Introduction
Helicobacter pylori is an important pathogen
in intestinal and diffuse non-cardia adenocarcinoma (1). Various virulence components are
associated with the pathogenicity of H. pylori, including
flagella, lipopolysaccharide, vacuolating cytotoxin VacA and
cytotoxin-associated gene pathogenicity island (2). Using the human whole genome
microarray, it was previously demonstrated that co-culture of the
H. pylori strain cagAvacAs1m1, isolated from patients with
gastric cancer, with gastric epithelial GES-1 cells resulted in
markedly increased expression of tumor necrosis factor
receptor-associated factor 1 (TRAF1), tumor necrosis factor
receptor superfamily member 9 [4-1BB/cluster of differentiation
(CD) 137], and chemokine interleukin (IL) −8 [a downstream target
of the nuclear factor (NF)-κB pathway] (3,4).
Silencing of TRAF1 using short hairpin RNA has been demonstrated to
inhibit the growth and induce the apoptosis of gastric cancer
BGC823 cells (5). Further clinical
studies have demonstrated that TRAF1 and 4-1BB are markedly
upregulated in intestinal metaplasia with atypical hyperplasia and
gastric cancer tissues, and these are associated with H.
pylori cagAvacAs1m1 infection (6,7).
These previous data indicate that the upregulation of TRAF1 and
4-1BB is associated with H. pylori cagAvacAs1m1 infection,
and contributes to the increased carcinogenicity of H.
pylori cagAvacAs1m1. However, the underlying mechanism remains
unclear.
Cytotoxin associated gene A (CagA) is one of the
most important virulence factors of H. pylori and serves a
key role in H. pylori-mediated tumorigenesis in gastric
cancer. A number of studies have demonstrated that infection with
CagA-positive H. pylori strains is associated with an
increased risk of non-cardia cancer, compared with infection with
CagA-negative H. pylori strains (8–10).
The upregulation of TRAF1 and 4-1BB, and the activation of the
NF-κB pathway following H. pylori cagAvacAs1m1 infection,
have led to the hypothesis that CagA protein may promote the
tumorigenesis of gastric cancer by increasing the expression of
TRAF1 and 4-1BB, in addition to activating the NF-κB pathway.
In infection experiments in vitro, complex
interactions occur between H. pylori and host cells.
Infection may induce numerous H. pylori-specific and
non-specific cellular responses. Therefore, it is difficult to
clarify which genes are directly affected by CagA following H.
pylori infection. In the present study, gene transfection of a
CagA eukaryotic expression plasmid in cells was used to overexpress
CagA protein. The results of the present study demonstrated that
ectopic expression of CagA markedly increased the expression of
TRAF1, 4-1BB and IL-8 in GES-1 cells.
Materials and methods
Reagents
SYBR Premix EX Taq™ was purchased from Takara
Biotechnology Co., Ltd. (Dalian, China). Lipofectamine 3000 and
TRIzol were obtained from Invitrogen (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The RevertAid First Strand cDNA Synthesis
kit was obtained from Thermo Fisher Scientific, Inc. The Annexin
V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit I was
purchased from BD Biosciences (Franklin Lakes, NJ, USA). The IL-8
ELISA kit (cat. no. SEA080Hu) was obtained from Wuhan Uscn Business
Co., Ltd. (Wuhan, China). Rabbit anti-CD137 polyclonal antibody was
purchased from Abcam (Cambridge, UK; cat. no. ab203391); rabbit
anti-TRAF1 (45D3) monoclonal antibody was purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA; cat. no. 4715);
rabbit anti-CagA (cat. no. sc-25766), goat anti-rabbit
immunoglobulin (Ig)G-horseradish peroxidase (HRP; cat. no. sc-2030)
and goat anti-mouse IgG-HRP (cat. no. sc-2302) antibodies were
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA);
and mouse anti-GAPDH monoclonal antibody (cat. no. MAB374) was
purchased from Merck KGaA (Darmstadt, Germany).
Cell line and plasmids
GES-1 cells and the empty vector pEGFP-C1 were
provided by the Cancer Research Institute of Central South
University (Changsha, China). The GES-1 cells were cultured in
RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT,
USA) containing 10% fetal bovine serum (Biological Industries
Israel Beit-Haemek, Ltd., Kibbutz Beit-Haemek, Israel). The CagA
eukaryotic expression plasmid p enhanced green fluorescent protein
(EGFP)-C1/CagA was provided by Professor Yongliang Zhu (Zhejiang
University, Hangzhou, China).
