Introduction
Thyroid cancer is the most prevalent malignant
tumors of the endocrine organs and accounts for one third of all
head and neck tumors. In the past several decades, the morbidity
and mortality of thyroid cancer has increased dramatically
worldwide (1,2). It is estimated that, in 2016, there
would be 64,300 new cases and 1,980 mortalities due to thyroid
cancer in the United States (2).
It is commonly accepted that genetic factors, environmental
exposure, epigenetic alteration, nodular disease of the thyroid and
family history serve important roles in thyroid cancer initiation
and progression (3,4). Thyroid cancer can be classified into
three pathology subtypes, including papillary thyroid carcinoma
(PTC), follicular thyroid carcinoma and anaplastic thyroid
carcinoma (5). PTC is the most
common type of thyroid cancer and accounts for 80–90% of all
thyroid cancer cases (6). The vast
majority of patients with PTC have a good prognosis (7). However, patients with local invasion
or distant metastases tend to have a poor prognosis, mainly due to
the poor response to standard treatments (8,9).
Therefore, it is of great significance to elucidate the critical
molecular mechanisms of PTC progression, in order to investigate
efficient therapeutic targets for patients with this disease.
MicroRNAs (miRNAs/miRs) are a group of short, single
strand, non-coding and naturally existing RNAs of ~18-23
nucleotides in length (10). They
maintain control of gene expression at the transcriptional and
post-transcriptional level, through binding to the 3′-untranslated
region (UTR) of their target genes, resulting in gene degradation
or translation inhibition (11).
Increasing studies have demonstrated that the expression levels of
miRNAs are significantly dysregulated between tumor and healthy
tissues, and function as oncogenes or tumor suppressor genes
(12,13). By affecting gene regulation, miRNAs
are involved in cancer carcinogenesis and progression through
regulation of a great deal of physiological and pathological
processes, including the cell cycle, and cellular proliferation,
invasion, migration, metastasis, differentiation and apoptosis
(14–16). For instance, in non-small cell lung
cancer, miR-361-3p inhibited cell growth, proliferation, colony
formation, invasion and migration in vitro, and suppressed
proliferation and metastasis in vivo via negative regulation
of SH2B adaptor protein 1 (17).
miR-14b-5p was upregulated in PTC, and restoration of miR-146b-5p
enhanced cellular migration and invasion via blockade of mothers of
decapentaplegic 4 (18).
The present study investigated the expression and
roles of miR-150 in PTC. It was demonstrated that miR-150 can
directly target Rho-associated protein kinase 1 (ROCK1) and
downregulate ROCK1 expression, thereby inhibiting PTC cell growth
and metastasis. These results were a useful addition to the current
understanding of the molecular mechanism of PTC progression.
Materials and methods
Tissue samples
PTC tissues and adjacent healthy thyroid tissues
were collected from 45 PTC patients (male, 18; female, 27; age
<55 years, 15; age ≥55 years, 30) at Weifang People's Hospital
(Weifang, China) from 2010 to 2014. None of these PTC patients had
received any preoperative treatment. All tissue samples were
immediately snap-frozen in liquid nitrogen, and stored at −80°C.
This study was approved by the Ethics Committee of Weifang People's
Hospital, and informed consent was also obtained from these
patients.
Cell lines, culture conditions and
transfection
The TPC-1, BCPAP, CGTH-W3 and HTH83 PTC-derived
thyroid cell lines and the HT-ori3 healthy human thyroid cell line
were purchased from American Type Culture Collection (Manassas, VA,
USA), and maintained in RPMI-1640 or Dulbecco's modified Eagle's
medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1%
antibiotics (all purchased from Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), at 37°C in 5% CO2.
Chemically synthesized miR-150 and negative control
(NC) mimics, and ROCK1 and NC small interfering RNA (siRNA) were
obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The
ROCK1 siRNA sequence was 5′-GGGUAACUCAUCUGGUAAATT-3′ and the NC
siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. Transfection of the
cells with miRNA mimics or siRNA was carried out with
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using
the miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany) following the
manufacturer's protocol. Concentration of total RNA was determined
using the NanoDrop 2000 (Thermo Fisher Scientific, Inc.). miR-150
expression levels were measured using an All-in-One™
miRNA qRT-PCR Detection kit (GeneCopoeia, Inc., Rockville, MD,
USA). For detection of ROCK1 mRNA expression levels, cDNA was
synthesized with a PrimeScript™ RT reagent kit (Takara
Bio, Inc., Otsu, Japan) and subjected to qPCR with a SYBR-Green PCR
Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The thermocycling conditions for qPCR were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min.
U6 and GADPH served as internal controls for miR-150 and ROCK1 mRNA
expression levels, respectively. The primers were designed as
follows: miR-150, 5′-GCTCTCCCAACCCTTGT-3′ (forward) and
5′-TGCGTGTCGTGGAGTC-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); ROCK1,
5′-AGGAAGGCGGACATATTGATCCCT-3′ (forward) and
5′-AGACGATAGTTGGGTCCCGGC-3′ (reverse); and GAPDH,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). Each sample was analyzed
in triplicate. The relative expression of miR-150 and ROCK1 was
calculated using the 2−ΔΔCq method (19).
3-(4,5-Dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation capacity was evaluated by MTT
assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Transfected
cells were collected, counted and plated into 96-well plates at a
density of 2,000 cells per well. After incubation for 24, 48, 72
and 96 h, MTT assay was performed. Briefly, 20 µl MTT solution (5
mg/ml) was added to each well of the 96-well plates, and cells were
then incubated at 37°C for additional 4 h. The culture medium was
removed carefully and 150 µl dimethyl sulfoxide was added to each
well. After incubation at 37°C for 30 min, the absorbance at 490 nm
was detected using an automatic multi-well spectrophotometer
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each sample was
performed in triplicate.
Transwell assay
The migration and invasion capacities of cells were
assessed by Transwell assay using Transwell chambers (Corning
Incorporated, Corning, NY, USA) and Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA)-coated Transwell chambers, respectively.
