Introduction
Hepatocellular carcinoma (HCC) is the most common
malignant primary liver cancer and accounts for 70–85% of the total
liver cancer cases worldwide (1).
It is the fifth most commonly diagnosed cancer and the third
leading cause of cancer-related death worldwide, with an estimated
over 500,000 new cases and 600,000 deaths due to HCC per year
(2–4). Development of HCC involves a variety
of etiological factors, including infection with hepatitis B virus
(HBV) and hepatitis C virus (HCV), chronic alcohol consumption,
intake of dietary aflatoxin B1, arsenic exposure, obesity, and
non-alcoholic fatty liver disease (5–7). On
the basis of tumor differentiation, HCC could be divided into four
histological grades, including well differentiated (G1), moderately
differentiated (G2), poorly differentiated (G3), and
undifferentiated (G4) types (8).
Currently, surgical resection and liver transplantation are the
major curative therapeutic strategies for patients with HCC
(9). Although great progress has
been achieved in the diagnostic techniques and treatments of HCC,
the prognosis remains unsatisfactory with an overall 5-year
survival rate of only 5% (10).
The poor prognosis of HCC cases is a results of recurrence even
after surgery, early invasion into blood vessels, intra-hepatic
metastases and extra-hepatic metastases (11). Therefore, it is necessary to fully
understand the molecular mechanisms underlying the progression of
HCC and explore novel efficacious therapeutic targets for this
disease.
microRNAs (miRNAs) are a type of endogenous, highly
conserved and non-coding short RNAs with approximately 19–25
nucleotides in length (12). By
pairing to complementary binding sites within the 3′untranslated
regions (3′UTRs) of their target genes, miRNAs suppressed their
protein translation or induced degradation of target mRNAs
(13). As regulators of gene
expression, miRNAs play a critical role in a wide range of
important biological process, such as cell proliferation,
differentiation, apoptosis, cell cycle, metabolism, invasion,
metastasis, angiogenesis, immunity, and development (14–16).
Abundant reports have demonstrated that the aberrant expression of
miRNAs contributes to carcinogenesis and progression of human
malignancies, also including HCC (17–19).
Indeed, miRNAs are frequently located in genomic breakpoint regions
and can act as tumor suppressors or oncogenes in human cancers,
depending on the roles of their target genes (20). For example, miR-548a-5p is
upregulated in HCC and targets a tumor suppressor, Tg737, to
promote cell proliferation and repress apoptosis (21); whereas miR-186 is downregulated in
HCC and inhibits cell proliferation, migration and invasion via
targeting Yes-associated protein 1 (22). Therefore, miRNAs could be promising
therapeutic targets for HCC patients.
In this study, we measured miR-187 expression in HCC
tissues and cell lines, investigated its functions on HCC cells
proliferation, migration and invasion, and explored its underlying
molecular mechanisms. Our results demonstrated that miR-187 was
downregulated in HCC and acted as a tumor suppressor through
directly targeting insulin-like growth factor 1 receptor
(IGF-1R).
Materials and methods
Ethic statement and tissues
samples
The present study was approved by the Ethics
Committee at Qianfoshan Hospital Affiliated to Shandong University
(Jinan, China). All HCC patients in this study had not been treated
with chemotherapy or radiotherapy prior to surgery section.
Experimental procedure were performed following to the Declaration
of Helsinki and the federal regulation on human research in the
People's Republic of China. Sixteen HCC tissues and adjacent
non-tumor tissues were collected at the Department of Hepatobiliary
Surgery, Qianfoshan Hospital Affiliated to Shandong University
(Jinan, China) between June 2013 and October 2014. All tissues
samples were immediately frozen in liquid nitrogen and stored at
−80°C.