Transient transfection of
plasmids
GES-1 cells were seeded in 6-well plates at a
density of 5×106 cells/well and incubated in a 5% CO2
humidified atmosphere at 37°C. When 50–60% confluence was reached,
the cells were transfected with 2.5 µg plasmid with 5 µl
Lipofectamine 3000 in 125 µl RPMI-1640 medium followed by the
addition of 1,875 µl complete 1640 medium. The cells were incubated
for 24, 48 and 72 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (2 µg) was extracted using TRIzol reagent,
according to the manufacturer's protocol, and reverse transcribed
in a 20-µl reaction system using a RevertAid First Strand cDNA
Synthesis kit. The qPCR reaction was performed using SYBR Premix
ExTaq™ reagents, according to the manufacturer's protocol. The
primer sequences were as follows: CagA forward,
5′-CGTCGCCGACATTGATCCTA-3′, CagA reverse,
5′-TAGCCACATTGTCGCCTTGT-3′; TRAF1 forward,
5′-TCCCGTAACACCTGATTAA-3′, TRAF1 reverse,
5′-ACAACTCCCAAACCATACAC-3′; 4-1BB forward,
5′-CGTGGTCTGTGGACCATCTC-3′, 4-1BB reverse,
5′-ACAACAGAGAAACGGAGCGT-3′; IL-8 forward,
5′-CCAGGAAGAAACCACCGGAA-3′, IL-8 reverse,
5′-TTCCTTGGGGTCCAGACAGA-3′; GAPDH forward,
5′-AACGGATTTGGTCGTATTGGG-3′, and GAPDH reverse,
5′-TCGCTCCTGGAAGATGGTGAT-3′. Conditions were as follows:
Pre-denaturation at 95°C for 3 min; and 40 cycles of 95°C for 10
sec and 60°C for 30 sec. The relative expression of CagA, TRAF1,
4-1BB and IL-8 was normalized to GAPDH; expression was calculated
using the 2−∆∆Cq method (11).
Western blot analysis
Total protein was extracted from cells using lysis
buffer, containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA,
1% Triton-X 100, 1% DTT and 1% protease inhibitor cocktail (Roche
Diagnostics, Basel, Switzerland). Nuclear and cytoplasmic proteins
were extracted using a nuclear and cytoplasmic protein extraction
kit (Beyotime Institute of Biotechnology, Haimen, China), according
to the manufacturer's protocol. The protein concentration was
measured using the BCA Protein assay kit (Thermo Fisher Scientific,
Inc.). Equal amounts of protein extracts (50 µg) were separated
using SDS-PAGE on a 10% gel and transferred onto a polyvinylidene
fluoride membrane. The membranes were blocked with 5% w/v non-fat
dried milk dissolved in TBS with Tween-20 (TBS-T; 0.1% Tween-20, pH
8.3) at room temperature for 1 h, and incubated with primary
antibodies at 4°C overnight. The following primary antibodies were
used: anti-CD137 (1:500); anti-GAPDH (1:5,000); anti-TRAF1 (1:500);
and anti-CagA (1:500). Following washing with TBS-T, the membranes
were incubated with HRP-labeled anti-rabbit or mouse IgG secondary
antibody (both 1:5,000) for 1 h at room temperature. Bands were
visualized using an enhanced chemiluminescence kit (EMD Millipore)
and ChemiDoc XRS system (Image Lab™ software version 4.0; Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
ELISA analysis
Following transfection for 24, 48 and 72 h, the cell
culture medium was collected and centrifuged at 94 × g for 20 min
at 4°C. The amount of IL-8 in the supernatant was detected using
the IL-8 ELISA kit, according to the manufacturer's protocol. The
optical density (OD) of each well was read using a microplate
reader at a wavelength of 450 nm.
Cell viability assay
Cell viability was determined using an MTT assay.
Cells were seeded at a density of 2×103 cells/well in 96-well
plates. At 24 h subsequent to seeding, cells were transfected with
plasmid for 24, 48, 72 and 96 h. At the indicated time points, 20
µl MTT solution (5 mg/ml) was added to each well, and the cells
were cultured for an additional 4 h. The culture medium was removed
and 150 µl dimethyl sulfoxide was added to dissolve the formazan.
Cell viability was quantified by measuring the absorbance at 490
nm, using a microplate spectrophotometer to calculate OD
values.
Cellular apoptosis assay
Cellular apoptosis was assayed using flow cytometry.
Cellular apoptosis was detected using a FITC Annexin V Apoptosis
Detection Kit I (BD Biosciences), according to the manufacturer's
instructions. Following transfection for 48 and 72 h, cells were
harvested and re-suspended in cold PBS. Subsequent to
centrifugation at 94 × g for 5 min at 4°C, the cells were
resuspended with 500 µl binding buffer and mixed with 5 µl annexin
V-FITC. The cells were subsequently incubated with 5 µl propidium
iodide (PI) in the dark at room temperature for 5-15 min.
Excitation was at 488 nm and the emission filters used were 515–545
BP (green, FITC) and 620 LP (red, PI). The samples were analyzed
with a FACSCanto-II flow cytometer (BD Biosciences) and FlowJo
software (version 7.6; Tree Star, Inc., Ashland, OR, USA). All
assays were performed in triplicate.
