CCP, n (%) | 45 (90) | – |
| AKA, n (%) | 11 (22) | – |
| DAS28a | 4.810±0.2476 | – |
| Medication |
| – |
| Steroids n
(%)b | 10 (20) | – |
| ≤10 mg/day, n
(%) | 6
(12) | – |
| >10 mg/day, n
(%) | 4 (8) | – |
| DMARDs
c, n (%) | 49 (98) | – |
| Anti-TNF
agentsd, n (%) | 11 (22) | – |
Sample handling and RNA
processing
Peripheral blood samples were obtained from each
subject and collected in tubes containing acid citrate dextrose
formula A. Erythrocytes were immediately lysedin lysis buffer for
20 min at 4°C and total RNA was extracted from the leukocytes using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently,
1 µg of RNA was reverse transcribed into cDNA using SuperScript II
reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.)
and oligo dT primers.
Quantitative polymerase chain reaction
(qPCR) analysis
To determine the quantity of mRNA, the cDNA was
amplified using qPCR analysis with SYBR-Green (SYBR Premix Ex Taq™
RT-PCR kit; Takara Biotechnology Co., Ltd., Dalian, China), and the
expression of ribosomal protein L13A (RPL13A) was determined as the
internal control. The SYBR Green assays were performed on a 7900HT
real-time instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Relative expression levels were calculated using the
2−ΔΔCq method (21).
The primers used were as follows: Dicer forward,
5′-AGGAAGAGGCTGACTATGAAG-3′ and reverse,
5′-GGTTGAAAAAGGAGAAAGAGA-3′; Ago2 forward,
5′-GTCTCTGAAGGCCAGTTCCA-3′ and reverse, 5′-ATACAGGCCTCACGGATGG-3′;
Drosha forward, 5′-CAGCTACGAACGGAGCAGT-3′ and reverse,
5′-TTTTTCTTCCTCCCAACGAG-3′; TNF-α forward,
5′-CCCAGGGACCTCTCTCTAATCA-3′ and reverse,
5′-GCTACAGGCTTGTCACTCGG-3′; RPL13A forward,
5′-CCTGGAGGAGAAGAGGAAAGAGA-3′ and reverse,
5′-TTGAGGACCTCTGTGTATTTGTCAA-3′. RT-qPCR analysis was performed at
95°C for 15 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 30 sec, and then at 95°C for 15 sec, 60°C for 15 sec and 95°C
for 15 sec. The results were analyzed using SDS software version
2.3 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Isolation of peripheral blood
mononuclear cells (PBMCs)
PBMCs were obtained from healthy volunteer donors.
The PBMCs were separated from heparinized whole blood using
density-gradient centrifugation at 800 × g for 20 min at 37°C with
LymphoprepFicoll-Paque Plus (GE Healthcare Life Sciences, Chalfont,
UK).
Cell culture
The HeLa cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and were maintained at 37°C
in an atmosphere of 5% CO2 in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.)
containing 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and
100 U/ml of penicillin/streptomycin. The purified PBMCs were
cultured in RPMI-1640 medium supplemented with 10% serum from
either a healthy control ora patients with RA, 100 U/ml of
penicillin and 100 U/ml of streptomycin at 37°C in an atmosphere of
5% CO2.
Transfection and stimulation
The HeLa cells were plated at a density of 5×104 in
culture dishes for 24 h and then transfected with 100 nM of Dicer
small interfering (si)RNA or control siRNA with Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. At 48 h post-transfection, the HeLa cells
were stimulated with lipopolysaccharide (LPS; 10 µg/ml) from
Escherichia coli strain K12; InvivoGen, San Diego, CA, USA) for 2 h
at 37°C in an atmosphere of 5% CO2. The siRNA sequences
were as follows: Control siRNA sense,
5′-CAGUACUUUUGUGUAGUACAAdTdT-3′ and antisense,
5′-TTGTACTACACAAAAGTACTGdTdT-3′; Dicer siRNA sense,
5′-ACUGCUUGAAGCAGCUCUGGAdTdT-3′ and antisense,
5′-UCCAGAGCUGCUUCAAGCAGUdTdT-3′.
Enzyme-linked immunosorbent assay
(ELISA)
The protein levels of TNF-α secreted into the cell
culture supernatantwere quantified using commercially available
ELISA kits (XiTang Biological Technology Co., Ltd., Shanghai,
China) according to the manufacturer's protocol.
Statistical analysis
Data were analyzed using Prism 4 software, version
5.01 (GraphPad Software, Inc., La Jolla, CA, USA). The
nonparametric Mann-Whitney test was used to compare between groups.
Spearman's correlation test was used for correlation analysis.
P<0.05 (two-tailed) was considered to indicate a statistically
significant difference.