Cell culture
HCC cell lines, HepG2, SMMC-7721, Hep3B, and Huh7
were obtained from the Cell Bank of Type Culture Collection of
Chinese Academy of Sciences (Shanghai, China), and cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA). The
immortalized normal liver epithelial cell line, THLE-3, was
purchased from ATCC (Manassas, VA, USA), and grown under the
conditions provided by the manufacturer. All cells were cultured at
37°C in a humidified incubator with 5% CO2.
Transient transfection
miR-187 mimics and miRNA mimics negative control
(NC) were obtained from GenePharma (Shanghai, China). The duplexes
of siRNA targeting IGF-1R (IGF-1R siRNA) and the negative control
(NC siRNA) were purchased from RIBOBIO (Guangzhou, China). Cells
were transfected with these oligonucleotide by Lipofectamine 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA), in accordance
with the manufacturer's instructions.
RNA extraction and quantitative
real-time reverse transcription-PCR (qRT-PCR)
Expressions levels of miR-187 and IGF-1R mRNA were
measured by using qRT-PCR. Total RNA was extracted from tissues or
cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse
transcription was done with M-MLV Reverse Transcriptase (Promega,
Madison, WI, USA), followed by real-time PCR with SYBR Premix Ex
Taq™ (Takara, Dalian, China). Relative miR-187 or IGF-1R mRNA
expression were calculated as 2−ΔΔCt and were normalized
to U6 or GAPDH, respectively.
3-(4,5-dimethylthiazolyl-2-yl)-2-5
diphenyl tetrazolium bromide (MTT) assay
HCC cells (3,000 cells/well) were seeded in 96-well
plates. At 24 h following transient transfection, cells were
cultured at 37°C in a humidified incubator with 5% CO2
for 1, 2, 3, and 4 days, respectively. At each time-point, MTT
assay (Sigma, St. Louis, MO, USA) was performed. In briefly, 20 µl
MTT solution (5 mg/ml) was placed in each well and cultured at 37°C
for additional 4 h. Subsequently, the culture medium was removed
and replaced with 150 µl DMSO (Sigma). Cells were incubated at room
temperature for 10 min, and the plates were shaken for 10 min with
an Eppendorf Mixmate (Eppendorf, GRE) for fully dissolution of MTT
crystals. The optical density (OD) at 490 nm was detected using a
microplate reader (Bio-Rad, Richmond, CA, USA).
Migration and invasion assay
Cell migration and invasion were measured using a
Transwell chamber model (8-µm pore size; Millipore, Billerica, MA,
USA). Transfected cells were harvested 48 h post-transfection and
suspended at a concentration of 3×105 cells/ml. For migration
assay, 150 µl of the suspension was added into the upper chambers.
For invasion assay, 150 µl of the suspension was placed into the
upper chambers pre-coated with Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA). Cells were then incubated at 37°C in a humidified
incubator with 5% CO2 for 48 h. Cells remaining in the
upper chambers were removed carefully with cotton wool. Migrated or
invaded cells through the membranes were fixed in 10% formalin and
stained with 0.5% crystal violet. Five visual fields (x200) were
randomly photographed from each membrane by using an inverted
microscope. All experiments were performed in triplicate.
miR-187 target predictions
The putative targets of miR-187 were analyzed by
using miRNA target prediction programs: miRDB (http://mirdb.org/cgi-bin/), and TargetScan (http://www.targetscan.org/).
Luciferase reporter assay
Luciferase reporter plasmids, pGL3-IGF-1R-3′UTR Wt
and pGL3-IGF-1R-3′UTR Mut, were constructed by GenePharma. For
luciferase reporter assay, HEK293T cells were seeded into 24-well
plates and grown to 60–70% confluence. Cells were co-transfected
with pGL3-IGF-1R-3′UTR Wt or pGL3-IGF-1R-3′UTR Mut, and miR-187
mimics or NC using Lipofectamine® 2000. Transfeted cells were
incubated at 37°C in a humidified incubator with 5% CO2
for 48 h and harvested for luciferase activities measurement with
Dual-luciferase reporter assay system (Promega). Firefly luciferase
activities were normalized to its corresponding Renilla
luciferase activities.
