Introduction
Solid pseudopapillary neoplasm (SPN) of the pancreas
is a low-grade malignant neoplasm with circumscribed, variegated,
hemorrhagic, solid and cystic features (1). SPN was first described by Frantz in
1959 (2), and in 2010 the World
Health Organization defined the cancer as solid pseudopapillary
neoplasm of the pancreas (3). SPN
accounts for ~5% of cystic pancreatic tumors and ~1–6% of all
exocrine pancreatic tumors (4).
Despite primarily occurring in younger women, patients with SPN
have been reported to range from 2–85 years old (5). SPN is currently treated by complete
surgical excision, and diagnosed either by imaging, using electron
microscopy, or histology, using immunohistochemistry. However, the
exact molecular pathology and pathogenesis of SPN remains unclear
(6).
SPN pathogenesis has been investigated extensively.
Activation of the Wnt-β-catenin signal pathway, associated with
mutations of exon 3 in the β-catenin gene, CTNNB1, may be involved
in the tumorigenesis of SPN (7–9).
β-catenin acts as a transcriptional activator in conjunction with T
cell factor and lymphoid enhancer factor in the Wnt-β-catenin
pathway, inducing the expression of target genes, and these may be
useful diagnostic molecular markers (10). Kang et al (11) demonstrated that expression of the
Wnt-β-catenin signaling pathway targets genes for matrix
metalloproteinase (MMP)-7, cyclin-D1 and c-Myc, and may result in
an unpredictable clinical course in SPN. β-catenin is also involved
in cell-cell adhesion, helping E-cadherin to link to the
cytoskeleton (12). Silencing of
E-cadherin mutations and nuclear translocation of β-catenin
following activation of mutations results in loss of adherens
junctions, and this same loss is commonly observed in patients with
SPN (13).
However, little is known about SPN besides the
activation of the Wnt-β-catenin signaling pathway. In order to
identify the molecular pathogenesis of SPN, microarray data were
downloaded and analyzed to identify differentially expressed genes
(DEGs) between SPN and non-neoplastic pancreatic tissues.
Significantly enriched pathways and functions were also screened,
followed by the functional annotation of DEGs based on
transcription factor and tumor-associated gene databases.
Resultantly, a protein-protein interaction (PPI) network of DEGs
was constructed and visualized.
Materials and methods
Obtaining and preprocessing of mRNA
expression profile data
The mRNA expression profiles of SPN and
non-neoplastic pancreatic tissues were obtained from the National
Center of Biotechnology Information Gene Expression Omnibus (GEO,
https://www.ncbi.nlm.nih.gov/geo/)
database. The access number was GSE43795 (14), and datasets from 14 SPN samples and
6 control samples were used for further analysis. The platform used
was Illumina Human HT-12 V4.0 Expression BeadChip (Illumina, Inc.,
San Diego, CA, USA). Original data were preprocessed with the Limma
package (version 3.2.2; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(15), AFFY package (version
1.32.0; http://www.bioconductor.org/packages/release/bioc/html/affy.html)
(16) and the org.Hs.eg.db package
using Bioconductor software (version 2.14; Fred Hutchinson Cancer
Research Center, Seattle, WA, USA). Preprocessing of the data
included background correction (17), quantile normalization and probe
summarization. The expression matrix was then obtained, with each
row representing the expression of a gene, and each column a
sample.
DEGs screening
Bayesian analysis was performed using the Limma
package (15), to identify DEGs
between SPN and control samples. FDR<0.01 and log2
FC≥3 were used as the thresholds.
Enrichment analysis of DEGs
To study DEGs at functional level, gene ontology
(GO, http://www.geneontology.org) functional
enrichment analysis (18) and
Kyoto Encyclopedia of Gene and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html)
pathway enrichment (19) were
performed using the Database for Annotation Visualization and
Integrated Discovery (DAVID; version 6.7) software, an online
biological tool (20). GO is a
collection of controlled vocabularies, and only the biological
process functions were enriched. P<0.01 was set as the cut-off
criterion for enrichment analysis.
Gene functional annotation
analysis
Functional annotation analysis of genes is an
important task, as it demonstrates associations between genes and
biological pathways (21).
According to the information on transcription factors provided by
TRANSFAC (version 11.2), the screened DEGs were further annotated.
In order to investigate the molecular mechanism of SPN, all known
oncogenes and tumor suppressor genes were extracted, based on the
Tumor Associated Genes (TAG) database (version 3.07) (22), the Tumor Suppressor Gene database
(version 2.0) (22) and the work
of Zhao et al (23).