Statistical analysis
All data were analyzed using SPSS software (version
17.0; SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean
± standard deviation. Comparisons were made between two groups
using independent sample t-tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
Ectopic expression of CagA increases
the expression of TRAF1 and 4-1BB in GES-1 cells
In order to assess the function of the H.
pylori CagA protein in the tumorigenesis of gastric cancer and
activation of the NF-κB pathway, CagA-containing eukaryotic
expression plasmid pEGFP-C1/CagA was used to overexpress CagA in
GES-1 cells. Transient transfection of the plasmid pEGFP-C1/CagA in
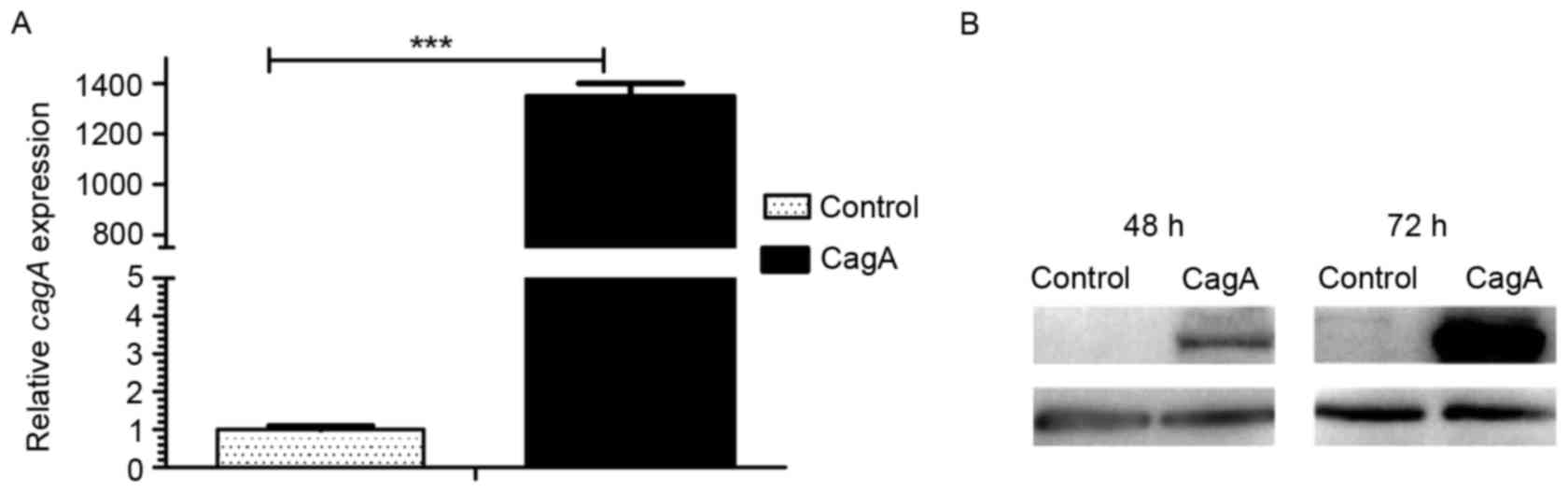
GES-1 cells for 48 h resulted in a 1,347-fold increase in the CagA
mRNA level compared with cells transfected with the control vector
pEGFP-C1 (Fig. 1A). CagA protein
expression was detected using immunoblotting following transient
transfection of the plasmid pEGFP-C1/CagA in GES-1 cells for 48 and
72 h. The results of the present study demonstrated that CagA
protein expression was undetectable in cells transfected with the
control vector pEGFP-C1, while the transfection of pEGFP-C1/CagA in
GES-1 cells resulted in apparent expression of CagA protein for 48