Protein extraction and western
blot
After transfection 72 h, cells were collected for
total protein extraction using radio-immunoprecipitation assay
(RIPA) buffer supplemented with protease inhibitors (Roche,
Shanghai, China). BCA Protein Assay kit (Santa Cruz Biotechnology,
Santa Cruz, CA, USA) was used to measure total protein
concentration. Equivalent proteins were separated on a 10%
SDS-polyacrylamide gel (PAGE) and transferred to
polyvinylidenefluoride (PVDF) membranes (Millipore, Boston, MA,
USA). In order to block non-specific binding, the membranes were
then incubated with 5% skim milk in Tris-buffered saline and 0.1%
Tween-20 (TBST) at room temperature for 1 h. Subsequently, the
membranes were probed with mouse anti-human IGF-1R monoclonal
primary antibody (1:1,000 dilution, ab985) and mouse anti-human
GADPH monoclonal primary antibody (1:1,000 dilution, ab127428)
(both from Abcam, Tokyo, Japan), at 4°C overnight. After washing
with TBST, goat anti-mouse horseradish peroxidase conjugated
secondary antibody (1:5,000 dilution; Abcam) were added for 2 h at
room temperature. Finally, the signal was visualized using enhanced
chemiluminescence (Millipore, Billerica, WI, USA).
Statistical analysis
All data were presented as mean ± SD., and compared
using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Double-tailed
P<0.05 was considered to be statistically significant.
Results
Expression of miR-187 was
downregulated in HCC tissues and cell lines
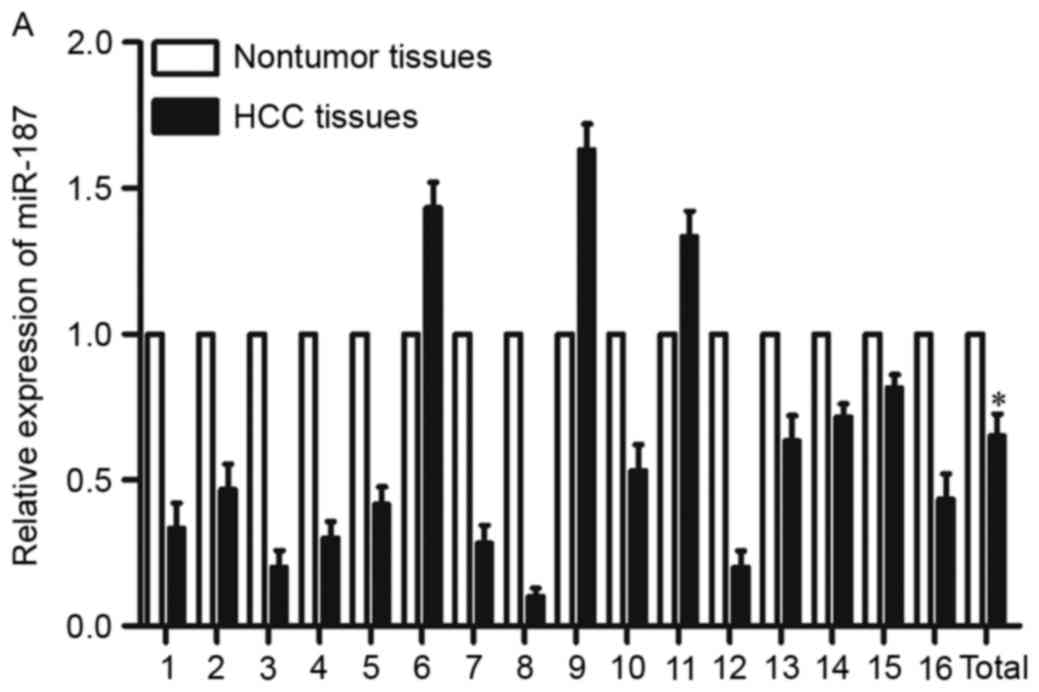
To investigate miR-187 expression in HCC, we
compared its expression levels between HCC tissues and their
adjacent nontumor tissues. Results of qRT-PCR showed that
expression of miR-187 was lower in HCC tissues than those in
adjacent nontumor tissues (Fig.