Results
mRNA expression of Dicer Ago2 and
Drosha
The demographic and baseline clinical data of the
study subjects are summarized in Table
I. The present study examined the mRNA expression levels of
Dicer, Ago2 and Drosha in samples obtained from 50 patients with RA
and 25 healthy controls using RT-qPCR analysis, as described above.
There were no significant differences in mean age or gender
distribution between the patients with RA and the healthy donors
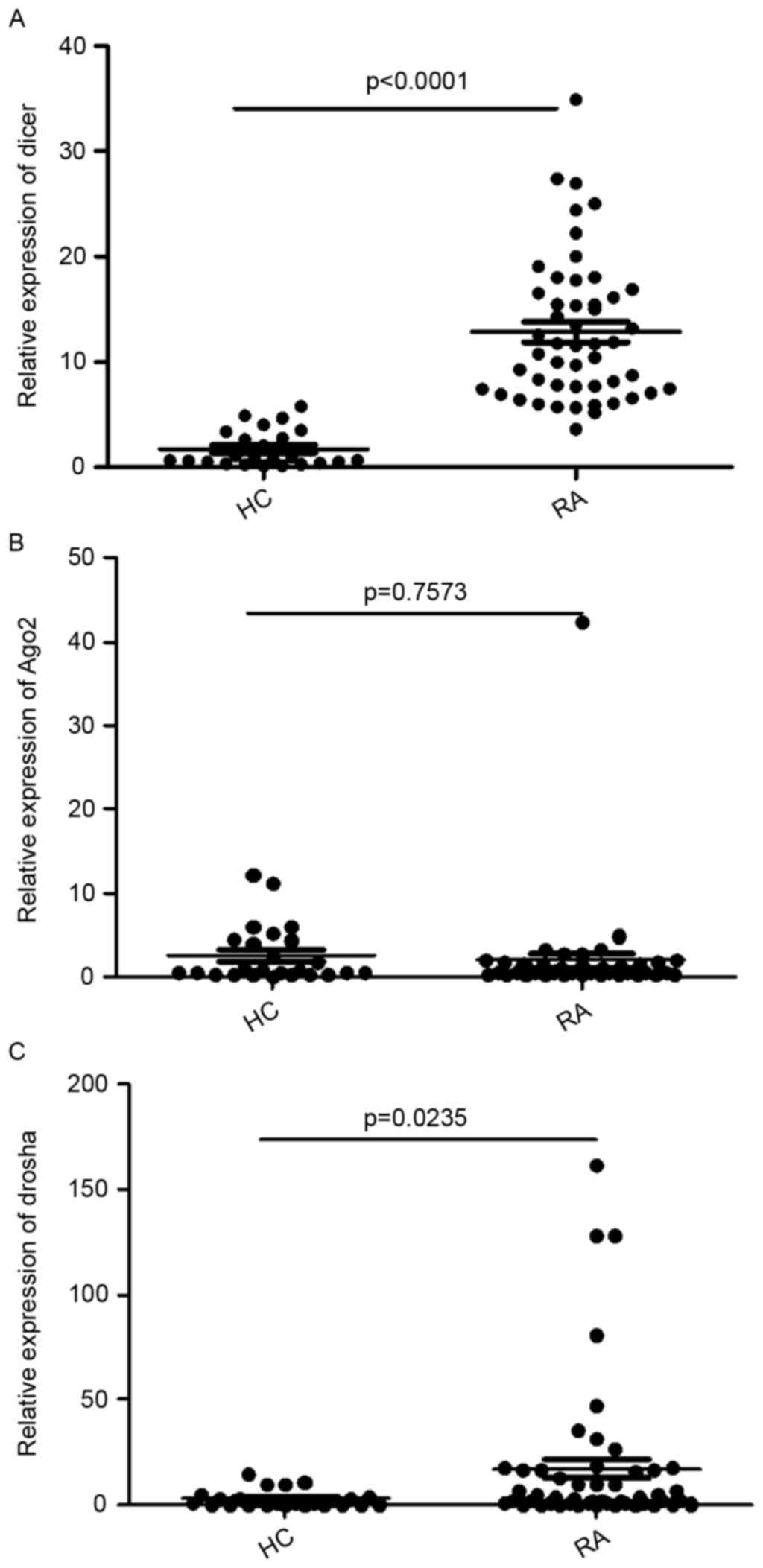
(Table I). As shown in Fig. 1A, the RA group had a higher mRNA
level of Dicer, compared with the healthy controls (12.86±6.83, vs.
1.727±1.713; P<0.0001). There was no significant difference in
the mRNA expression of Ago2 between the patients with RA and
healthy controls (1.886±5.944, vs. 2.549±3.3889; P=0.7573; Fig. 1B). The RA group had a higher mRNA
level of Drosha, compared with the healthy controls (17.14±34.76,
vs. 2.824±4.037 P=0.0235; Fig.