Transfected cells were harvested, counted, re-suspended and
5×104 cells in 200 ml FBS-free culture medium were
plated into the upper chamber. A total of 500 µl culture medium
supplemented with 20% FBS was added into the lower chamber. After
incubation for 24 h at 37°C in 5% CO2, the cells
remaining on the surface membranes of upper chamber were removed
carefully, while cells on the lower surface were fixed with 100%
methanol (Beyotime Institute of Biotechnology, Haimen, China),
stained with 0.5% crystal violet (Beyotime Institute of
Biotechnology), washed with PBS (Gibco; Thermo Fisher Scientific,
Inc.), and imaged under an inverted microscope (Olympus
Corporation, Tokyo, Japan).
Western blotting
Transfected cells were collected, washed with PBS
and lysed in radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology). Following cell lysis, the lysates were
centrifuged at 4°C for 10 min at 24,000 × g and the supernatants
were obtained for analysis of the protein concentration. The
concentration of total protein was determined using a Bicinchoninic
Acid protein assay kit (Pierce Biotechnology; Thermo Fisher
Scientific, Inc.). Equal amounts of protein were separated by 10%
SDS-PAGE and electrophoretically transferred onto polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). After
blocking with 5% skim milk in TBS with Tween-20 (TBST), the
membranes were incubated with primary antibodies at 4°C overnight.
The primary antibodies used in the present study were: Mouse
anti-human monoclonal ROCK1 (cat. no. sc-365628; 1:1,000 dilution;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse
anti-human monoclonal β-actin (cat. no. sc-47778; 1:1,000 dilution;
Santa Cruz Biotechnology, Inc.). Membranes were subsequently washed
with TBST three times and incubated with a goat anti-mouse
horseradish peroxidase-conjugated IgG secondary antibody (1:2,000
dilution; Santa Cruz Biotechnology, Inc.) at room temperature for 1
h, followed by developing with Enhanced Chemiluminescence Plus
reagents (Pierce; Thermo Fisher Scientific, Inc.). β-actin served
as an internal control for ROCK1 expression.
Luciferase reporter assay
To identify how miR-150 exerts its tumor suppressive
roles in PTC, its target genes were investigated using TargetScan
(www.targetscan.org) and miRanda
(www.microrna.org). The entire human ROCK1 3′-UTR
harboring miR-150 target sequence and the seed-sequence mutated
version were synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). For the luciferase reporter assay, cells were
seeded into 24-well plates (3×105 cells/well) and
transfected with miR-150 mimics or NCs, together with
psiCHECK2-ROCK1-3′-UTR wild-type (Wt) or psiCHECK2-ROCK1-3′-UTR
mutant (Mut) using Lipofectamine 2000, according to the
manufacturer's protocol. After incubation for 48 h at 37°C in 5%
CO2, cells were harvested, and firefly and Renilla
luciferase activities were detected using a Dual-Luciferase
Reporter Assay system (Promega, Corporation, Madison, WI, USA). The
assay was performed in duplicate in three independent
experiments.
Statistical analysis
Data are expressed as the mean ± standard deviation,
and were analyzed using SPSS 15.0 software (SPSS, Inc., Chicago,
IL, USA). A paired Student's t-test or one-way analysis of
variance, followed by a Student-Newman-Keuls multiple comparison
test, were performed for analysis. Correlations were analyzed using
the Chi-squared test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-150 is downregulated in PTC
tissues and cell lines
To investigate the roles of miR-150 in human PTC,
its expression levels in PTC tissues and adjacent normal thyroid
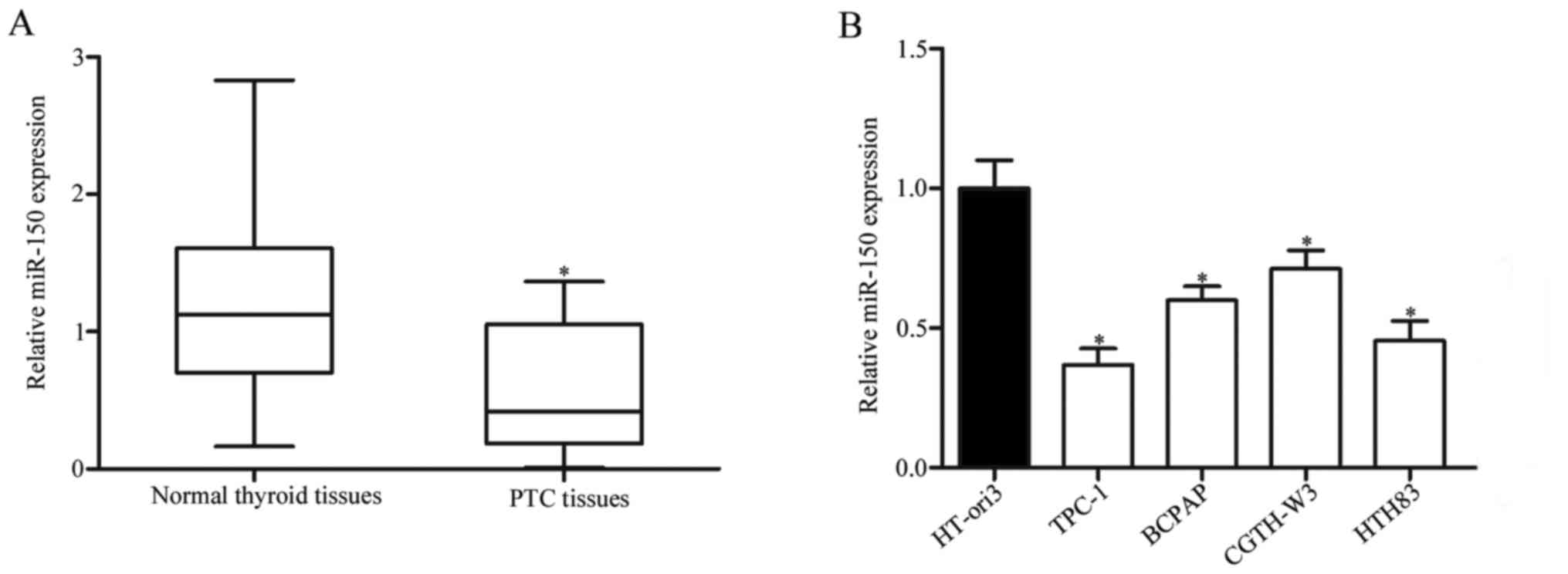
tissues were examined. As presented in Fig. 1A, miR-150 levels were significantly
reduced in PTC tissues compared with adjacent normal thyroid
tissues (P<0.05). miR-150 expression in the TPC-1, BCPAP,
CGTH-W3 and HTH83 PTC cell lines, and in the HT-ori3 healthy human
thyroid cell line. Compared with HT-ori3, miR-150 was markedly
downregulated in all the PTC cell lines, but expression levels
varied between them (Fig. 1B;
P<0.05). These findings suggested that downregulation of miR-150
may be involved in PTC.