1A, P<0.05). Furthermore, the miR-187 expression was also
measured in HCC cell lines as well as immortalized normal liver
epithelial cells THLE-3. The results showed that miR-187 was
downregulated in all HCC cell lines compared with THLE-3 (Fig. 1B, P<0.05). HepG2 and Hep3B cell
lines, expressed relatively lower expression of miR-187, were
chosen as representatives for further functional experiments.
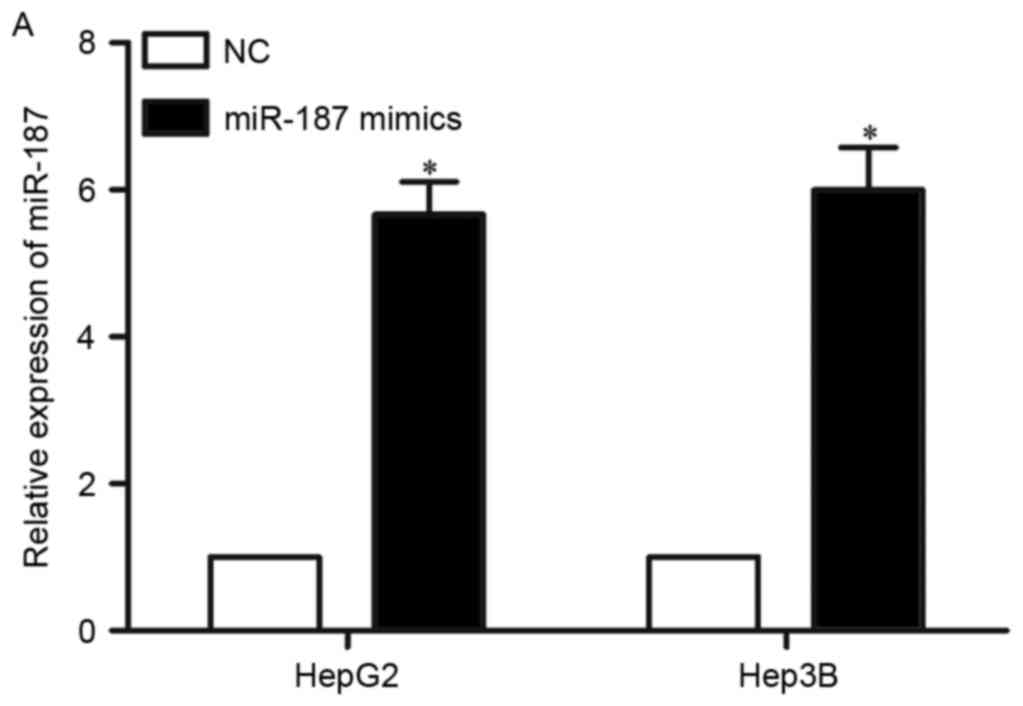
miR-187 overexpression inhibited cell
proliferation, migration and invasion of HCC
To investigate the biological functions of miR-187
in HCC, miR-187 mimics was used to increase its expression in HepG2
and Hep3B cells (Fig. 2A,
P<0.05). MTT assay showed that upregulation of miR-187
significantly inhibited the HepG2 and Hep3B cells proliferation
(Fig. 2B, P<0.05). The
migration and invasion abilities of HepG2 and Hep3B cells were also
dramatically reduced by miR-187 overexpression (Fig. 2C, P<0.05). These results
suggested that miR-187 may act as a tumor suppressor in HCC.