PPI network construction and
sub-network detection
PPI network analysis is necessary to comprehensively
understand the intracellular process. The Search Tool for the
Retrieval of Interacting Gene/Proteins (STRING) database (24) has been widely used to construct PPI
networks. To begin, PPI data (verified through experiments, text
mining and co-expression analysis) were downloaded (2014.05.09)
from STRING (version 10.0; http://string-db.org/). All DGEs were mapped to this
dataset and a threshold of combined score ≥0.9 was applied to
screen the interaction pairs. Finally, selected pairs were
visualized using Cytoscape software (version 3.2.0; National
Institute of General Medical Sciences, Bethesda, MD, USA).
The identification of significantly differentially
expressed sub-networks within a large network is the primary task
when a PPI network is constructed. The BioNet package (version 2.1)
(25) was employed for sub-network
analysis, and FDR<0.0001 was set as the cut-off criterion. KEGG
enrichment analysis was also performed at the sub-network
level.
Results
DEG screening
Bayesian analysis was performed on the mRNA
expression profile data with the criteria FDR<0.01 and
|log2FC|≥3. Based on these criteria a total of 1,420
DEGs were screened out, among which 710 DEGs corresponding to 751
transcripts were upregulated and 710 DEGs corresponding to 746
transcripts were downregulated.
Enrichment analysis of DEGs
KEGG pathway enrichment analysis indicated that the
710 upregulated DEGs were enriched in 10 pathways, including
pancreatic secretion, maturity onset diabetes of the young, protein
digestion and absorption, while the 710 downregulated DEGs were
enriched in 17 pathways, including the Wnt signaling pathway,
melanogenesis, axon guidance, protein digestion and absorption
(P<0.01). The top ten pathways are listed in Table I.
 | Table I.KEGG pathway analysis of
differentially expressed genes. |
Table I.
KEGG pathway analysis of
differentially expressed genes.
| Pattern | KEGG pathway | Gene counts | P-value |
|---|
| Down | Pancreatic
secretion | 28 | 2.55E-15 |
|
| Maturity onset
diabetes of the young | 12 | 1.97E-10 |
|
| Protein digestion
and absorption | 19 | 2.12E-09 |
|
| Drug
metabolism-cytochrome P450 | 14 | 3.98E-06 |
|
| Proximal tubule
bicarbonate reclamation | 7 | 4.99E-05 |
|
| Metabolism of
xenobiotics by cytochrome P450 | 12 | 7.35E-05 |
|
| Fat digestion and
absorption | 8 | 9.77E-04 |
|
| Glutathione
metabolism | 8 | 1.72E-03 |
|
| Tyrosine
metabolism | 7 | 2.26E-03 |
|
| Starch and sucrose
metabolism | 8 | 2.84E-03 |
| Up | Wnt signaling
pathway | 15 | 3.92E-04 |
|
| Melanogenesis | 11 | 1.16E-03 |
|
| Axon guidance | 12 | 2.77E-03 |
|
| Protein digestion
and absorption | 9 | 2.82E-03 |
|
| Leukocyte
transendothelial migration | 11 | 3.54E-03 |
|
| Cell adhesion
molecules (CAMs) | 12 | 3.56E-03 |
|
| Basal cell
carcinoma | 7 | 3.84E-03 |
|
| Pathways in
cancer | 22 | 4.23E-03 |
|
| Arrhythmogenic
right ventricular cardiomyopathy | 8 | 5.66E-03 |
|
| Tight junction | 11 | 9.33E-03 |
GO functional enrichment analysis demonstrated that
the 710 upregulated DEGs were enriched in 47 functions, including
digestion, secretion and the cellular response to zinc ions, and
the 710 downregulated DEGs were enriched in 88 pathways, including
nervous system development, cell differentiation and neuron
differentiation (P<0.01). The top ten pathways are listed in
Table II.
 | Table II.Significantly enriched biological
process function of differentially expressed genes. |
Table II.
Significantly enriched biological
process function of differentially expressed genes.