h and increased expression of CagA protein for 72 h (Fig. 1B). The results of the present study
demonstrated stable and high expression of CagA protein following
the transient transfection of plasmid pEGFP-C1/CagA in GES-1
cells.
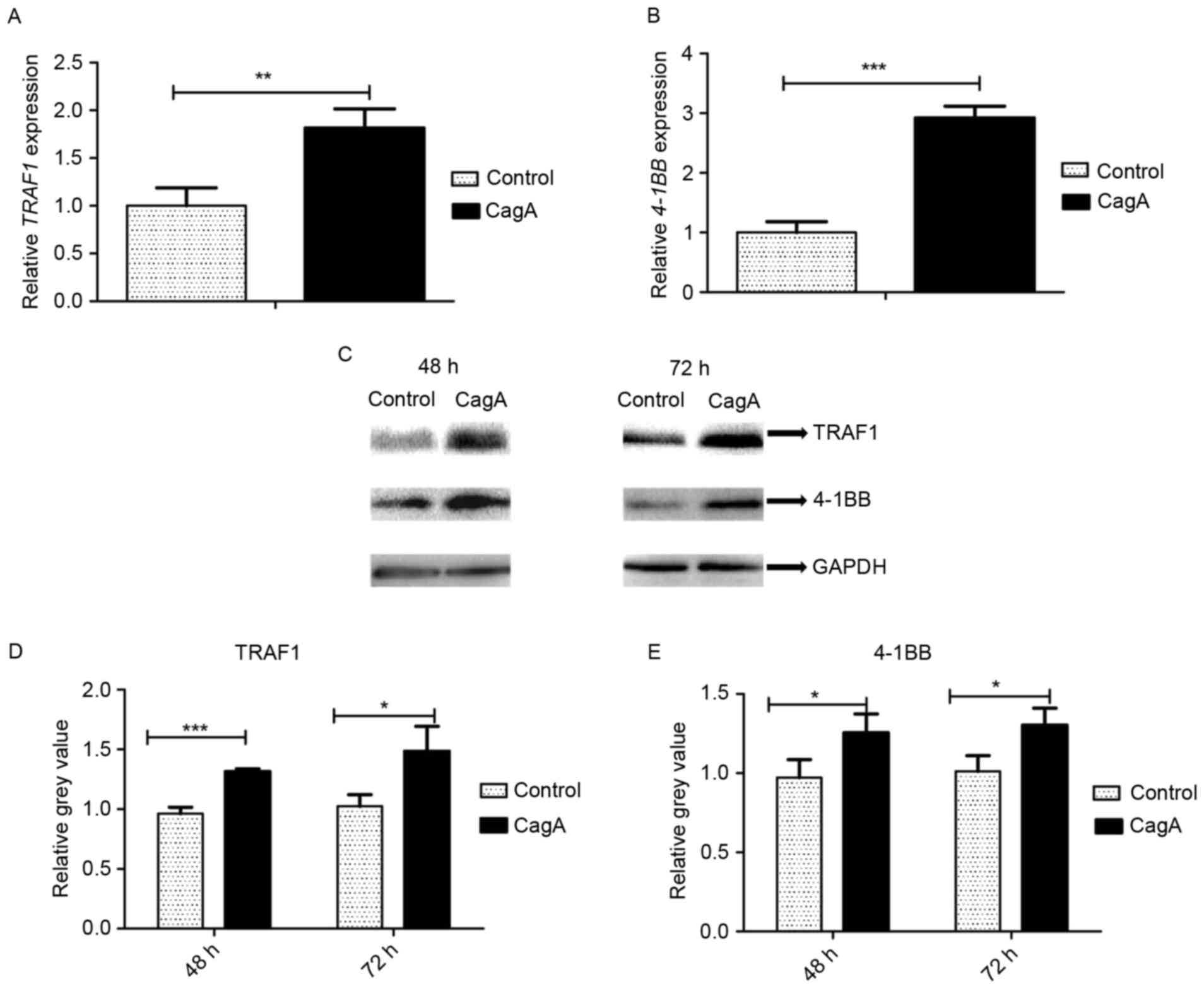
In order to determine the effect of CagA on TRAF1
and 4-1BB expression, GES-1 cells were transfected with
pEGFP-C1/CagA in parallel with pEGFP-C1 for 48 h. TRAF1 and 4-1BB
mRNA levels were determined by RT-qPCR. The transfection of
pEGFP-C1/CagA resulted in the upregulation of TRAF1 by 1.8-fold
(Fig. 2A), and of 4-1BB by
2.9-fold (Fig. 2B), compared with
cells transfected with pEGFP-C1. A statistically significant
different was observed in TRAF1 and 4-1BB mRNA levels between
pEGFP-C1/CagA- and pEGFP-C1-transfected cells (P<0.01 and
P<0.001, respectively). Consistent with the alteration in mRNA
expression, western blot analysis demonstrated that the
transfection of pEGFP-C1/CagA significantly increased TRAF1
(Fig. 2C and D) and 4-1BB
(Fig. 2C and E) protein levels
compared with cells transfected with pEGFP-C1. The results of the
present study demonstrated that the expression of CagA upregulated
the expression of TRAF1 and 4-1BB in GES-1 cells.