1C).
Clinical associations
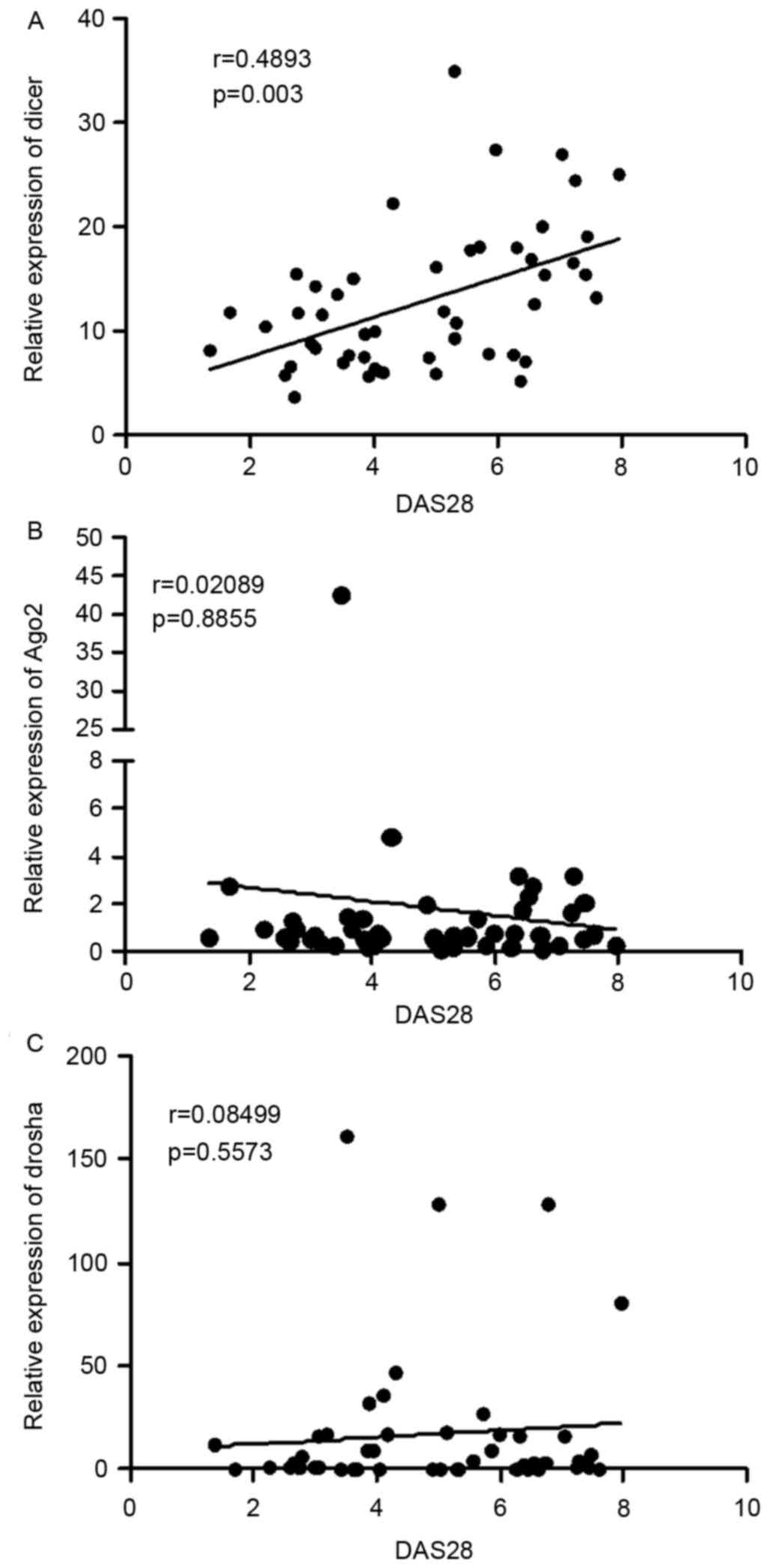
The present study performed analysis to determine
the correlation between the mRNA levels of Dicer, Ago2 and
Droshaand clinical features. As shown in Fig. 2A, a direct positive correlation was
observed between the mRNA levels of Dicer and DAS28 scores
(r=0.4893; P=0.003). However, there was no correlation between the
mRNA level of Ago2 (r=0.02089; P=0.8855) and DAS28 scores or the
level of Drosha and DAS28 scores (r=0.08499; P=0.5573), as shown in
Fig. 2B and C.