| Pattern | GO ID | Term | Gene counts | P-value |
|---|
| Down | GO:0007586 | Digestion | 31 | 1.11E-16 |
|
| GO:0046903 | Secretion | 72 | 6.84E-10 |
|
| GO:0071294 | Cellular response
to zinc ion |
8 | 9.69E-10 |
|
| GO:0031018 | Endocrine pancreas
development | 15 | 1.10E-09 |
|
| GO:0035270 | Endocrine system
development | 22 | 1.16E-08 |
|
| GO:0001525 | Angiogenesis | 39 | 4.71E-08 |
|
| GO:0010038 | Response to metal
ion | 28 | 9.86E-08 |
|
| GO:0030001 | Metal ion
transport | 48 | 4.77E-07 |
|
| GO:0042593 | Glucose
homeostasis | 20 | 9.07E-07 |
|
| GO:0071248 | Cellular response
to metal ion | 15 | 1.11E-06 |
| Up | GO:0007399 | Nervous system
development | 119 | 5.82E-10 |
|
| GO:0030154 | Cell
differentiation | 168 | 8.72E-10 |
|
| GO:0030182 | Neuron
differentiation | 79 | 3.39E-09 |
|
| GO:0001501 | Skeletal system
development | 42 | 5.94E-09 |
|
| GO:0043392 | Negative regulation
of DNA binding |
9 | 1.01E-05 |
|
| GO:0046189 | Phenol-containing
compound biosynthetic process |
7 | 6.87E-05 |
|
| GO:0060412 | Ventricular septum
morphogenesis |
7 | 8.76E-05 |
|
| GO:0007268 | Synaptic
transmission | 46 | 9.26E-05 |
|
| GO:0002720 | Positive regulation
of cytokine production involved in immune response |
6 | 1.02E-04 |
|
| GO:0007155 | Cell adhesion | 59 | 3.00E-04 |
Gene functional annotation
analysis
To investigate the molecular mechanisms of SPN, the
function of DEGs as transcriptional factors and TAGs were also
analyzed. A total of 74 DEGs were transcriptional factors, among
which 31 were downregulated and 43 were upregulated; and 124 DEGs
were TAGs, among which 73 were downregulated and 51 were
upregulated (Table III).
Additionally, through comparison with data collected by Schriml
et al (26), membrane
metallo-endopeptidase (MME), MMP-2 and MMP-9 were identified as
DEGs associated with proliferative diabetic retinopathy.
 | Table III.Functional statistics of
differentially expressed genes between solid pseudopapillary
neoplasm and control samples. |
Table III.
Functional statistics of
differentially expressed genes between solid pseudopapillary
neoplasm and control samples.
| Pattern | TF counts | TF genes | TAG counts | TAG genes |
|---|
| Down | 31 | CDX2, EHF, ELF3,
FOSB, FOXA2, FOXA3, FOXC1, FOXQ1, GATA4, HEYL, HHEX, HNF4G | 73 | Oncogene: CD24,