Expression of CagA upregulates the
expression of chemokine IL-8 in GES-1 cells
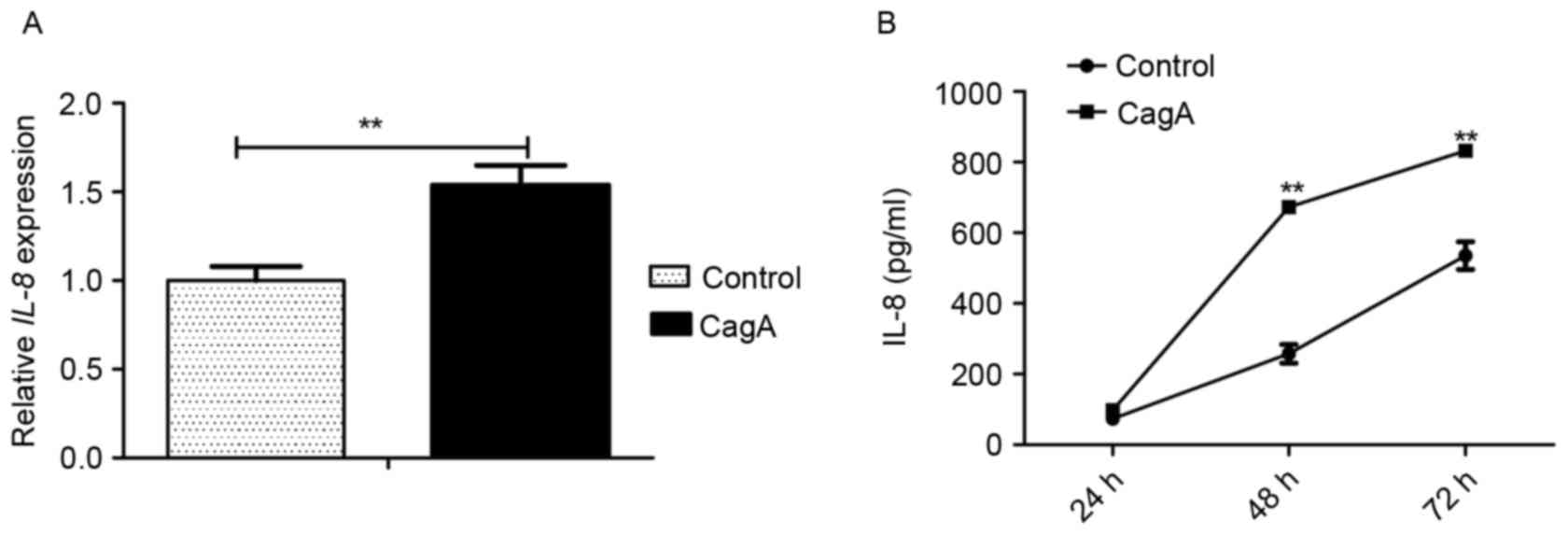
GES-1 cells were transfected with pEGFP-C1/CagA in
parallel with pEGFP-C1 for 48 h, and the mRNA and protein
expression levels of IL-8, a downstream target of NF-κB signaling,
were determined using RT-qPCR and ELISA analysis, respectively. The
transfection of the plasmid pEGFP-C1/CagA for 48 h significantly
increased the mRNA level of IL-8 compared with cells transfected
with the control vector, pEGFP-C1 (Fig. 3A). The analysis of IL-8 in the cell
culture supernatant using ELISA revealed that the transfection of
the plasmid pEGFP-C1/CagA in GES-1 cells significantly induced the
release of IL-8 compared with cells transfected with the control
vector, pEGFP-C1, for 48 or 72 h (Fig.
3B). The results of the present study demonstrated that the
expression of CagA led to the upregulation of IL-8 in GES-1
cells.
Expression of CagA promotes
proliferation and inhibits apoptosis in GES-1 cells
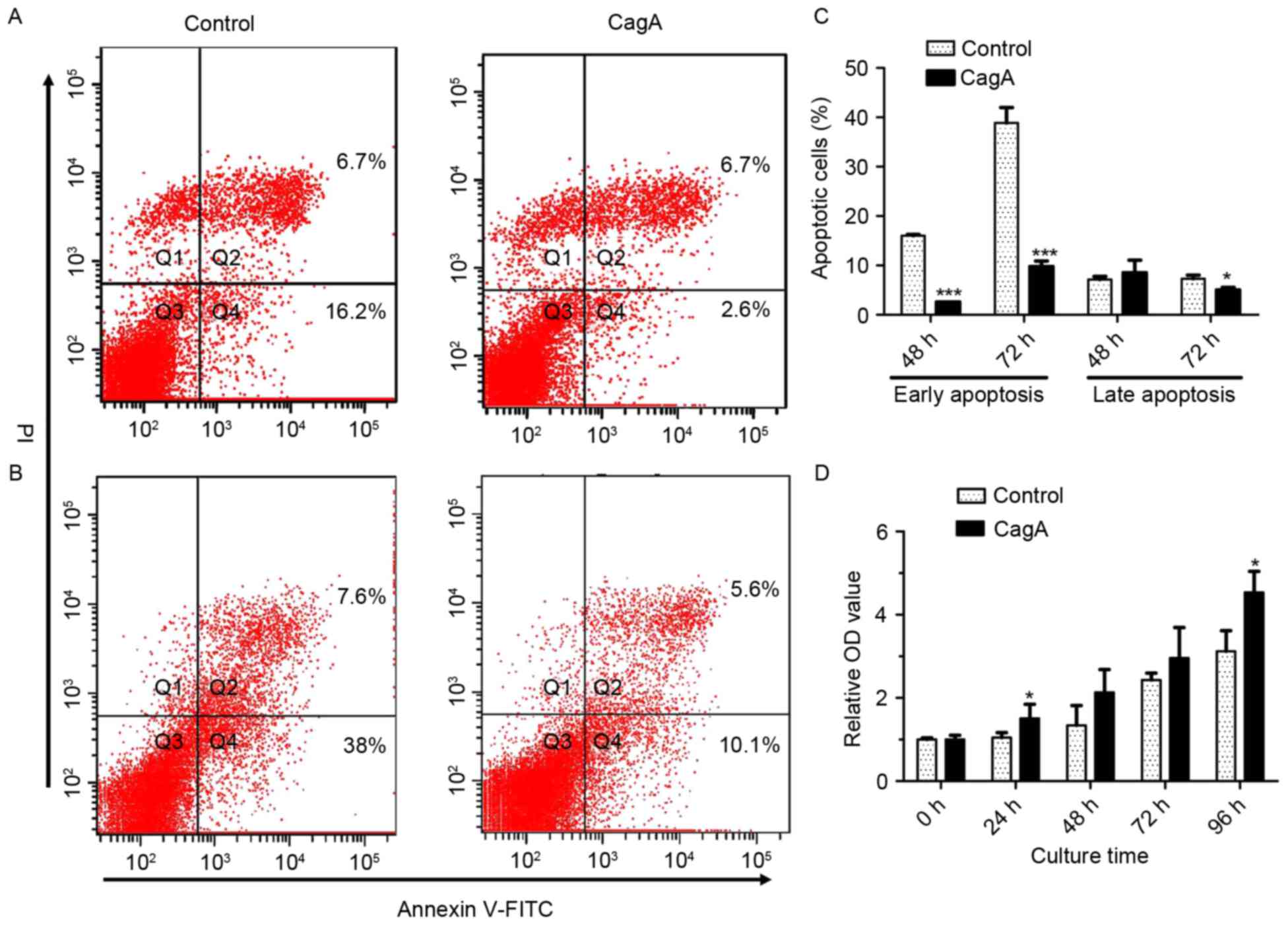
In order to assess the cell viability of GES-1 cells
following the expression of CagA, GES-1 cells were transfected with
pEGFP-C1/CagA in parallel with pEGFP-C1 at different time points.