IGF-1R was a target gene of
miR-187
It is generally believed that miRNAs play critical
roles in biological process through negatively regulation of their
target genes via directly binding to the 3′UTR. To understand the
mechanism underlying the tumor suppressive roles of miR-187 in HCC,
we searched for its downstream target genes. Bioinformatic analysis
revealed that IGF-1R may be a direct target gene of miR-187
(Fig. 3A). First, we detected
IGF-1R mRNA expression in HCC tissues and their adjacent nontumor
tissues by using qRT-PCR. Interestingly, IGF-1R mRNA was
significantly upregulated in HCC tissues in comparison with
adjacent nontumor tissues (Fig.
3B, P<0.05). Moreover, Spearman's correlation analysis found
an inverse correlation between miR-187 and IGF-1R mRNA expression
in HCC tissues (r=−0.7719, P=0.0005, Fig. 3C).
To further confirm this hypothesis, the
pGL3-IGF-1R-3′UTR Wt or pGL3-IGF-1R-3′UTR Mut were transfected into
HEK293T cells together with miR-187 mimics or NC. The results
showed that the relative luciferase activities of pGL3-IGF-1R-3′UTR
Wt was obviously decreased in miR-187 mimics group as compared with
NC group (Fig. 3D, P<0.05).
However, the luciferase activities of pGL3-IGF-1R-3′UTR Mut was
unaffected by co-transfection of miR-187 mimics.
The downregulation of IGF-1R protein in HepG2 and
Hep3B after transfection with miR-187 mimics further confirmed that
IGF-1R was a direct target gene of miR-187 (Fig. 3E, P<0.05). Moreover,
upregulation of miR-187 had no regulation effect on IGF-1R mRNA
expression, indicating miR-187 regulated IGF-1R expression at
post-transcriptional level (Fig.
3F, P>0.05). These results demonstrated that IGF-1R was a
direct target of miR-187.
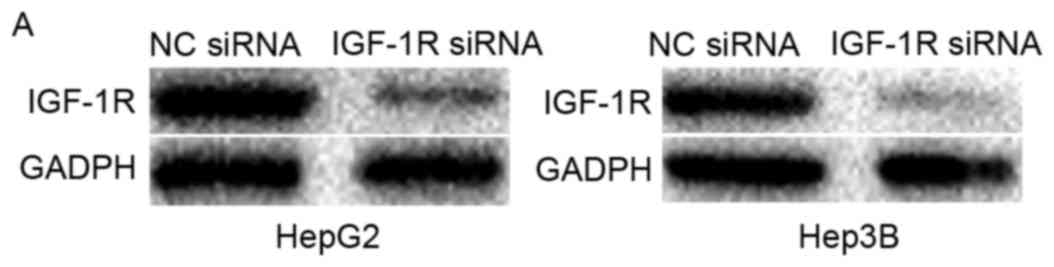
IGF-1R knockdown inhibited cell
proliferation, migration and invasion of HCC
To determine whether IGF-1R was a functional target
of miR-187 in HCC, HepG2 and Hep3B cells were injected with IGF-1R
siRNA or NC siRNA. Western blot analysis showed that IGF-1R was
reduced both in HepG2 and Hep3B cells following to transfection
with IGF-1R siRNA (Fig. 4A,
P<0.05). Functionally, IGF-1R knockdown could simulate the
effects of miR-187 overexpression on HCC, resulting in a
significant inhibition of cell proliferation (Fig. 4B, P<0.05), migration and
invasion (Fig. 4C, P<0.05).
These results provided further evidences that IGF-1R was a
functional target of miR-187 in HCC.
Discussion
The dysregulation of miR-187 occurs frequently in
many human cancers. miR-187 expression was observed to be
downregulated in both renal cell carcinoma tumor tissue and plasma.
Reduced miR-187 was closely correlated with higher tumor grade and
stage. Moreover, renal cell carcinoma patients with low miR-187
level have poor prognosis compared with patients with high miR-187
expression (23). In colorectal
cancer, expression level of miR-187 was lower in tumor tissues and
cell lines. Kaplan-Meier analysis revealed that reduced miR-187
expression was associated with shorter overall survival and
relapse-free survival of patients with colorectal cancer (24). Consistent with these results,
downregulation of miR-187 was also found in prostate cancer
(25) and lung cancer (26,27).