CXCL1, EGFR, ELF3, ERBB3, FGFR1, FGFR3, GATA4, GFI1, GPX2, JUN,
LCN2, MEIS1, MYC, SPHK1 |
|
|
| INSM1, ISL1,
KCNIP3, KLF5, LMO3, MEIS1, NKX2.2, NKX2.5, NR4A2, NR5A2, ONECUT1,
PAX6, PBX3, PDX1, PKNOX2, PLAGL1, SOX9, TEAD4, XBP1 |
| Tumor suppressor:
WNK2, VIL1, UCHL1, TPM1, TFPI2, SYT13, STEAP3, SRPX, SIK1,
SFRP5, SERPINI2, RAP1GAP, RAB25, PTPRK, PRKCDBP, PLK2, PLAGL1,
PDX1, PDGFRL, PAX6, ONECUT1, NRCAM, MUC1, MTUS1, MT1G, MEG3, LPL,
KLF5, KLF10, ID4, GNMT, GAS1, FOXC1, FOXA2, ERRFI1, EPHA1, ENC1,
EHF, DEFB1, DAPK1, CLDN23, CEBPA, CDH1, C2orf40, BTG2, BMP2, BIN1,
ADAMTS9 Other: TACC2, SLC43A1, RRAS2, PBX3, NR4A2, MAP3K5, GRB7,
CHRM3, CDX2, CD44 |
| Up | 43 | TWIST2, TFAP2C,
TCF7, TBX3, T, SOX11, SIM2, SHOX2, RUVBL1 | 51 | Oncogene: RUNX2,
NRAS, NOV, NET1, MME, MLLT11, MAP3K8, MAFG, LAMC2, GNA12, FYN,
FGF20 |
|
|
| RUNX2, REST,
PRDM1, PITX2, PGR, NR0B1, NFAT5, MAFG, MAF, LEF1, KLF12, HOXC9,
HOXC8, HOXC6, HOXC5, HOXC4, HOXB8, HOXB7, HOXB3, HEY2, HEY1, HAND2,
HAND1, GTF2H2, GLI2 |
| Tumor Suppressor:
ZBTB7C, WNT5A, WIF1, TWIST2, TMEM127, TMEFF2, THSD1, SOX11,
RASL10B, PTPRG, PRDM1, PPP1R1B, MIR185, MCPH1, LSAMP, ISG15, HPGD,
GLIPR1, FANCD2, DKK3, CSMD1, CNTNAP2, CDKN2D, CDH11, CABLES2,
C10orf90, BIK, AXIN2, ARHGAP29, ARHGAP20 |
|
|
| GATA1, FUBP1,
ETV5, ESRRG, EMX2, DR1, DBP, BARX2, AR |
| Other: WNT2B,
TPH1, TPD52L1, TFAP2C, PITPNA, OGG1, MCC, MAF, HOXC6 |
PPI network construction and
sub-network detection
A PPI network of DEGs was constructed based on the
STRING database. The top 6 genes with degree >5 were epidermal
growth factor receptor (EGFR), proto-oncogene tyrosine protein
kinase Fyn (FYN), c-JUN (JUN), glucagon (GCG), c-Myc (MYC) and CD44
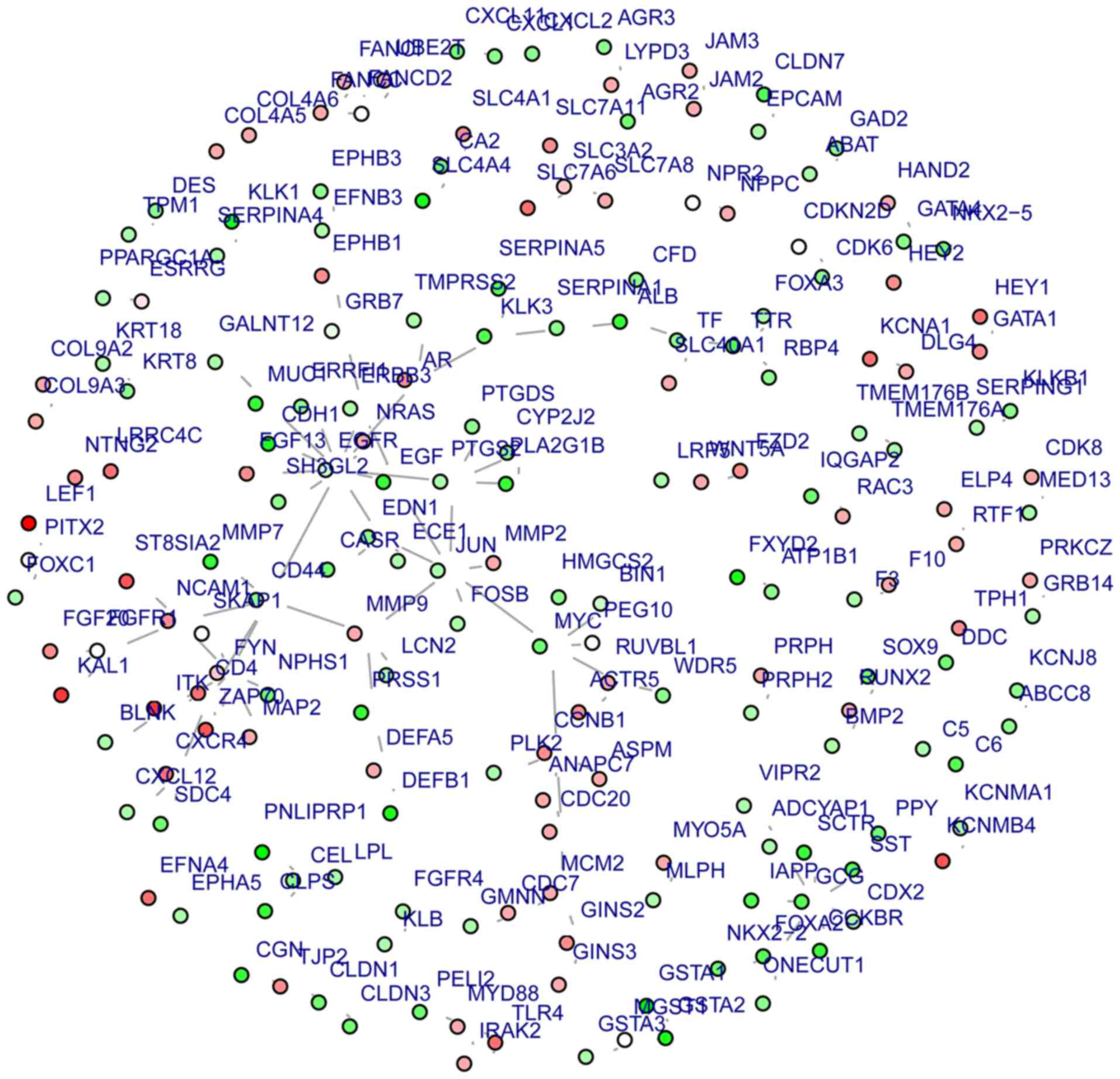
(Fig. 1). A sub-network involving
70 gene nodes was identified with EGFR (degree=12) as the central
gene (Fig. 2). Genes in this
sub-network primarily participate in various types of cancer and
cancer-associated processes, including signaling pathways [such as
the epidermal growth factor receptor (ErbB) and
gonadotropin-releasing hormone (GnRH) signaling pathways], and
immune response pathways (Table
IV).
 | Table IV.Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of differentially expressed
genes in the identified sub-network. |
Table IV.
Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of differentially expressed
genes in the identified sub-network.
| KEGG pathway | Gene counts | P-value | Gene |
|---|
| Bladder cancer | 7 | 2.11E-08 | MYC, EGFR, EGF,
CDH1, MMP9, NRAS, MMP2 |
| Endometrial
cancer | 5 | 4.19E-05 | MYC, EGFR, EGF,
CDH1, NRAS |
| Melanoma | 5 | 0.0001884 | EGFR, EGF, CDH1,
NRAS, FGF13 |
| Cell cycle | 6 | 0.0003266 | CDC20, CCNB1,
CDC7, MYC, ANAPC7, MCM2 |
| Prostate
cancer | 5 | 0.0005422 | AR, KLK3, EGFR,
EGF, NRAS |
| ErbB signaling
pathway | 6 | 4.57E-05 | JUN, MYC, EGFR,
EGF, ERBB3, NRAS |
| GnRH signaling
pathway | 5 | 0.0009664 | JUN, EGFR,
PLA2G1B, NRAS, MMP2 |
| T cell receptor
signaling pathway | 6 | 0.0001534 | JUN, CD4, FYN,
ZAP70, NRAS, ITK |
| Axon guidance | 7 | 4.76E-05 | CXCR4, FYN,
EFNB3, CXCL12, EPHB1, NRAS, EPHB3 |
| Pathways in
cancer | 12 | 3.841E-06 | AR, JUN, KLK3,
MYC, EGFR, EGF, CDH1, MMP9, PTGS2, NRAS, MMP2, FGF13 |
Discussion
The SPN is a grossly solid or solid and cystic
malignant epithelial neoplasm, where poorly cohesive cells
surrounding delicate blood vessels form degenerative pseudopapillae
(27). The present study aimed to
investigate the potential mechanisms of SPN, and identify genes to
use as diagnostic markers and understand tumor phenotype and
behavior, aiding in the development of molecularly-targeted
therapy. A total of 1,420 DEGs were identified between SPN and
control samples. Following PPI network analysis, EGFR, FYN, JUN,
GCG, MYC and CD44 were identified. GO functional enrichment
analysis and KEGG pathway enrichment analysis indicated that these
were predominantly enriched in the ErbB, GnRH and Wnt signaling
pathways.
MME, MMP-2 and MMP-9 were upregulated and identified
to be associated with proliferative diabetic retinopathy. MME, also
termed CD10, encodes MME, which is a 100-kD type II transmembrane
glycoprotein. CD10 is associated with various types of cancer,
including gastric (28), breast
(29), colorectal (30) and pancreatic cancer (31). Ikenaga et al (31) demonstrated that CD10+
pancreatic stellate cells promote the invasion of pancreatic cancer
cells and secrete MMP-3, contributing to the progression of
pancreatic cancer. Therefore, CD10 may be an optimal therapeutic
target in the treatment of SPN. MMP-2 and MMP-9 both encode members
of the MMP family, a major family of proteases involved in
remodeling the extracellular matrix. Activation of MMP-2 and MMP-9
has been demonstrated to be associated with the metastasis process
and local recurrence rate (32).
Inhibiting MMP activation blocks the metastasis process and is an
effective therapeutic approach (33). El-Ghlban et al (34) demonstrated that the fusion form of
chlorotoxin (CTX), which is formed by CTX and the human lgG-Fc
domain, may be an effective treatment for pancreatic cancer, as it
binds to MMP-2 and suppresses its expression. Thus, MMP-2 also has
the potential to be used as therapeutic target in the treatment of
SPN.