Cell viability was determined using an MTT assay, and annexin
V-FITC staining coupled with flow cytometry analysis was used to
assess apoptosis. Apoptosis analysis by annexin V-FITC staining
coupled with flow cytometry revealed that the transfection of the
plasmid pEGFP-C1/CagA significantly inhibited early apoptosis for
48 and 72 h, and late apoptosis for 72 h, compared with cells
transfected with pEGFP-C1 (Fig.
4A-C). The MTT assay demonstrated that the transfection of the
plasmid pEGFP-C1/CagA increased cell viability at 24 and 96 h
compared with the control group, and the difference was
statistically significant (Fig.
4D). The results of the present study demonstrated that the
expression of CagA resulted in an enhancement of cell
proliferation, while inhibiting cellular apoptosis in GES-1
cells.
Discussion
In the present study, it was observed that ectopic
expression of CagA significantly increased the expression of TRAF1
and 4-1BB in GES-1 cells. IL-8 was upregulated by CagA in GES-1
cells. In addition, CagA significantly promoted the proliferation
and inhibited the apoptosis of GES-1 cells. The results of the
present study demonstrated that CagA may promote cell proliferation
and inhibit apoptosis by activating the NF-κB signaling pathway via
the upregulation of TRAF1/4-1BB.
Proteins in the TRAF family were first identified as
signaling molecules that directly interact with the cytoplasmic
regulatory domain of the tumor necrosis factor receptor (TNFR)
(12). The TRAF protein contains
an N-terminal zinc domain, followed by a number of different zinc
fingers (13). TRAF1 is an
important scaffold protein, and regulates the TNFR2 signaling
pathway in regulatory T cells through direct interaction with TRAF2
(14). TRAF1 serves an important
role in the regulation of T cell activation by limiting
NF-κB-inducing kinase (NIK) activation in activated T cells, and
additionally by promoting the 4-1BB-mediated activation of the
NF-κB classical pathway (15). The
direct binding of TRAF1 with NIK results in the disruption of its
association with ubiquitin E3 ligase TRAF2-cIAP2 and subsequent
NF-κB pathway activation (16). It
has been additionally reported that binding of the TRAF1/TRAF2
oligomeric complex with the NF-κB inhibitory protein A20 results in
the inhibition of the NF-κB signaling pathway (17). In addition, TRAF1 is an
indispensable downstream target of the 4-1BB signaling pathway, and
serves an important role in the regulation of the pro-apoptotic
Bcl-2-like protein 11 and CD8+ T cell viability
(18,19). 4-1BB is a member of the TNFR family
that recruits TRAF1 and TRAF2, which in turn leads to the
activation of downstream c-Jun N-terminal kinase, p38 and NF-κB
signaling pathways (15);
therefore, lymphocyte cycle progression is promoted via the
induction of cytokine secretion and the expression of anti-cell
death and anti-apoptotic genes (20). In the present study, it was
observed that the ectopic expression of CagA increased TRAF1 and
4-1BB expression. CagA is a key virulence factor of H.
pylori. The results of the present study demonstrated that
CagA-positive H. pylori may enhance tumorigenesis by
simulating the activation of 4-1BB-TRAF1 signaling. Future studies
are required to assess TRAF2 expression following ectopic CagA
expression.
CagA is able to activate the NF-κB signaling pathway
through various mechanisms. By enhancing the interaction between
3-phosphoinositide-dependent protein kinase 1 and Rac-α
serine/threonine protein kinase (AKT), CagA increases the
phosphorylation of AKT, thereby leading to the subsequent
activation of the NF-κB signaling pathway (21). Additionally, CagA activates the
NF-κB signaling pathway by binding to mitogen-activated protein
kinase kinase kinase 7 (TAK1) and promoting TRAF6-mediated TAK1
ubiquitination at lysine 63 (22).