However, others have reported that miR-187 was upregulated in
certain tumor types. In ovarian cancer, expression of miR-187 was
higher in tumor tissues, and its expression was obviously
correlated with better overall survival and recurrence-free
survival. Further, multivariate analysis indicated that miR-187
served as an independent prognostic factor for patients with
ovarian cancer (28). These
conflicting findings suggest that miR-187 has a tumor and
tissue-specific expression pattern. In our current study, miR-187
was found to be significantly downregulated in HCC tissues and cell
lines, which provides additional information about the expression
level of miR-187 in human cancer.
Several lines of evidence implicate the miR-187 in
cancer development. Zhao et al demonstrated that ectopic of
miR-187 expression suppressed cell growth and motility in
vitro, and decreased cell growth in vivo trough directly
targeting B7-H3 (23). In
non-small cell lung cancer, upregulation of miR-187 inhibited cell
proliferation, colony formation, migration, invasion, and induced
apoptosis via suppression of BCL-6 expression (27). Wang et al found that miR-187
repressed cell proliferation, migration, invasion, and promoted
cell apoptosis of colorectal cancer by negatively regulation of
CD276 (24). Study by Lynam-Lennon
et al reported that miR-187 overexpression improved
sensitivity of esophageal adenocarcinoma cells to X-ray radiation
and cisplatin (29). These studies
provided evidences to suggest that miR-187 functions as a tumor
suppressor; however, others have reported that miR-187 may play an
oncogenic role in other human cancers. For example, restoration of
miR-187 promoted ovarian cancer cells proliferation, migration and
epithelial-to-mesenchymal transition via blockade of Disabled
homolog-2 (28). These conflicting
observations also indicate that functions of miR-187 has a tumor
and tissue-specific dependence. In this study, for the first time,
we demonstrated that enforced miR-187 expression in HCC cells
inhibited cell proliferation, migration and invasion. These studies
suggested that miR-187 play significant roles in tumor
development.
Furthermore, our data also verified that IGF-1R was
a direct functional target of miR-187 in HCC. There are several
lines of evidence to demonstrate this. First, bioinformatic
analysis showed that 3′UTR of IGF-1R contained potential binding
sites of miR-187. Second, IGF-1R mRNA was upregulated in clinical
HCC tissues and inversed correlated with miR-187 expression. Third,
upregulation of miR-187 suppressed luciferase activities of
pGL3-IGF-1R-3′UTR Wt, and this suppressive effect could be
abolished by mutation of the miR-187 seed binding sites, suggesting
miR-187 could directly targeted the 3′UTR of IGF-1R. Fourth,
miR-187 overexpression decreased IGF-1R protein expression, but not
IGF-1R mRNA expression of HCC, indicating miR-187 regulated IGF-1R
expression at posttranscriptional level. Finally, the suppressive
effect of IGF-1R knockdown on HCC cells were similar with those
induced by miR-187 overexpression. These results demonstrated that
the tumor suppressive roles of miR-187 was partly mediated by
downregulation of IGF-1R.
IGF-1R, a member of the insulin receptor family of
receptor tyrosine kinases, has been reported to be upregulated in
multiple types of cancer, such as HCC (30), prostate cancer (31), lung cancer (32), gastric cancer (33), bladder cancer (34) and so on.
Significant amount of studies demonstrated that
IGF-1R is capable of mediating both IGF-I and IGF-II signaling and
regulates a great deal of cellular processes including cell cycle,
migration, metabolism, survival, proliferation, and differentiation
(31,35–37).
Increasing studies suggested that IGF-1R knockdown could inhibit
cell growth (38,39), invasion (40), and improve cell apoptosis (38,40)
and chemo-sensitivity (39). These
findings provide sufficient evidence that IGF-IR plays important
roles in hepatocarcinogenesis and HCC progression, and therefore
may be a promising therapeutic target.