Correlation between
clinicopathological features and miR-150 expression levels in PTC
patients
To investigate whether miR-150 expression levels
were correlated with clinicopathological features in PTC cases, the
Chi-squared test was used. As presented in Table I, analysis revealed that reduced
miR-150 expression was significantly negatively correlated with TNM
stage (P=0.001) and lymph node metastasis (P=0.015), which are both
indicators of poor prognosis. However, there was no significant
correlation between miR-150 expression and other
clinicopathological factors, including age (P=0.714), sex (P=0.057)
and tumor size (P=0.094).
 | Table I.Correlation between miR-150
expression levels and clinicopathological features in papillary
thyroid cancer patients. |
Table I.
Correlation between miR-150
expression levels and clinicopathological features in papillary
thyroid cancer patients.
|
|
| miR-150
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | N | Low | High | P-value |
|---|
| Sex |
|
|
| 0.714 |
|
Male | 18 | 9 | 9 |
|
|
Female | 27 | 15 | 12 |
|
| Age |
|
|
| 0.057 |
| <55
years | 15 | 11 | 4 |
|
| ≥55
years | 30 | 13 | 17 |
|
| Tumor size
(cm) |
|
|
| 0.094 |
| <3
cm | 24 | 10 | 14 |
|
| ≥3
cm | 21 | 14 | 7 |
|
| TNM stage |
|
|
| 0.001 |
|
T1-T2 | 32 | 12 | 20 |
|
|
T3-T4 | 13 | 12 | 1 |
|
| Lymph node
metastasis |
|
|
| 0.015 |
| No | 33 | 14 | 19 |
|
|
Yes | 12 | 10 | 2 |
|
Suppression of PTC cell proliferation,
migration and invasion by miR-150
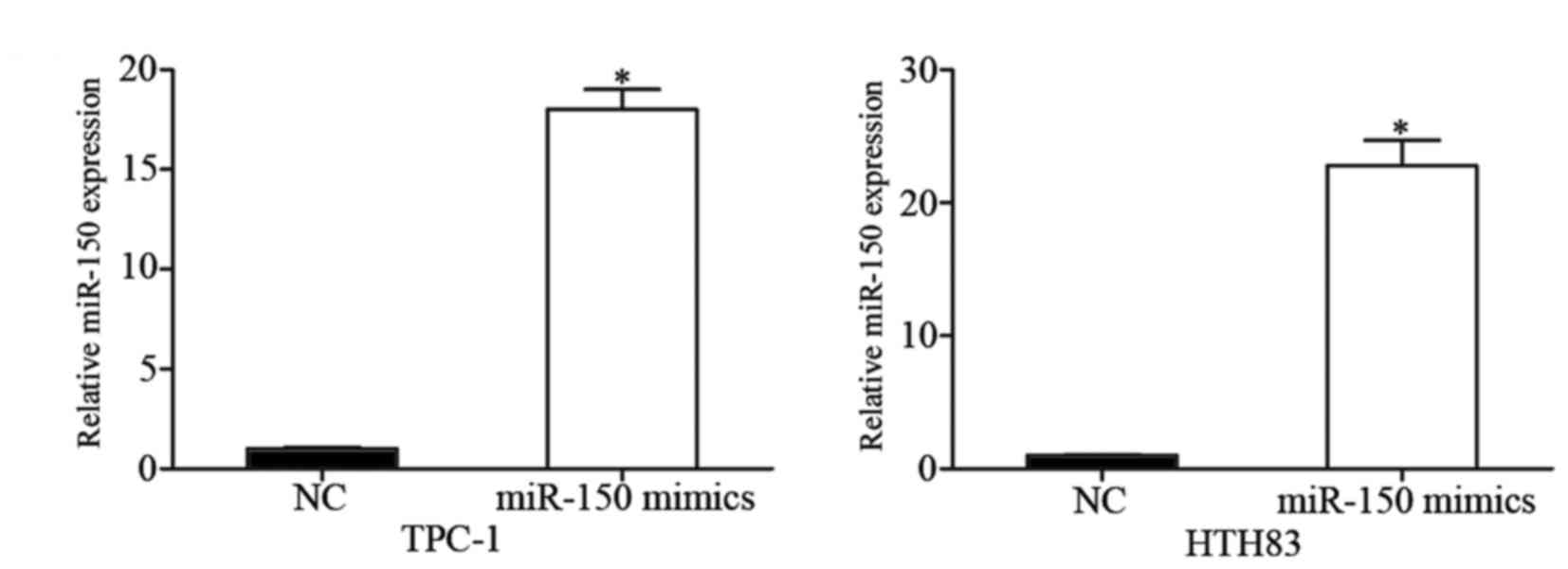
To investigate the potential effect of miR-150 on
PTC, miR-150 mimics were transfected into TPC-1 and HTH83 cells,
and overexpression of miR-150 in these cells was confirmed by
RT-qPCR (Fig. 2; P<0.05).