It was reported that CagA promoted NF-κB-mediated inflammation
following H. pylori infection by activating the hepatocyte
growth factor receptor-phosphatidylinositol 3-kinase (PI3K)-AKT
signaling pathway (23). It has
been previously reported that the persistent activation of the
NF-κB signaling pathway serves a role in the early stages of H.
pylori-mediated transition from chronic gastritis to oncogenic
transformation (24). H.
pylori infection in gastric mucosa is associated with the
increased nuclear accumulation of NF-κB p65 and the expression of
IL-8 (25). The increased
expression of IL-8 stimulates neutrophil infiltration into the
gastric mucosa, leading to inflammatory responses and chronic
gastritis. In the present study, the expression of CagA induced the
upregulation of IL-8, which was consistent with previous reports
(26,27). These data demonstrate that CagA
increases the expression of IL-8 by activating the NF-κB
pathway.
The function of CagA in regulating cell
proliferation and apoptosis remains controversial. Handa et
al (28) reported that CagA
promotes cell growth by activating the SH2-containing phosphatase 2
signaling pathway, while inhibiting cell cycle progression by
suppressing the nuclear factor of activated T cells pathway. Yoon
et al (29) observed that
CagA enhances cell cycle progression and cell proliferation by
activating the NF-κB and PI3K signaling pathways; however, it was
additionally observed that the gastric tissues of patients infected
with CagA-positive H. pylori strains exhibit an increased
expression of anti-apoptotic proteins, including Bcl-2-like protein
1 and apoptosis regulator Bcl-2, and the reduced expression of the
pro-apoptotic protein apoptosis regulator BAX. Buti et al
(30) reported that CagA inhibits
cellular tumor antigen p53 (p53) -induced apoptosis by forming
CagA-apoptosis stimulating protein of p53 protein 2-p53
heterotrimeric complexes. The results of the present study
demonstrated that CagA promoted the proliferation and inhibited the
apoptosis of GES-1 cells. Increased proliferation and evasion of
apoptosis are hallmarks of cancer cells. The results of the present
study demonstrated an important mechanism underlying the role of
bacterial oncoproteins, including CagA, in tumorigenesis.
In conclusion, the results of the present study
indicated that CagA upregulated the expression of TRAF1/4-1BB,
which activated the NF-κB/IL-8 signaling axis, thereby promoting
cell proliferation and inhibiting apoptosis. The present study
elucidated an important mechanism of CagA in H. pylori
infection associated with gastric carcinogenesis. The results of
the present study suggested that TRAF1/4-1BB may be a potential
target for the development of anticancer drugs, providing treatment
for CagA-positive H. pylori-associated gastric cancers.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81541053
and 81670509), and the Natural Science Foundation of Hunan Province
(grant no. 14JJ2036). The abstract and Fig. 3 were presented at Digestive Disease
Week 21–24 May 2016 in San Diego, CA, USA and published as abstract
no. Mo1274 in Gastroenterology 150 (4), Supplement 1: 2016.
References
|
1
|
Hirata Y, Ohmae T, Shibata W, Maeda S,
Ogura K, Yoshida H, Kawabe T and Omata M: MyD88 and TNF
receptor-associated factor 6 are critical signal transducers in
Helicobacter pylori-infected human epithelial cells. J Immunol.
176:3796–3803. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamaoka Y: Mechanisms of disease:
Helicobacter pylori virulence factors. Nat Rev Gastroenterol
Hepatol. 7:629–641. 2010.PubMed/NCBI
|
|
3
|
Wang F, Luo LD, Pan JH, Huang LH, Lv HW,
Guo Q, Xu CX and Shen SR: Comparative genomic study of gastric
epithelial cells co-cultured with Helicobacter pylori. World J
Gastroenterol. 18:7212–7224. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang F, Pan J, Luo L, Huang L, Lu H, Guo
Q, Xu C and Shen S: Chronic Helicobacter pylori infection induces
the proliferation and apoptosis in gastric epithelial cells and
gastric precancerosis in Mongolian gerbils. Zhong Nan Da Xue Xue
Bao Yi Xue Ban. 36:865–871. 2011.(In Chinese). PubMed/NCBI
|
|
5
|
Wang F, Yang Y, Feng Q, Bu G, Huang L, Lu
H, Guo Q, Xu C and Shen S: The effect of silencing of TRAF1 by
shRNA on the biological functions of gastric cancer cells. In:
Proceedings of Central South University Medical Edition.
37:876–882. 2012.(In Chinese).
|
|
6
|
Wang F, Wu X, Liu Z, Bu G, Li X, Qu N,
Peng J, Xu C, Shen S and Yuan Y: Association between Virulence
Factors and TRAF1/4-1BB/Bcl-xL expression in gastric mucosa
infected with Helicobacter pylori. Gastroenterol Res Pract.
2015:6484792015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang F, Bu G, Feng Q, Liu Z, Xu C, Shen S
and Yuan Y: The expression level of TRAF1 in human gastric mucosa
is related to virulence genotypes of Helicobacter pylori. Scand J
Gastroenterol. 49:925–932. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lamb A and Chen LF: Role of the
Helicobacter pylori-induced inflammatory response in the
development of gastric cancer. J Cell Biochem. 114:491–497. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hatakeyama M: SagA of CagA in Helicobacter
pylori pathogenesis. Curr Opin Microbiol. 11:30–37. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peek RM Jr: Orchestration of aberrant
epithelial signaling by Helicobacter pylori CagA. Sci STKE.