In conclusion, we demonstrated that miR-187 was
downregulated in HCC tissues and cell lines. Overexpression of
miR-187 inhibited HCC growth and metastasis through directly
targeting IGF-1R. These findings provide insights into the
pathogenesis and development of HCC, which could be used to explore
novel therapeutic strategies for HCC treatments.
References
|
1
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schwartz M, Roayaie S and Konstadoulakis
M: Strategies for the management of hepatocellular carcinoma. Nat
Clin Pract Oncol. 4:424–432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Poon RT and Fan ST: Hepatectomy for
hepatocellular carcinoma: Patient selection and postoperative
outcome. Liver Transpl. 10 2 Suppl 1:S39–S45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology. 127 5
Spuul 1:S72–S78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park KU, Seo YS, Lee YH, Park J, Hwang I,
Kang KJ, Nam J, Kim SW and Kim JY: Altered microRNA expression
profile in hepatitis B virus-related hepatocellular carcinoma.
Gene. 573:278–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han DH, Choi GH, Kim KS, Choi JS, Park YN,
Kim SU, Park JY, Ahn SH and Han KH: Prognostic significance of the
worst grade in hepatocellular carcinoma with heterogeneous
histologic grades of differentiation. J Gastroenterol Hepatol.
28:1384–1390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bosch FX, Ribes J, Diaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127:(5 Suppl 1): S5–S16. 2004. View Article : Google Scholar
|
|
10
|
Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu
C, Zou L, Li Z, Zhao J and Lin N: MicroRNA-101 suppresses
SOX9-dependent tumorigenicity and promotes favorable prognosis of
human hepatocellular carcinoma. FEBS Lett. 586:4362–4370. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harlan LC, Parsons HM, Wiggins CL, Stevens
JL and Patt YZ: Treatment of hepatocellular carcinoma in the
community: Disparities in standard therapy. Liver Cancer. 4:70–83.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Papaconstantinou I, Karakatsanis A,
Gazouli M, Polymeneas G and Voros D: The role of microRNAs in liver
cancer. Eur J Gastroenterol Hepatol. 24:223–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rottiers V, Najafi-Shoushtari SH, Kristo
F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N,
Mostoslavsky R and Näär AM: MicroRNAs in metabolism and metabolic
diseases. Cold Spring Harb Symp Quant Biol. 76:225–233. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q,
Yang W, Yao Y, Liu Q and Tu K: MicroRNA-212 suppresses tumor growth
of human hepatocellular carcinoma by targeting FOXA1. Oncotarget.
6:13216–13228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z
and Jiang J: miR-214 inhibits cell growth in hepatocellular
carcinoma through suppression of β-catenin. Biochem Biophys Res
Commun. 428:525–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao G, Wang T, Huang QK, Pu M, Sun W,
Zhang ZC, Ling R and Tao KS: MicroRNA-548a-5p promotes
proliferation and inhibits apoptosis in hepatocellular carcinoma
cells by targeting Tg737. World J Gastroenterol. 22:5364–5373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ruan T, He X, Yu J and Hang Z:
MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo
signaling and tumorigenesis in hepatocellular carcinoma. Oncol
Lett. 11:2941–2945. 2016.PubMed/NCBI
|
|
23
|
Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y,
Fan S and Liu Y: MicroRNA-187, down-regulated in clear cell renal
cell carcinoma and associated with lower survival, inhibits cell
growth and migration though targeting B7-H3. Biochem Biophys Res
Commun. 438:439–444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL,
Yu MH and Qin J: MicroRNA-187 inhibits tumor growth and invasion by
directly targeting CD276 in colorectal cancer. Oncotarget.