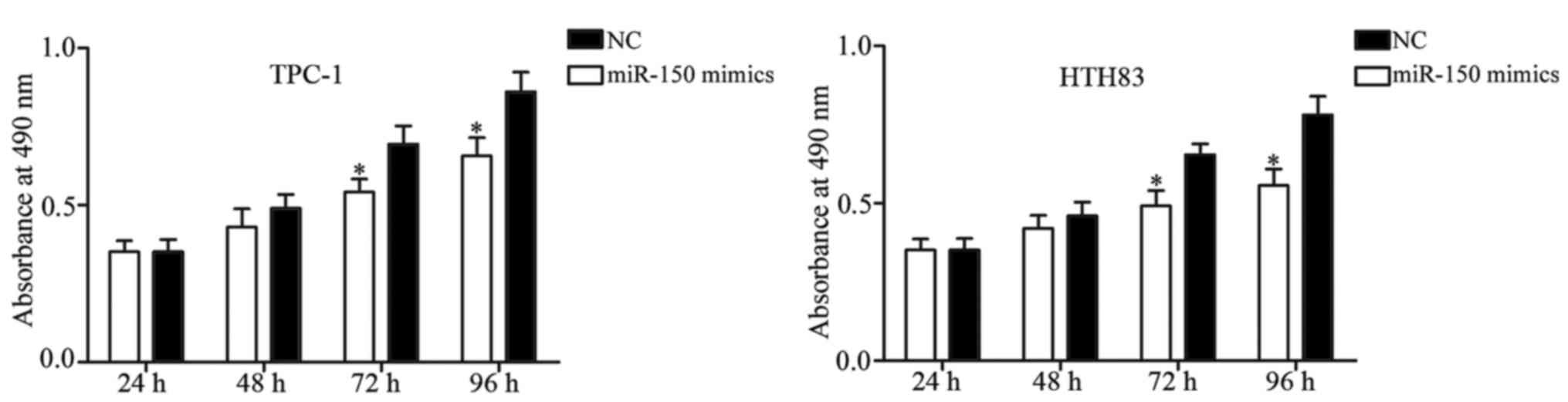
MTT assay was performed to evaluate the effect of
miR-150 on PTC cell proliferation. The results of the MTT assay
revealed that cell proliferation was significantly suppressed in
miR-150-transfected TPC-1 and HTH83 cells in comparison with the NC
groups (Fig. 3; P<0.05). Next,
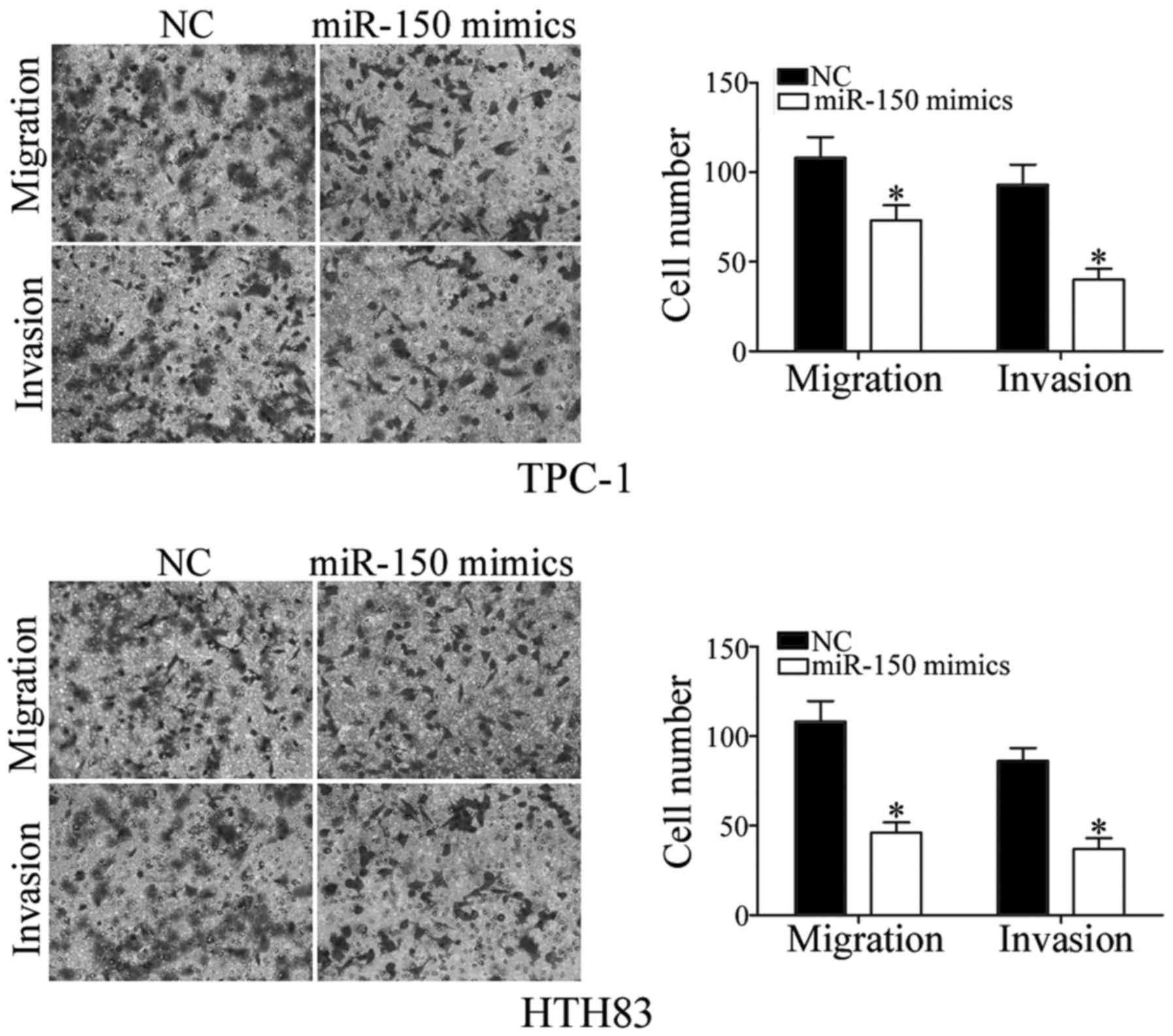
the role of miR-150 in regulating PTC cell migration and invasion
was investigated. As demonstrated by Transwell assay, increased
expression of miR-150 reduced the migration and invasiveness of
TPC-1 and HTH83 cells (Fig. 4;
P<0.05). Taken together, these results suggested that miR-150
has a tumor suppressor function in PTC cells.
miR-150 decreases ROCK1 expression by
directly binding to its 3′-UTR
To identify how miR-150 exerts its tumor suppressive
roles in PTC, its target genes were investigated using TargetScan
and miRanda. The analysis predicted that ROCK1 may be a potential
target of miR-150 (Fig. 5A).