PPI network analysis demonstrated that the
expression levels of EGFR, FYN, JUN, GCG, MYC and CD44 were
significantly increased in SPN samples compared with controls,
indicating that these genes are associated with SPN. EGFR encodes
the transmembrane glycoprotein epidermal growth factor, a member of
the protein kinase superfamily (35). It induces receptor dimerization and
tyrosine autophosphorylation, and is overexpressed in pancreatic
cancer (36,37). Phosphorylation of EGFR initiates
modules including the mitogen-activated protein kinase (MAPK)
pathway, the phosphatidylinositol 3-kinase/Akt pathway and
MAPK/extracellular signal-related kinase (ERK) pathway, all of
which have been proven to affect cell survival, metastasis,
proliferation, invasion and induction of cancer (38). JUN encodes c-Jun, a proto-oncogene
and basic region-leucine zipper transcription factor involved in
multiple cellular processes through the formation of various
dimeric complexes (39). The
direct combination of JUN transcriptional activation and cyclin D1
provides a molecular link between growth factor signaling and the
changes in cell cycle proteins that drive the G1/S
transition. Previous studies have demonstrated that cyclin D1
activates the MAPK/ERK pathway and induces cancer (40,41).
MYC encodes c-Myc, an avian myelocytomatosis viral oncogene homolog
that participates in apoptosis, adhesion, differentiation, growth
and migration (42).
Overexpression of MYC in pancreatic cancer (43,44)
has been demonstrated previously. MYC activation results in
upregulation of G1-specific cyclins and cyclin-dependent
kinases, and inhibits negative regulatory factors of cell cycle
progression. Cells were therefore able to pass through the
restriction point and progress from the G1 to the S
phase (45).
It has been previously demonstrated that the Wnt
signaling pathway is involved in the tumorigenesis of SPN (7), and KEGG pathway analysis of all the
upregulated DEGs indicated enrichment of the Wnt signaling pathway.
The ErbB and GnRH signaling pathway were also demonstrated to be
significantly enriched. The ErbB protein family contains four
structurally-associated receptor tyrosine kinases including
ErbB-1/HER1/EGFR, ErbB-2/HER2, ErbB-3/HER3 and ErbB-4/HER4.
Excessive ErbB signaling is associated with the development of
various types of solid tumor (46). Previous clinical studies have
demonstrated that ErbB-1 and ErbB-2 expression is altered in
numerous types of human cancer, and the resultant excessive
signaling may be critical factors in tumor etiology and progression
(47). It has been previously
demonstrated that ErbB-1 induces cancer (48), and ErbB-2 homodimers alone may
contribute to malignancy (49).
However, a number of observations suggest that ErbB-2 may
potentiate ErbB-1 signaling (47).
GnRH encodes a pre-prohormone, consisting of a
23-amino-acid signal peptide. The GnRH receptor (GnRH-R) is
currently treated as a molecular target in the treatment of
hormone-dependent tumors. GnRH-R activation, coupled to
Gαq/11-Gβγ proteins, leads to elevation of intracellular
Ca2+ levels, altered cytoskeletal function and changes
in protein kinase activity, including protein kinase C, mitogen
activated serine/threonine kinases and stress-activated kinases
(50). Sikora and Vali (51) previously demonstrated that, in
addition to the Wnt-β-catenin pathway, additional pathways
intervening with growth factor signaling, key kinases and inherent
converging points in the signaling machinery also affect SPN.
To conclude, in order to illustrate the pathological
mechanisms of SPN, gene expression profiles of 19 samples were
downloaded and analyzed. Gene functional annotation analysis
demonstrated that the genes MME, MMP-2 and MMP-9, which are
involved in proliferative diabetic retinopathy, are also involved
in SPN. Through PPI network and module analysis, the genes EGFR,
FYN, JUN, GCG, MYC and CD44 were identified as potential key SPN
genes. In addition, the ErbB and GnRH signaling pathways may be
involved with SPN progression. Furthermore, the above DEGS might
function as potential targets for the further gene treatment of
SPN.