2005:pe142005.PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ha H, Han D and Choi Y: TRAF-mediated
TNFR-family signaling. Curr Protoc Immunol Chapter. 11:Unit11
19D2009.
|
|
13
|
Xie P: TRAF molecules in cell signaling
and in human diseases. J Mol Signal. 8:72013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim CM, Choi JY, Bhat EA, Jeong JH, Son
YJ, Kim S and Park HH: Crystal structure of TRAF1 TRAF domain and
its implications in the TRAF1-mediated intracellular signaling
pathway. Sci Rep. 6:255262016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McPherson AJ, Snell LM, Mak TW and Watts
TH: Opposing roles for TRAF1 in the alternative versus classical
NF-κB pathway in T cells. J Biol Chem. 287:23010–23019. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choudhary S, Kalita M, Fang L, Patel KV,
Tian B, Zhao Y, Edeh CB and Brasier AR: Inducible tumor necrosis
factor (TNF) receptor-associated factor-1 expression couples the
canonical to the non-canonical NF-κB pathway in TNF stimulation. J
Biol Chem. 288:14612–146123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song HY, Rothe M and Goeddel DV: The tumor
necrosis factor-inducible zinc finger protein A20 interacts with
TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci
USA. 93:6721–6725. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sabbagh L, Pulle G, Liu Y, Tsitsikov EN
and Watts TH: ERK-dependent Bim modulation downstream of the
4-1BB-TRAF1 signaling axis is a critical mediator of CD8 T cell
survival in vivo. J Immunol. 180:8093–8101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sabbagh L, Srokowski CC, Pulle G, Snell
LM, Sedgmen BJ, Liu Y, Tsitsikov EN and Watts TH: A critical role
for TNF receptor-associated factor 1 and Bim down-regulation in CD8
memory T cell survival. Proc Natl Acad Sci USA. 103:18703–18708.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Wang Z, Li T, Tsitsikov EN and
Ding HF: NF-kappaB2 mutation targets TRAF1 to induce
lymphomagenesis. Blood. 110:743–751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang BG, Hu L, Zang MD, Wang HX, Zhao W,
Li JF, Su LP, Shao Z, Zhao X, Zhu ZG, et al: Helicobacter pylori
CagA induces tumor suppressor gene hypermethylation by upregulating
DNMT1 via AKT-NFκB pathway in gastric cancer development.
Oncotarget. 7:9788–39800. 2012.
|
|
22
|
Lamb A, Yang XD, Tsang YH, Li JD, Higashi
H, Hatakeyama M, Peek RM, Blanke SR and Chen LF: Helicobacter
pylori CagA activates NF-kappaB by targeting TAK1 for
TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 10:1242–1249. 2012.
View Article : Google Scholar
|
|
23
|
Suzuki M, Mimuro H, Kiga K, Fukumatsu M,
Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D,
et al: Helicobacter pylori CagA phosphorylation-independent
function in epithelial proliferation and inflammation. Cell Host
Microbe. 5:23–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yanai A, Maeda S, Shibata W, Hikiba Y,
Sakamoto K, Nakagawa H, Ohmae T, Hirata Y, Ogura K, Muto S, et al:
Activation of IkappaB kinase and NF-kappaB is essential for
Helicobacter pylori-induced chronic gastritis in Mongolian gerbils.
Infect Immun. 76:781–787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Isomoto H, Mizuta Y, Miyazaki M, Takeshima
F, Omagari K, Murase K, Nishiyama T, Inoue K, Murata I and Kohno S:
Implication of NF-kappaB in Helicobacter pylori-associated
gastritis. Am J Gastroenterol. 95:2768–2776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brandt S, Kwok T, Hartig R, König W and
Backert S: NF-kappaB activation and potentiation of proinflammatory
responses by the Helicobacter pylori CagA protein. Proc Natl Acad
Sci USA. 102:9300–9305. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu K, Zhang W and Wang J: Helicobacter
cytotoxin promotes gastric secretion of interleukin-8. World J
Gastroent. 10:907–911. 2002.
|
|
28
|
Handa O, Naito Y and Yoshikawa T: CagA
protein of Helicobacter pylori: A hijacker of gastric epithelial
cell signaling. Biochem Pharmacol. 73:1697–1702. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yoon JH, Seo HS, Choi SS, Chae HS, Choi
WS, Kim O, Ashktorab H, Smoot DT, Nam SW, Lee JY and Park WS:
Gastrokine 1 inhibits the carcinogenic potentials of Helicobacter
pylori CagA. Carcinogenesis. 35:2619–2629. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Buti L, Spooner E, van der Veen AG,
Rappuoli R, Covacci A and Ploegh HL: Helicobacter pylori
cytotoxin-associated gene A (CagA) subverts the
apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor
pathway of the host. Proc Natl Acad Sci USA. 108:9238–9243. 2011.
View Article : Google Scholar : PubMed/NCBI
|