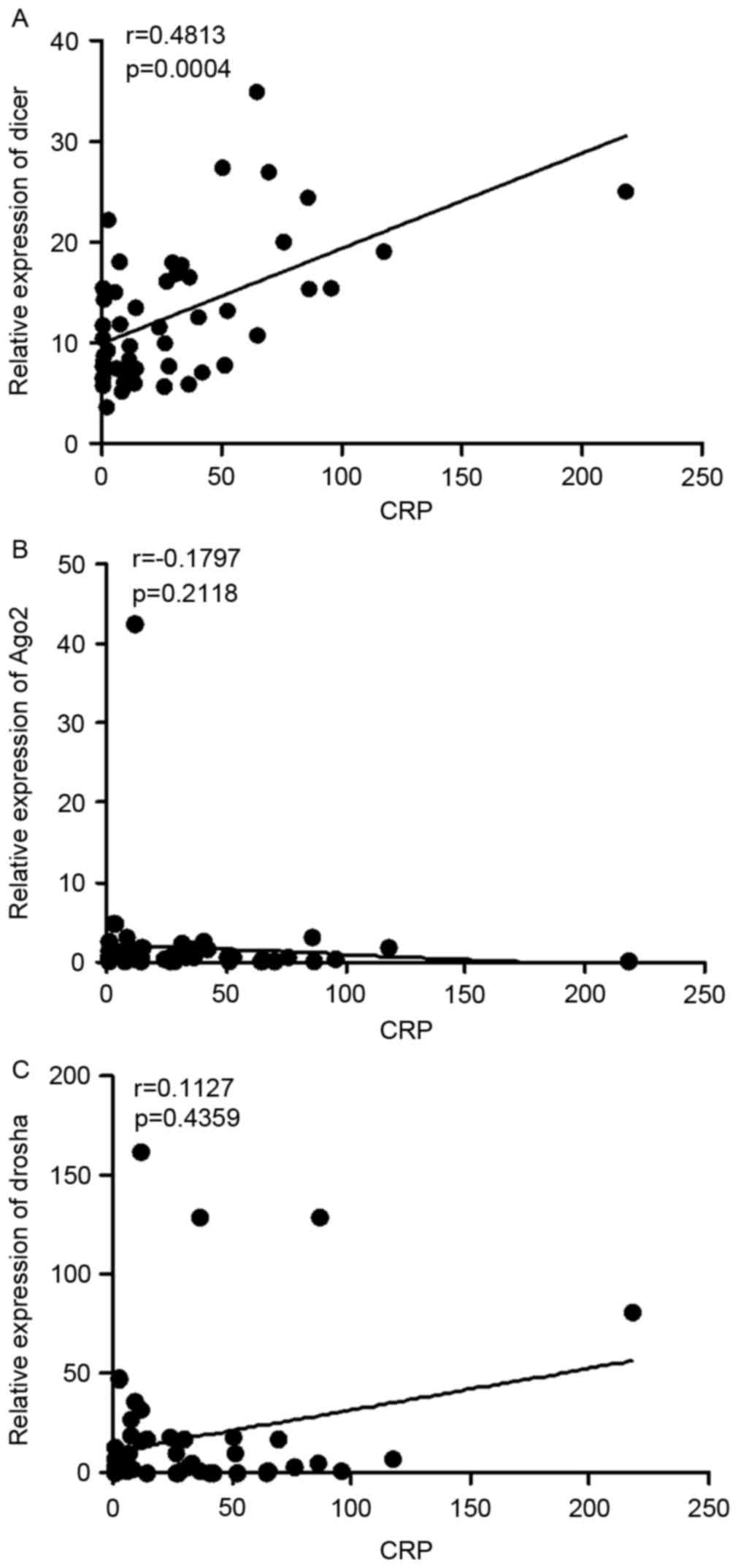
The levels of C-reactive protein (CRP) are also an
indicator of disease activity. High CRP levels are often observed
in patients with RA with active disease. Further analysis in the
present study revealed that the mRNA expression levels of Dicer
were correlated with the levels of CRP (r=0.4813; P=0.0004;
Fig. 3A), whereas the mRNA levels
of Ago2 (r=0.1793; P=0.2118) and Drosha (r=0.1127; P=0.4359) were
not (Fig. 3B and C). Taken
together, these results indicated that the expression levels of
Dicer were positively correlate with RA disease activity.