Glossary
Abbreviations
Abbreviations:
|
SPN
|
solid pseudopapillary neoplasm
|
|
DEG
|
differentially expressed gene
|
|
PPI
|
protein-protein interaction
|
|
TAG
|
tumor associated gene
|
|
GEO
|
Gene Expression Omnibus
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
DAVID
|
The Database for Annotation,
Visualization and Integrated Discovery
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Gene/Proteins
|
|
MMP
|
matrix metalloproteinase
|
|
ECM
|
extracellular matrix
|
|
PKC
|
protein kinase C
|
References
|
1
|
Patil TB, Shrikhande SV, Kanhere HA, Saoji
RR, Ramadwar MR and Shukla PJ: Solid pseudopapillary neoplasm of
the pancreas: A single institution experience of 14 cases. HPB
(Oxford). 8:148–150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frantz VK: Tumors of the pancreasAnonymous
Atlas of Tumor Pathology. Armed Forces Institute of Pathology;
Washington, DC: pp. 32–33. 1959
|
|
3
|
Martin RC, Klimstra DS, Brennan MF and
Conlon KC: Solid-pseudopapillary tumor of the pancreas: A surgical
enigma? Ann Surg Oncol. 9:35–40. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reindl BA, Lynch DW and Jassim AD:
Aggressive variant of a solid pseudopapillary neoplasm: A case
report and literature review. Arch Pathol Lab Med. 138:974–978.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cao D, Maitra A, Saavedra JA, Klimstra DS,
Adsay NV and Hruban RH: Expression of novel markers of pancreatic
ductal adenocarcinoma in pancreatic nonductal neoplasms: Additional
evidence of different genetic pathways. Mod Pathol. 18:752–761.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cavard C, Audebourg A, Letourneur F,
Audard V, Beuvon F, Cagnard N, Radenen B, Varlet P, Vacher-Lavenu
MC and Perret C: Gene expression profiling provides insights into
the pathways involved in solid pseudopapillary neoplasm of the
pancreas. J pathol. 218:201–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abraham SC, Klimstra DS, Wilentz RE, Yeo
CJ, Conlon K, Brennan M, Cameron JL, Wu TT and Hruban RH:
Solid-pseudopapillary tumors of the pancreas are genetically
distinct from pancreatic ductal adenocarcinomas and almost always
harbor beta-catenin mutations. Am J Pathol. 160:1361–1369. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kobayashi T, Ozasa M, Miyashita K, Saga A,
Miwa K, Saito M, Morioka M, Takeuchi M, Takenouchi N, Yabiku T, et
al: Large solid-pseudopapillary neoplasm of the pancreas with
aberrant protein expression and mutation of β-catenin: A case
report and literature review of the distribution of β-catenin
mutation. Intern Med. 52:2051–2056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
11
|
Kang CM, Kim HK, Kim H, Choi GH, Kim KS,
Choi JS and Lee WJ: Expression of Wnt target genes in solid
pseudopapillary tumor of the pancreas: A pilot study. Pancreas.
38:e53–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aberle H, Schwartz H and Kemler R:
Cadherin-catenin complex: Protein interactions and their
implications for cadherin function. J Cell Biochem. 61:514–523.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang WW, Stelter AA, French S, Shen S, Qiu
S, Venegas R, Wen J, Wang HQ and Xie J: Loss of cell-adhesion
molecule complexes in solid pseudopapillary tumor of pancreas. Mod
Pathol. 20:509–513. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
21
|
Theodosiou T, Angelis L, Vakali A and
Thomopoulos GN: Gene functional annotation by statistical analysis
of biomedical articles. Int J Med Inform. 76:601–613. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen JS, Hung WS, Chan HH, Tsai SJ and Sun
HS: In silico identification of oncogenic potential of fyn-related
kinase in hepatocellular carcinoma. Bioinformatics. 29:420–427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao M, Sun J and Zhao Z: TSGene: A web
resource for tumor suppressor genes. Nucleic acids Res.
41:D970–D976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beisser D, Klau GW, Dandekar T, Muller T
and Dittrich MT: BioNet: An R-Package for the functional analysis
of biological networks. Bioinformatics. 26:1129–1130. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schriml LM, Arze C, Nadendla S, Chang YW,
Mazaitis M, Felix V, Feng G and Kibbe WA: Disease Ontology: A
backbone for disease semantic integration. Nucleic acids Res.
40:D940–D946. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi C, Daniels JA and Hruban RH: Molecular
characterization of pancreatic neoplasms. Adv Anat Pathol.
15:185–195. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang WB, Zhou XJ, Chen JY, Zhang LH, Meng
K, Ma HH and Lu ZF: CD10-positive stromal cells in gastric
carcinoma: Correlation with invasion and metastasis. Jap J Clin
Oncol. 35:245–250. 2005. View Article : Google Scholar
|
|
29
|
Makretsov NA, Hayes M, Carter BA, Dabiri
S, Gilks CB and Huntsman DG: Stromal CD10 expression in invasive
breast carcinoma correlates with poor prognosis, estrogen receptor
negativity, and high grade. Mod Pathol. 20:84–89. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ogawa H, Iwaya K, Izumi M, Kuroda M,
Serizawa H, Koyanagi Y and Mukai K: Expression of CD10 by stromal
cells during colorectal tumor development. Hum Pathol. 33:806–811.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ikenaga N, Ohuchida K, Mizumoto K, Cui L,
Kayashima T, Morimatsu K, Moriyama T, Nakata K, Fujita H and Tanaka
M: CD10+ pancreatic stellate cells enhance the progression of
pancreatic cancer. Gastroenterology. 139:1041–1051, 1051.e1-8.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Koshiba T, Hosotani R, Wada M, Miyamoto Y,
Fujimoto K, Lee JU, Doi R, Arii S and Imamura M: Involvement of
matrix metalloproteinase-2 activity in invasion and metastasis of
pancreatic carcinoma. Cancer. 82:642–650. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Destouches D, Huet E, Sader M, Frechault
S, Carpentier G, Ayoul F, Briand JP, Menashi S and Courty J:
Multivalent pseudopeptides targeting cell surface nucleoproteins
inhibit cancer cell invasion through tissue inhibitor of
metalloproteinases 3 (TIMP-3) release. J Biol Chem.