Following this, whether ROCK1 was a direct and specific target of
miR-150 was validated using a luciferase reporter assay. TPC-1 and
HTH83 cells were transfected with psiCHECK2-ROCK1-3′-UTR Wt or
psiCHECK2-ROCK1-3′-UTR Mut, together with miR-150 mimics or NCs.
The results demonstrated that miR-150 decreased the luciferase
activities of psiCHECK2-ROCK1-3′-UTR Wt, but not
psiCHECK2-ROCK1-3′-UTR Mut in both TPC-1 and HTH83 cells (Fig. 5B; P<0.05). Additionally, ectopic
miR-150 expression decreased ROCK1 mRNA (Fig. 5C; P<0.05) and protein (Fig. 5D; P<0.05) expression levels in
TPC-1 and HTH83 cells. Taken together, these results demonstrated
that ROCK1 was directly and negatively regulated by miR-150 in
PTC.
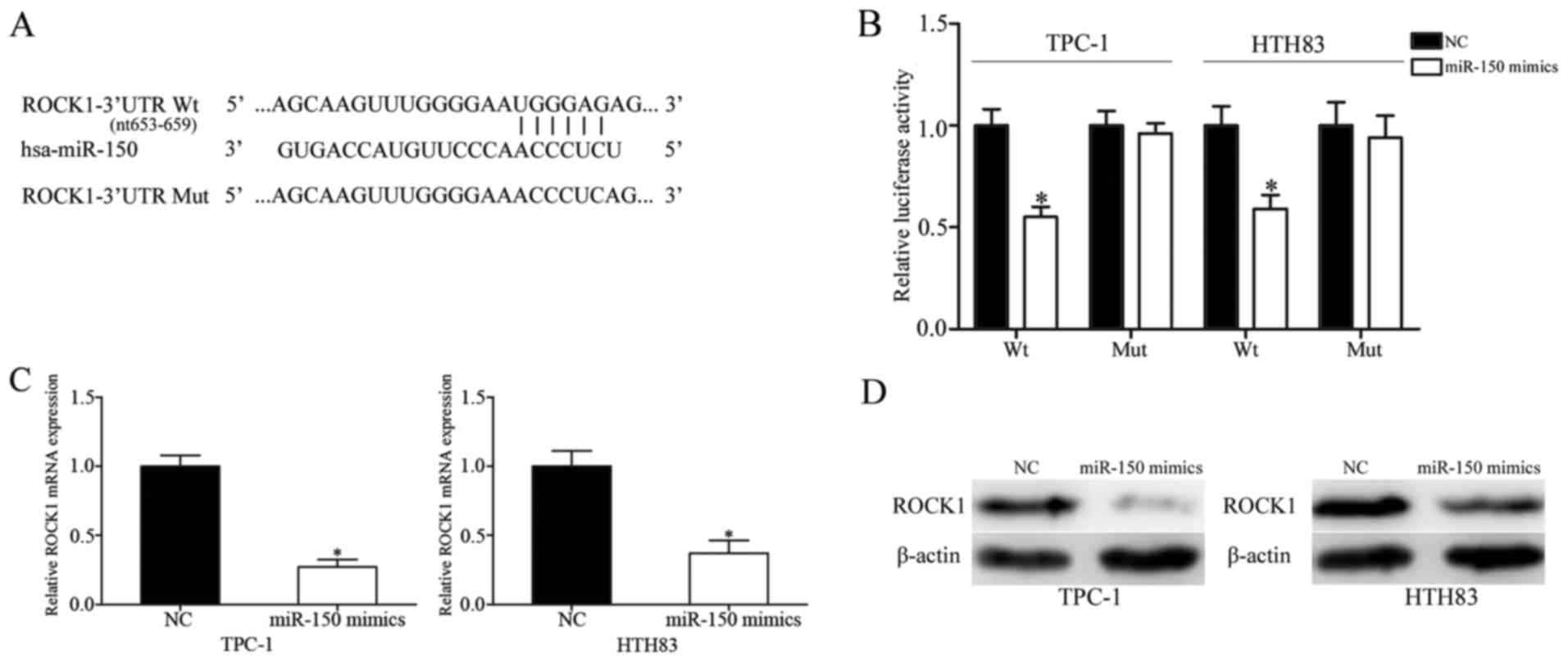 | Figure 5.ROCK1 is a direct target of miR-150
in papillary thyroid cancer. (A) Predicted consequential pairing of
the target 3′-UTR region of ROCK1 (Wt and Mut) and miR-150
sequence. (B) Luciferase reporter assay of TPC-1 and HTH83 cells
transfected with psiCHECK2-ROCK1-3′-UTR Wt or
psiCHECK2-ROCK1-3′-UTR Mut, along with miR-150 mimics or NC. ROCK1
(C) mRNA and (D) protein expression levels, as assessed by reverse
transcription-quantitative polymerase chain reaction and western
blot analysis, respectively, in TPC-1 and HTH83 cells transfected
with miR-150 mimics or NC. Data are presented as the mean ±
standard deviation. *P<0.05 vs. NC in same group. NC, negative
control; miR, microRNA; Wt, wild-type; Mut, mutant; UTR,
untranslated region; ROCK1, Rho-associated protein kinase 1. |
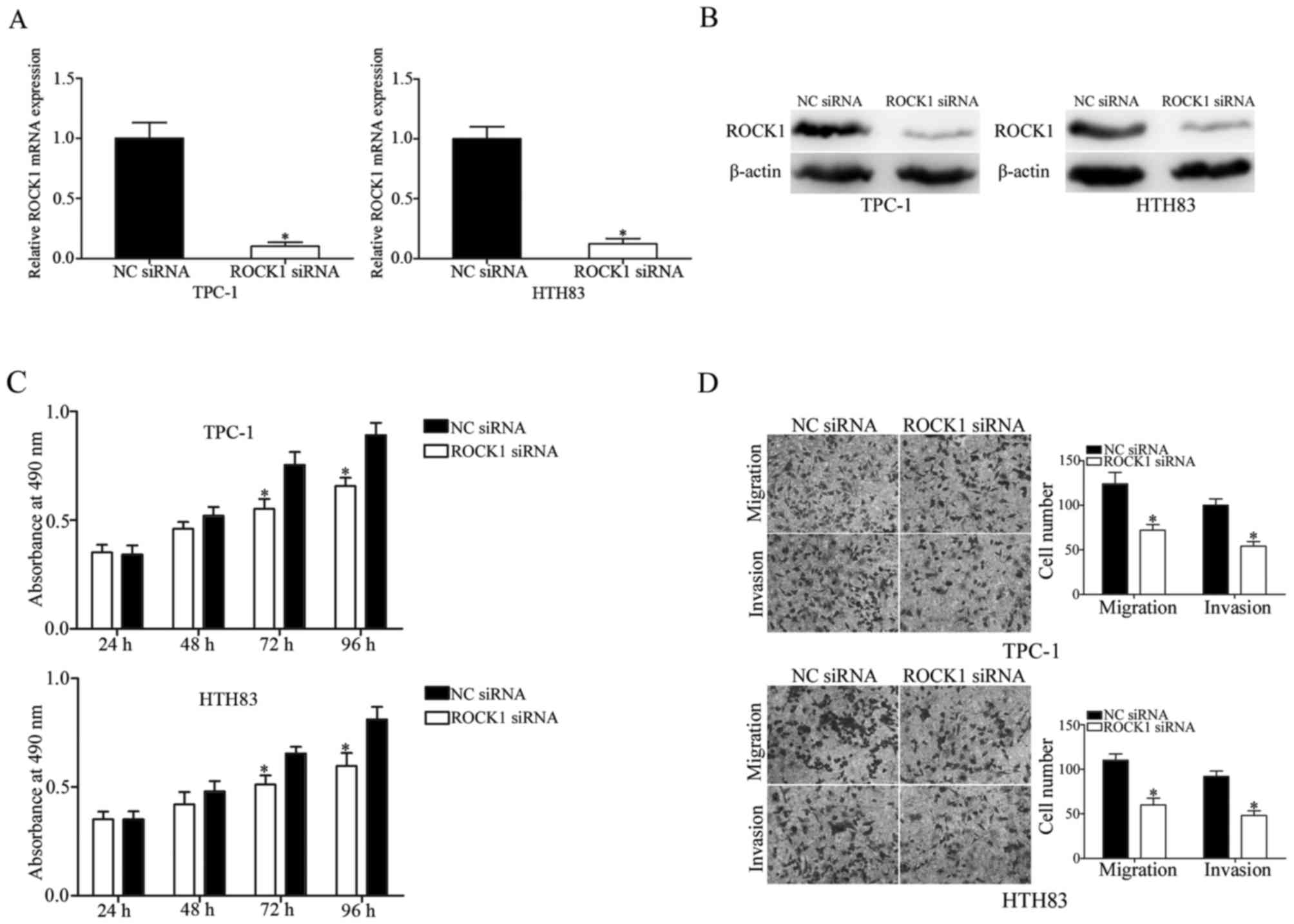
ROCK1 suppresses cell proliferation,
migration and invasion of PTC cells
ROCK1 was identified as a direct target of miR-150.
Therefore, it was hypothesized that the tumor suppressive roles
induced by miR-150 overexpression in PTC cells may be achieved by
regulation of ROCK1. To investigate this, TPC-1 and HTH83 cells
were transfected with ROCK1 siRNA or NC siRNA, and downregulation
of ROCK1 in these cells was determined by RT-qPCR (Fig. 6A; P<0.05) and western blot