Manipulation of Dicer function alters
the expression of TNF-α
TNF-α is one of the major proinflammatory cytokines
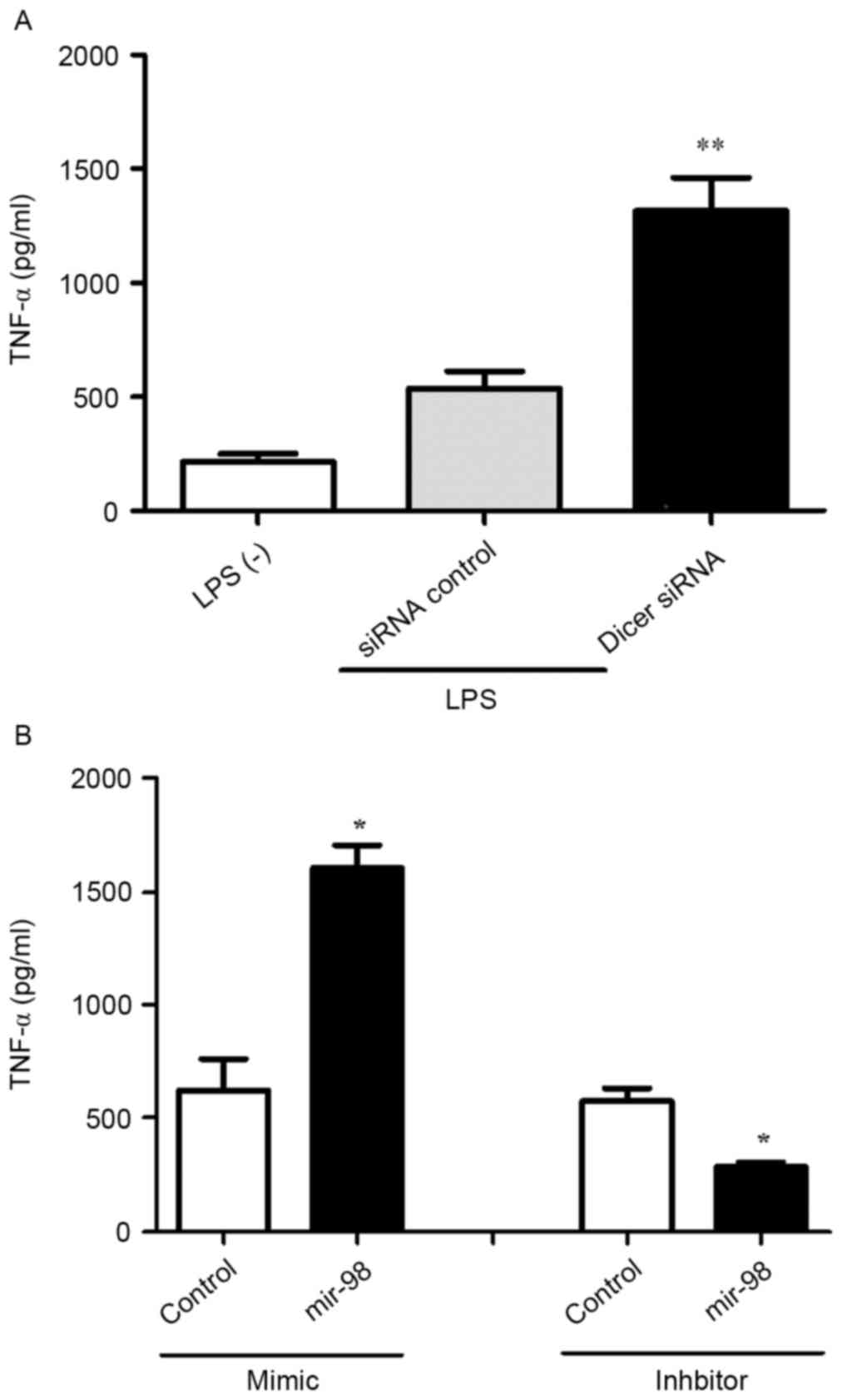
involved in the pathogenesis of RA (22). In order to examine the effect of
Dicer on TNF-α, the present study treated HeLa cells with Dicer
siRNA or control siRNA. At 48 h post-transfection, the HeLa cells
were stimulated with LPS (10 µg/ml) for 2 h, following which the
protein level of TNF-α was detected using ELISA. Transfection with
Dicer siRNA increased the expression of TNF-α (Fig. 4A), suggesting that the expression
of TNF-α was regulated by Dicer at the protein level.
The expression of Dicer can be regulated by the
let-7/miR-98 family via mRNA degradation and translational
repression (23). In the present
study, the expression of Dicer was altered by manipulation of
miR-98 and the effect on the expression of TNF-α was examined. HeLa
cells were treated with miR-98 mimic or inhibitor for 48 h.
Following transfection, the HeLa cells were stimulated with LPS (10
µg/ml) for 2 h and the protein expression of TNF-α was examined
using ELISA. The miR-98 mimic, which downregulated the expression
of Dicer, increased the protein expression levels of TNF-α, whereas
the miR-98 inhibitor, which upregulated the expression of Dicer,
decreased theexpression of TNF-α (Fig.
4B), suggesting that the expression of TNF-α was regulated by
the manipulation of Dicer.