287:43685–43693. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El-Ghlban S, Kasai T, Shigehiro T, Yin HX,
Sekhar S, Ida M, Sanchez A, Mizutani A, Kudoh T, Murakami H and
Seno M: Chlorotoxin-Fc fusion inhibits release of MMP-2 from
pancreatic cancer cells. Biomed Res Int. 2014:1526592014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang T, Yang J, Xu J, Li J, Cao Z, Zhou L,
You L, Shu H, Lu Z, Li H, et al: CHIP is a novel tumor suppressor
in pancreatic cancer through targeting EGFR. Oncotarget.
5:1969–1986. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tzeng CW, Frolov A, Frolova N, Jhala NC,
Howard JH, Vickers SM, Buchsbaum DJ, Heslin MJ and Arnoletti JP:
EGFR genomic gain and aberrant pathway signaling in pancreatic
cancer patients. J Surg Res. 143:20–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stock AM, Hahn SA, Troost G, Niggemann B,
Zänker KS and Entschladen F: Induction of pancreatic cancer cell
migration by an autocrine epidermal growth factor receptor
activation. Exp cell Res. 326:307–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wisdom R, Johnson RS and Moore C: c-Jun
regulates cell cycle progression and apoptosis by distinct
mechanisms. EMBO J. 18:188–197. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peeper DS, Upton TM, Ladha MH, Neuman E,
Zalvide J, Bernards R, DeCaprio JA and Ewen ME: Ras signalling
linked to the cell-cycle machinery by the retinoblastoma protein.
Nature. 386:177–181. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Woods D, Parry D, Cherwinski H, Bosch E,
Lees E and McMahon M: Raf-induced proliferation or cell cycle
arrest is determined by the level of Raf activity with arrest
mediated by p21Cip1. Mol Cell Biol. 17:5598–5611. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meyer N and Penn LZ: Reflecting on 25
years with MYC. Nat Rev Cancer. 8:976–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li YJ, Wei ZM, Meng YX and Ji XR:
Beta-catenin up-regulates the expression of cyclinD1, c-myc and
MMP-7 in human pancreatic cancer: Relationships with carcinogenesis
and metastasis. World J Gastroenterol. 11:2117–2123. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
He C, Jiang H, Geng S, Sheng H, Shen X,
Zhang X, Zhu S, Chen X, Yang C and Gao H: Expression and prognostic
value of c-Myc and Fas (CD95/APO1) in patients with pancreatic
cancer. Int J Clin Exp Pathol. 7:742–750. 2014.PubMed/NCBI
|
|
45
|
Amati B, Alevizopoulos K and Vlach J: Myc
and the cell cycle. Front Biosci. 3:d250–d268. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hynes NE and Lane HA: ERBB receptors and
cancer: The complexity of targeted inhibitors. Nat Rev Cancer.
5:341–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Olayioye MA, Neve RM, Lane HA and Hynes
NE: The ErbB signaling network: Receptor heterodimerization in
development and cancer. EMBO J. 19:3159–3167. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dimova I, Raicheva S, Dimitrov R, Doganov
N and Toncheva D: Coexistence of copy number increases of c-Myc,
ZNF217, CCND1, ErbB1 and ErbB2 in ovarian cancers. Onkologie.
32:405–410. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Muthuswamy SK, Li D, Lelievre S, Bissell
MJ and Brugge JS: ErbB2, but not ErbB1, reinitiates proliferation
and induces luminal repopulation in epithelial acini. Nat Cell
Biol. 3:785–792. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Morgan K, Meyer C, Miller N, Sims AH,
Cagnan I, Faratian D, Harrison DJ, Millar RP and Langdon SP: GnRH
receptor activation competes at a low level with growth signaling
in stably transfected human breast cell lines. BMC Cancer.
11:4762011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sikora SS and Vali S: Solid
pseudopapillary tumor and wnt signaling pathway Way to go!? J
Gastroenterol Hepatol. 26:215–217. 2011. View Article : Google Scholar : PubMed/NCBI
|