analysis (Fig. 6B; P<0.05). MTT
assay revealed that downregulation of ROCK1 significantly inhibited
TPC-1 and HTH83 cell proliferation at 72 and 96 h compared with the
NC siRNA group (Fig. 6C;
P<0.05). Transwell assay was performed to evaluate the migration
and invasiveness of TPC-1 and HTH83 cells transfected with ROCK1 or
NC siRNA. The results demonstrated that ROCK1 knockdown suppressed
migration and invasion in TPC-1 and HTH83 cells (Fig. 6D; P<0.05). These findings
indicated that miR-150 inhibits cell growth and metastasis of PTC
via negative regulation of ROCK1.
Discussion
In the present study, miR-150 was demonstrated to
serve tumor suppressive roles in inhibiting the growth and
metastasis of PTC cells. Firstly, RT-qPCR analysis revealed that
miR-150 expression levels were reduced in PTC tissues and cell
lines compared with adjacent healthy thyroid tissues and a healthy
human thyroid cell line, respectively.
In addition, reduced miR-150 expression was
obviously correlated with TNM stage and lymph node metastasis in
PTC patients. Functional analysis revealed that overexpression of
miR-150 inhibited cell proliferation, migration and invasion in
PTC. Notably, ROCK1 was validated as a direct target gene of
miR-150 in PTC. ROCK1 knockdown may mimic the tumor suppressive
functions induced by miR-150 overexpression in PTC. All these
findings demonstrated that miR-150 serves substantial roles in PTC
inhibition and may serve as a therapeutic target in patients with
PTC.
Altered miR-150 expression has been identified in
various kinds of human cancer. miR-150 was reported to be
downregulated in pancreatic cancer (20), osteosarcoma (21), esophageal squamous cell carcinoma
(22), colorectal cancer (23), hepatocellular carcinoma (24,25),
ovarian cancer (26) and malignant
lymphoma (27). However,
upregulation of miR-150 was also identified in prostate (28), cervical (29), non-small cell lung (30), breast (31) and gastric cancer (32). These conflicting studies indicated
that expression levels of miR-150 in human cancers have tissue
specificity.