7:44266–44276. 2016.PubMed/NCBI
|
|
25
|
Casanova-Salas I, Masiá E, Armiñán A,
Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ
and López-Guerrero JA: miR-187 targets the androgen-regulated gene
ALDH1A3 in prostate cancer. PLoS One. 10:e01255762015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Azad F Mirzadeh, Naeli P, Malakootian M,
Baradaran A, Tavallaei M, Ghanei M and Mowla SJ: Two lung
development-related microRNAs, miR-134 and miR-187, are
differentially expressed in lung tumors. Gene. 577:221–226. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F
and Li D: MicroRNA-187-3p mitigates non-small cell lung cancer
(NSCLC) development through down-regulation of BCL6. Biochem
Biophys Res Commun. 471:82–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chao A, Lin CY, Lee YS, Tsai CL, Wei PC,
Hsueh S, Wu TI, Tsai CN, Wang CJ, Chao AS, et al: Regulation of
ovarian cancer progression by microRNA-187 through targeting
disabled homolog-2. Oncogene. 31:764–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lynam-Lennon N, Bibby BA, Mongan AM,
Marignol L, Paxton CN, Geiersbach K, Bronner MP, O'Sullivan J,
Reynolds J and Maher SG: Low miR-187 expression promotes resistance
to chemoradiation therapy in vitro and correlates with treatment
failure in patients with esophageal adenocarcinoma. Mol Med.
22:2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
E C, Li J, Shao D, Zhang D, Pan Y, Chen L
and Zhang X: The insulin-like growth factor-I receptor inhibitor
picropodophyllin-induced selective apoptosis of hepatocellular
carcinoma cell through a caspase-dependent mitochondrial pathway.
Oncol Res. 21:103–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y, Cheng Q, Ren Z, Xu L, Zhao Y, Sun J,
Hu S and Xiao W: Induction of IGF-1R expression by EGR-1
facilitates the growth of prostate cancer cells. Cancer Lett.
317:150–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wei YH, Tang HX, Liao YD, Fu SL, Xu LQ,
Chen G, Zhang C, Ju S, Liu ZG, You LK, et al: Effects of
insulin-like growth factor 1 receptor and its inhibitor AG1024 on
the progress of lung cancer. J Huazhong Univ Sci Technolog Med Sci.
35:834–841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gryko M, Kisluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie QX, Lin XC, Zhang MF, Han CX and Guo
YH: Expression of IGF-I and IGF-IR in bladder cancer. Ai Zheng.
23:707–709. 2004.(In Chinese). PubMed/NCBI
|
|
35
|
Singh RK, Gaikwad SM, Jinager A, Chaudhury
S, Maheshwari A and Ray P: IGF-1R inhibition potentiates cytotoxic
effects of chemotherapeutic agents in early stages of
chemoresistant ovarian cancer cells. Cancer Lett. 354:254–262.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen HX and Sharon E: IGF-1R as an
anti-cancer target-trials and tribulations. Chin J Cancer.
32:242–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Singh I, Amin H, Rah B and Goswami A:
Targeting EGFR and IGF 1R: A promising combination therapy for
metastatic cancer. Front Biosci (Schol Ed). 5:231–246. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yue L, Wang Y, Wang H, Gao H, Liang J, Sui
A, Xiang J, Zhou F, Xu C, Zhao W, et al: Inhibition of
hepatocellular carcinoma cell growth by an anti-insulin-like growth
factor-I receptor monoclonal antibody. Oncol Rep. 28:1453–1460.
2012.PubMed/NCBI
|
|
39
|
Zhang YW, Yan DL, Wang W, Zhao HW, Lu X,
Wu JZ and Zhou JR: Knockdown of insulin-like growth factor I
receptor inhibits the growth and enhances chemo-sensitivity of
liver cancer cells. Curr Cancer Drug Targets. 12:74–84. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yao WF, Liu JW, Sheng GL and Huang DS:
Blockade of IGF-IR exerts anticancer effects in hepatocellular
carcinoma. Mol Med Rep. 4:719–722. 2011.PubMed/NCBI
|