Expression of Dicer and TNF-α are
enhanced following LPS treatment or by supplementation with 10%
serum from patients with RA
The excessive stimulation of innate immunity,
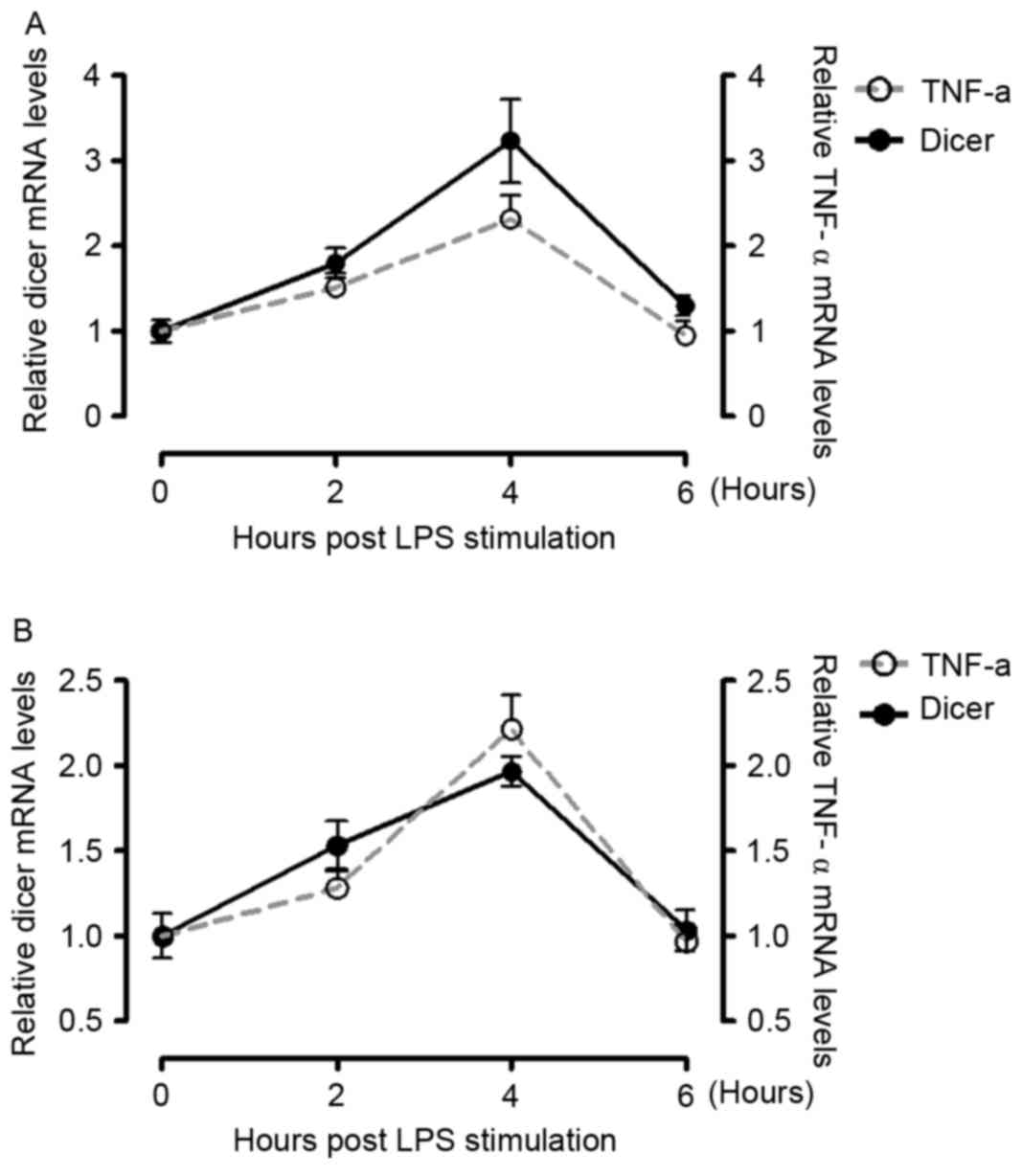
including LPS, can lead to the overproduction of TNF-α (24). In the present study, PBMCs
werefreshly purified from healthy donors and stimulated LPS (10
µg/ml from E. coli strain K12), following which the mRNA levels of
TNF-α and Dicer were measured using RT-qPCR analysis at different
times post-LPS stimulation. It was found that the mRNA levels of
TNF-α and Dicer were enhanced, with a peak 4 h, following LPS
treatment (Fig. 5A). This process
was mimicked by supplementing the cells with 10% serum from
patients with RA (Fig. 5B).
Discussion
RNAi, which was initially recognized as an ancient
defense strategy protecting organisms from RNA virus infection
(25), is an evolutionarily
conserved mechanism of post-transcriptional gene silencing in a
sequence-specific manner (26). It
has been shown that gene silencing by RNAi has emerged as a useful
tool for genetic analysis and treatment of disease.