Expression levels of miR-150 in caners have been
reported to be correlated with clinicopathological features and
prognosis. For example, in non-small cell lung cancer, miR-150
levels were significantly increased compared with matched
non-cancerous tissues. miR-150 expression was obviously associated
with lymph node metastasis, distant metastasis and clinical TNM
stage in non-small cell lung cancer. Kaplan-Meier analysis revealed
that the cumulative 5-year overall survival rate was 40.8% in the
high expression group, and 69.2% in the low expression group
(33). In prostate cancer, miR-150
was also upregulated, and high miR-150 expression was correlated
with tumor recurrence and metastasis. Prostate cancer patients with
high miR-150 expression had significantly poorer overall and
disease-free survival compared with those with low miR-150
expression. Multivariate Cox regression analysis revealed that the
expression of miR-150 was an independent prognostic predictor for
prostate cancer patients (28).
Jin et al (26) reported
that expression level of miR-150 was reduced in epithelial ovarian
cancer, and low miR-150 expression was associated with aggressive
clinicopathological factors of epithelial ovarian cancer patients,
including high clinical stage and pathological grade, and shorter
overall and progression-free survivals. More importantly, the
multivariate analysis validated that miR-150 expression was an
independent prognostic biomarker in epithelial ovarian cancer.
Combined with the findings of the present study, miR-150 may be a
useful prognosis marker in human cancers.
Accumulated evidences have demonstrated that
deregulated expression of miR-150 contributes to cancer initiation
and progression. Srivastava et al (20) reported that miR-150 repressed
pancreatic cancer cell growth and malignant behavior via directly
targeting MUC4. A study by Huang et al (31) revealed that upregulation of miR-150
enhanced proliferation and clonogenicity, and decreased apoptosis
in breast cancer cells via blockade of PX27. It also has been
demonstrated that miR-150 targets C-C motif chemokine 20 and C-C
chemokine receptor type 6 to inhibit invasion and metastasis of
advanced cutaneous T-cell lymphoma (34). In non-small cell lung cancer,
miR-150 knockdown inhibited cell proliferation and induced
apoptosis by directly targeting brassinosteroid insensitive
1-associated receptor kinase 1 in vitro (30). Furthermore, in colorectal cancer,
enforced miR-150 expression inhibited cell proliferation and
motility, and improved cell apoptosis and G1 arrest via negative
regulation of c-Myb and mucin-4 (35,36).
However, the biological roles and underling
molecular mechanisms of miR-150 in PTC remain largely unknown. The
present study demonstrated that miR-150 functioned as a tumor
suppressor in PTC cells, via inhibiting tumor cell proliferation,
migration and invasion.
To investigate the molecular mechanisms underlying
miR-150-mediated inhibition of proliferation, migration and
invasion in PTC, ROCK1 was selected for further study as it was
predicted by TargetScan and miRanda to be a potential direct target
of miR-150. ROCK1, located at chromosome 18 (18q11.1) (37), has been identified to be
upregulated in various types of human cancers. Previous studies
have demonstrated that ROCK1 is correlated with cancer progression,
metastasis and poor prognosis (38–40).
In the present study, an important molecular association between
miR-150 and ROCK1 was revealed. Luciferase reporter assays, RT-qPCR
and western blot analysis demonstrated that miR-150 could directly
target the 3′-UTR of ROCK1, and thereby decrease ROCK1 expression
at both mRNA and protein levels. ROCK1 knockdown was demonstrated
to result in miR-150-induced inhibition of PTC cell proliferation,
migration and invasion, demonstrating that ROCK1 serves as a
functionally relevant target of miR-150 in PTC.
In conclusion, miR-150 served as a tumor suppressor
of PTC cells by inhibiting growth and metastasis, partially by
regulating the expression of the downstream target gene ROCK1.
These findings suggested that miR-150 may be an effective
therapeutic target for the treatment of PTC.
References
|
1
|
Davies L, Morris LG, Haymart M, Chen AY,
Goldenberg D, Morris J, Ogilvie JB, Terris DJ, Netterville J, Wong
RJ, et al: American association of clinical endocrinologists and
American college of endocrinology disease state clinical review:
The increasing incidence of thyroid cancer. Endocr Pract.
21:686–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng H, Wang M, Jiang L, Chu H, Hu J,
Ning J, Li B, Wang D and Xu J: BRAF-activated long noncoding RNA
modulates papillary thyroid carcinoma cell proliferation through
regulating thyroid stimulating hormone receptor. Cancer Res Treat.
48:698–707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schneider AB and Sarne DH: Long-term risks
for thyroid cancer and other neoplasms after exposure to radiation.
Nat Clin Pract Endocrinol Metab. 1:82–91. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho BY, Choi HS, Park YJ, Lim JA, Ahn HY,
Lee EK, Kim KW, Yi KH, Chung JK, Youn YK, et al: Changes in the
clinicopathological characteristics and outcomes of thyroid cancer
in Korea over the past four decades. Thyroid. 23:797–804. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geraldo MV, Fuziwara CS, Friguglieti CU,
Costa RB, Kulcsar MA, Yamashita AS and Kimura ET: MicroRNAs
miR-146-5p and let-7f as prognostic tools for aggressive papillary
thyroid carcinoma: A case report. Arq Bras Endocrinol Metabol.
56:552–557. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sherman SI: Thyroid carcinoma. Lancet.
361:501–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vasko VV and Saji M: Molecular mechanisms
involved in differentiated thyroid cancer invasion and metastasis.
Curr Opin Oncol. 19:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Q, Ji M, Guan H, Shi B and Hou P:
Shikonin inhibits thyroid cancer cell growth and invasiveness
through targeting major signaling pathways. J Clin Endocrinol
Metab. 98:E1909–E1917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Srivastava A, Goldberger H, Dimtchev A,
Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen
LN, et al: MicroRNA profiling in prostate cancer-the diagnostic
potential of urinary miR-205 and miR-214. PLoS One. 8:e769942013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hodge LS, Elsawa SF, Grote DM,
Price-Troska TL, Asmann YW, Fonseca R, Gertz MA, Witzig TE, Novak
AJ and Ansell SM: MicroRNA expression in tumor cells from
Waldenstrom's macroglobulinemia reflects both their normal and
malignant cell counterparts. Blood Cancer J. 1:e242011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shah NR and Chen H: MicroRNAs in
pathogenesis of breast cancer: Implications in diagnosis and
treatment. World J Clin Oncol. 5:48–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fernandez S, Risolino M, Mandia N, Talotta
F, Soini Y, Incoronato M, Condorelli G, Banfi S and Verde P:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 34:3240–3250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY,
Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al: MicroRNA-30b functions
as a tumour suppressor in human colorectal cancer by targeting
KRAS, PIK3CD and BCL2. J Pathol. 232:415–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen W, Wang J, Liu S, Wang S, Cheng Y,
Zhou W, Duan C and Zhang C: MicroRNA-361-3p suppresses tumor cell
proliferation and metastasis by directly targeting SH2B1 in NSCLC.
J Exp Clin Cancer Res. 35:762016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lima CR, Geraldo MV, Fuziwara CS, Kimura
ET and Santos MF: MiRNA-146b-5p upregulates migration and invasion
of different papillary thyroid carcinoma cells. BMC Cancer.
16:1082016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, Chen L, Wang W, Meng FB, Zhao RT and
Chen Y: MicroRNA-150 inhibits cell invasion and migration and is
downregulated in human osteosarcoma. Cytogenet Genome Res.
146:124–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yokobori T, Suzuki S, Tanaka N, Inose T,
Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H and Kuwano
H: MiR-150 is associated with poor prognosis in esophageal squamous
cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci.
104:48–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pizzini S, Bisognin A, Mandruzzato S,
Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M,
Pilati P, et al: Impact of microRNAs on regulatory networks and
pathways in human colorectal carcinogenesis and development of
metastasis. BMC Genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu F, Lu Z, Chen B, Dong P and Zheng J:
microRNA-150: A promising novel biomarker for hepatitis B
virus-related hepatocellular carcinoma. Diagn Pathol. 10:1292015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun W, Zhang Z, Wang J, Shang R, Zhou L,
Wang X, Duan J, Ruan B, Gao Y, Dai B, et al: MicroRNA-150
suppresses cell proliferation and metastasis in hepatocellular
carcinoma by inhibiting the GAB1-ERK axis. Oncotarget.
7:11595–11608. 2016.PubMed/NCBI
|
|
26
|
Jin M, Yang Z, Ye W, Xu H and Hua X:
MicroRNA-150 predicts a favorable prognosis in patients with
epithelial ovarian cancer, and inhibits cell invasion and
metastasis by suppressing transcriptional repressor ZEB1. PLoS One.
9:e1039652014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Watanabe A, Tagawa H, Yamashita J, Teshima
K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T,
et al: The role of microRNA-150 as a tumor suppressor in malignant
lymphoma. Leukemia. 25:1324–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dezhong L, Xiaoyi Z, Xianlian L, Hongyan
Z, Guohua Z, Bo S, Shenglei Z and Lian Z: miR-150 is a factor of
survival in prostate cancer patients. J BUON. 20:173–179.
2015.PubMed/NCBI
|
|
29
|
Li J, Hu L, Tian C, Lu F, Wu J and Liu L:
microRNA-150 promotes cervical cancer cell growth and survival by
targeting FOXO4. BMC Mol Biol. 16:242015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu XY, Wang J, Luo YZ, Du Q, Li RR, Shi H
and Yu TP: Down-regulation of miR-150 induces cell proliferation
inhibition and apoptosis in non-small-cell lung cancer by targeting
BAK1 in vitro. Tumour Biol. 35:5287–5293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang S, Chen Y, Wu W, Ouyang N, Chen J,
Li H, Liu X, Su F, Lin L and Yao Y: miR-150 promotes human breast
cancer growth and malignant behavior by targeting the pro-apoptotic
purinergic P2×7 receptor. PLoS One. 8:e807072013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yin QW, Sun XF, Yang GT, Li XB, Wu MS and
Zhao J: Increased expression of microRNA-150 is associated with
poor prognosis in non-small cell lung cancer. Int J Clin Exp
Pathol. 8:842–846. 2015.PubMed/NCBI
|
|
34
|
Ito M, Teshima K, Ikeda S, Kitadate A,
Watanabe A, Nara M, Yamashita J, Ohshima K, Sawada K and Tagawa H:
MicroRNA-150 inhibits tumor invasion and metastasis by targeting
the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma.
Blood. 123:1499–1511. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin
H and Wang Y: miR-150 functions as a tumour suppressor in human
colorectal cancer by targeting c-Myb. J Cell Mol Med. 18:2125–2134.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang WH, Chen J, Zhao F, Zhang BR, Yu HS,
Jin HY and Dai JH: MiR-150-5p suppresses colorectal cancer cell
migration and invasion through targeting MUC4. Asian Pac J Cancer
Prev. 15:6269–6273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lock FE, Ryan KR, Poulter NS, Parsons M
and Hotchin NA: Differential regulation of adhesion complex
turnover by ROCK1 and ROCK2. PLoS One. 7:e314232012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou X, Wei M and Wang W: MicroRNA-340
suppresses osteosarcoma tumor growth and metastasis by directly
targeting ROCK1. Biochem Biophys Res Commun. 437:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Oellers P, Schroer U, Senner V, Paulus W
and Thanos S: ROCKs are expressed in brain tumors and are required
for glioma-cell migration on myelinated axons. Glia. 57:499–509.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lochhead PA, Wickman G, Mezna M and Olson
MF: Activating ROCK1 somatic mutations in human cancer. Oncogene.
29:2591–2598. 2010. View Article : Google Scholar : PubMed/NCBI
|