Dicer is important in antiviral responses. It
inhibits human immunodeficiency virus type 1 replication in PBMCs
(27), and knockdown of Dicer
leads to increased virus production and accelerated apoptosis of
influenza-A virus-infected cells (28). In vivo, Dicer is essential
for normal skeletal growth. Dicer is critical in the regulation of
chondrocyte proliferation and differentiation during skeletal
development (29). Dicer
deficiency in osteoclasts suppresses osteoclastic bone resorption
(30). A previous study showed
that Anti-Su autoantibodies from patients with RA and systemic
lupus erythematosusrecognize the Ago2 and Dicer proteins (31). However, the components of the RNAi
machinery in RA remain to be fully elucidated.
In the present study, the mRNA levels of Dicer, Ago2
and Drosha were examined, and the association between their
expression levels and specific clinical features of RA were
investigated. It was found that the mRNA expression levels of Dicer
and Drosha were upregulated in patients with RA, compared with
healthy controls, and the mRNA expression of Dicer was correlated
with the activity of disease. These results demonstrated that Dicer
mRNA can be used as a marker of RA disease activity.
TNF-α is a multifunctional cytokine involved in
important biological processes, including cell survival,
differentiation, proliferation and death (32). It is also crucial in the
pathogenesis of RA. Anti-TNF therapy has shown to benefit patients
with RA. To evaluate the effect of the manipulation Dicer on the
expression of TNF-α, the present study performed in vitro
experiments to determine whether Dicer RNAi alters the expression
of TNF-α. As shown in Fig. 4A,
transfection of the cells with Dicer siRNA increased the expression
of TNF-α. The miR-98 mimic, which downregulated the expression of
Dicer, increased the protein expression levels of TNF-α, whereas
the miR-98 inhibitor, which upregulated the expression of Dicer,
decreased the expression of TNF-α (Fig. 4B). These results suggested that the
expression of TNF-α, was regulated by the manipulation of Dicer. A
previous study showed that the nuclear factor (NF)-κB-dependent
transcription of Dicer is important for TNF-α homeostasis in
hepatocytes. The expression of Dicer and TNF-α were induced in
response to the activation of NF-κB (33). The present study showed that the
mRNA levels of Dicer and TNF-α were enhanced following LPS
treatment orsupplementation with 10% serum from patients with RA.
Dicer and TNF-α were activated in the serum of patients with RA. In
addition, activation of Dicer inhibited the production of TNF-α.
The outcome of these regulatory mechanisms was the balanced
production of TNF-α in RA.
In conclusion, the present study demonstrated that
the mRNA expression levels of Dicer and Drosha were upregulated in
RA, and that the increased level of Dicer was correlated with
disease activityin patients with RA. Therefore, Dicer can be used
as a marker of RA disease activity. Dicer and TNF-α were also
activated in the serum of patients with RA. The activation of Dicer
suppressed the production of TNF-α in RA, and the outcome of these
regulatory mechanisms was the balanced production of TNF-α in
RA.
Acknowledgements
This study was supported by a grantfrom the Natural
Science Foundation of Shandong Province (grant no.
ZR2013HQ044).
References
|
1
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sevignani C, Calin GA, Siracusa LD and
Croce CM: Mammalian microRNAs: A small world for fine-tuning gene
expression. Mamm Genome. 17:189–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McManus MT and Sharp PA: Gene silencing in
mammals by small interfering RNAs. Nat Rev Genet. 3:737–747. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meister G, Landthaler M, Patkaniowska A,
Dorsett Y, Teng G and Tuschl T: Human Argonaute2 mediates RNA
cleavage targeted by miRNAs and siRNAs. Mol Cell. 15:185–197. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Halder J, Kamat AA, Landen CN Jr, Han LY,
Lutgendorf SK, Lin YG, Merritt WM, Jennings NB, Chavez-Reyes A,
Coleman RL, et al: Focal adhesion kinase targeting using in vivo
short interfering RNA delivery in neutral liposomes for ovarian
carcinoma therapy. Clin Cancer Res. 12:4916–4924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Landen CN Jr, Chavez-Reyes A, Bucana C,
Schmandt R, Deavers MT, Lopez-Berestein G and Sood AK: Therapeutic
EphA2 gene targeting in vivo using neutral liposomal small
interfering RNA delivery. Cancer Res. 65:6910–6918. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McInnes IB and Schett G: The pathogenesis
of rheumatoid arthritis. N Engl J Med. 365:2205–2219. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang RY, Huang QC and Burgering BM: Novel
insight into the role of α-actinin-1 in rheumatoid arthritis.
Discov Med. 17:75–80. 2014.PubMed/NCBI
|
|
12
|
Salemi S, Biondo MI, Fiorentino C, Argento
G, Paolantonio M, Di Murro C, Malagnino VA, Canzoni M, Diamanti AP
and D'Amelio R: Could early rheumatoid arthritis resolve after
periodontitis treatment only? Case report and review of the
literature. Medicine (Baltimore). 93:e1952014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ursini F, Russo E, Hribal M Letizia, Mauro
D, Savarino F, Bruno C, Tripolino C, Rubino M, Naty S and Grembiale
RD: Abatacept improves whole-body insulin sensitivity in rheumatoid
arthritis: An observational study. Medicine (Baltimore).
94:e8882015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miao CG, Yang YY, He X, Xu T, Huang C,
Huang Y, Zhang L, Lv XW, Jin Y and Li J: New advances of microRNAs
in the pathogenesis of rheumatoid arthritis, with a focus on the
crosstalk between DNA methylation and the microRNA machinery. Cell
Signal. 25:1118–1125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu T, Huang C, Chen Z and Li J:
MicroRNA-323-3p: A new biomarker and potential therapeutic target
for rheumatoid arthritis. Rheumatol Int. 34:721–722. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murata K, Furu M, Yoshitomi H, Ishikawa M,
Shibuya H, Hashimoto M, Imura Y, Fujii T, Ito H, Mimori T and
Matsuda S: Comprehensive microRNA analysis identifies miR-24 and
miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS
One. 8:e691182013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abou-Zeid A, Saad M and Soliman E:
MicroRNA 146a expression in rheumatoid arthritis: Association with
tumor necrosis factor-alpha and disease activity. Genet Test Mol
Biomarkers. 15:807–812. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Philippe L, Alsaleh G, Suffert G, Meyer A,
Georgel P, Sibilia J, Wachsmann D and Pfeffer S: TLR2 expression is
regulated by microRNA miR-19 in rheumatoid fibroblast-like
synoviocytes. J Immunol. 188:454–461. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagata Y, Nakasa T, Mochizuki Y, Ishikawa
M, Miyaki S, Shibuya H, Yamasaki K, Adachi N, Asahara H and Ochi M:
Induction of apoptosis in the synovium of mice with
autoantibody-mediated arthritis by the intraarticular injection of
double-stranded MicroRNA-15a. Arthritis Rheum. 60:2677–2683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakasa T, Nagata Y, Yamasaki K and Ochi M:
A mini-review: MicroRNA in arthritis. Physiol Genomics. 43:566–570.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feldmann M: Translating molecular insights
in autoimmunity into effective therapy. Annu Rev Immunol. 27:1–27.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tokumaru S, Suzuki M, Yamada H, Nagino M
and Takahashi T: let-7 regulates Dicer expression and constitutes a
negative feedback loop. Carcinogenesis. 29:2073–2077. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin WJ and Yeh WC: Implication of
Toll-like receptor and tumor necrosis factor alpha signaling in
septic shock. Shock. 24:206–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Waterhouse PM, Wang MB and Lough T: Gene
silencing as an adaptive defence against viruses. Nature.
411:834–842. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meister G and Tuschl T: Mechanisms of gene
silencing by double-stranded RNA. Nature. 431:343–349. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Triboulet R, Mari B, Lin YL, Chable-Bessia
C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P,
Baillat V, et al: Suppression of microRNA-silencing pathway by
HIV-1 during virus replication. Science. 315:1579–1582. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matskevich AA and Moelling K: Dicer is
involved in protection against influenza A virus infection. J Gen
Virol. 88:2627–2635. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kobayashi T, Lu J, Cobb BS, Rodda SJ,
McMahon AP, Schipani E, Merkenschlager M and Kronenberg HM:
Dicer-dependent pathways regulate chondrocyte proliferation and
differentiation. Proc Natl Acad Sci USA. 105:1949–1954. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mizoguchi F, Izu Y, Hayata T, Hemmi H,
Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y and Noda M:
Osteoclast-specific Dicer gene deficiency suppresses osteoclastic
bone resorption. J Cell Biochem. 109:866–575. 2010.PubMed/NCBI
|
|
31
|
Jakymiw A, Ikeda K, Fritzler MJ, Reeves
WH, Satoh M and Chan EK: Autoimmune targeting of key components of
RNA interference. Arthritis Res Ther. 8:R872006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen G and Goeddel DV: TNF-R1 signaling: A
beautiful pathway. Science. 296:1634–1655. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guan Y, Yao H, Wang J, Sun K, Cao L and
Wang Y: NF-κB-DICER-miRs axis regulates TNF-α expression in
responses to endotoxin stress. Int J Biol Sci. 11:1257–1268. 2015.
View Article : Google Scholar : PubMed/NCBI
|