Introduction
OSCC is the most common type of oral cancer and is
the eleventh most common malignancy in humans worldwide (1). Although chemotherapeutic agents have
been widely used to treat a broad range of human malignancies,
their application is limited in OSCC due to tumor cell resistance.
One of these drugs is doxorubicin, whose application in OSCC
treatment is limited due to cancer cell resistance. Therefore, an
improved understanding regarding the molecular basis of doxorubicin
resistance is required, in order to develop strategies to increase
the sensitivity of OSCC cells and promote the use of this reagent
in the treatment of OSCC.
MicroRNAs (miRNAs/miR) are small noncoding RNAs
(22–26 nt) that post-transcriptionally regulate gene expression
through base-pair matching with the 3′ untranslated region (3′UTR)
of target mRNA transcripts (2).
miRNAs are involved in various cellular processes, including cell
differentiation, proliferation and apoptosis, and serve critical
roles in carcinogenesis (3).
miR-221 belongs to the miR-221/222 family, which is located as a
cluster in the short arm of the X chromosome (4). miR-221 has been reported to be
involved in cancer development and drug resistance (4–6). Wei
et al (7) reported that
exosomal miR-221/222 mediated tamoxifen resistance in recipient
estrogen receptor-positive breast cancer cells. Zhao et al
(8) demonstrated that inhibition
of miR-21 and miR-221 in tumor-initiating stem-like pancreatic
cancer cells reduced chemoresistance against gemcitabine and
5-fluorouracil. Furthermore, inhibition of miR-221 in SNU449 liver
cancer cells increased doxorubicin-induced cell apoptosis through
upregulating caspase-3 activity (9). Previous studies have indicated that
aberrant expression of miR-221 may have important roles in the
development of OSCC (5,10). Therefore, the present study aimed
to investigate whether miR-221 is involved in the chemoresistance
of OSCC to doxorubicin.
Tissue inhibitor of metalloproteinase-3 (TIMP3),
which is a member of the TIMP family, acts as an inhibitor of
matrix metalloproteinases and is involved in extracellular matrix
degradation (11). TIMP3 has been
identified as a target of miR-221/222 and is involved in regulating
sensitivity to chemotherapeutic agents in numerous types of cancer.
Gan et al (12) reported
that downregulation of miR-221/222 may enhance the sensitivity of
MCF-7 and MDA-MB-231 breast cancer cells to tamoxifen via
upregulation of TIMP3. In addition, Garofalo et al (13) demonstrated that, in non-small cell
lung cancer (NSCLC) and hepatocarcinoma cells, miR-221/222, by
targeting phosphatase and tensin homolog (PTEN) and TIMP3, induced
TNF-related apoptosis-inducing ligand (TRAIL) resistance and
enhanced cellular migration.
The present study investigated whether the
miR-221/TIMP3 axis is involved in regulating the sensitivity of
OSCC to doxorubicin. The results demonstrated that inhibition of
miR-221 restored sensitivity of the SCC4 and SCC9 OSCC cell lines
to doxorubicin via upregulation of TIMP3.
Materials and methods
Cell lines and culture
The SCC4 and SCC9 OSCC cell lines were obtained from
the Beijing Institute for Cancer Research (Beijing, China). The
cells were cultured in Dulbecco's modified Eagle's medium/F12
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Wuhan Boster Biological
Technology, Ltd., Wuhan, China) at 37°C in a humidified atmosphere
containing 5% CO2. Doxorubicin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was dissolved in dimethyl sulfoxide
(DMSO) at 50 mg/ml and further diluted to various concentrations
(0.1, 1.0 and 5.0 µM) in the culture medium. Cells were treated
with doxorubicin at the indicated concentrations for 24 h at 37°C
and then used for analysis.
Transfection of cells with TIMP3 small
interfering (si)RNA and anti-miR-221 oligonucleotides
Cells were plated in 6-well plates at a density of
2×105 cells/well. When cells reached 70% confluence,
they were transfected with siRNA oligonucleotides targeting human
TIMP3 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) or with a non-targeting control siRNA (Invitrogen; Thermo
Fisher Scientific, Inc.) at a final concentration of 50 nM, using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The anti-miR-221 and non-targeting scramble
oligonucleotides were obtained from Qiagen, Inc. (Valencia, CA,
USA). The miR-221 inhibitor (final concentration, 100 nM) or
control oligonucleotides were transfected into the cells using
Lipofectamine® 2000 according to the manufacturer's
protocol. Total RNA and cell lysates were collected 48 h
post-transfection and functional studies were conducted 48 h
post-transfection.
MTT assay
Cell viability was measured using an MTT assay (Cell
Viability kit 1; Roche Applied Science, Penzberg, Germany) as
previously described (14).
Briefly, cells were seeded in 96-well plates at 5,000 cells/well in
triplicate. After 1 day, cells were administered the treatments or
DMSO, as indicated. To measure cell viability, 10 µl MTT solution
was added directly to each well, and the plates were incubated for
4 h. Solubilization buffer (100 µl) was then added to each well
without removing the medium. The plates were then incubated at 37°C
overnight and absorbance was measured at 595 nm using a microplate
reader. This experiment was run in triplicate for each sample.
Relative cell viability was calculated as the ratio of absorbance
at 595 nm of the treatment group to absorbance of the control
cells. Results are presented as the mean ± standard deviation of
three independent experiments.
Annexin V-fluorescein isothiocyanate
(FITC)/Hoechst double staining for apoptosis
Cell apoptosis was detected using Annexin V-FITC
apoptosis detection reagent (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) according to the manufacturer's protocol, with
modifications. Briefly, cells were treated as indicated and were
then harvested by trypsin digestion. After collection, cells were
washed twice with PBS and centrifuged at 300 × g for 5 min. Cells
were then resuspended in 1X staining buffer and cell density was
adjusted to 1×105 cells/100 µl. To 100 µl of cell
suspension, 5 µl Annexin V-FITC was added and the cells were
incubated at room temperature for 15 min followed by the addition
of 400 µl 1X staining buffer. Prior to sample loading, 8 µl Hoechst
33258 was added. Cell apoptosis was then detected using a flow
cytometer (LSR II; BD Biosciences, Franklin Lakes, NJ, USA) and
data were analyzed using the FlowJo software version 9.0 (FlowJo,
LLC, Ashland, OR, USA).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) for
TIMP3 expression
Total RNA was extracted from the cells using the
RNeasy kit (Qiagen, Inc.) according to the manufacturer's protocol.
For each sample, 1.5 µg total RNA was reverse transcribed into cDNA
using the iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) according to the manufacturer's protocol. TIMP3
expression was then detected by RT-qPCR as previously described
(15). Briefly, total cDNA was
diluted 10-fold and 2 µl diluted cDNA was used as a template for
RT-qPCR. RT-qPCR was conducted using a CFX Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc.) using the iQ™
SYBR®-Green Supermix (Bio-Rad Laboratories, Inc.)
according to the manufacturer's protocol. RT-qPCR cycling
conditions were as follows: Initial denaturation at 95°C for 3 min,
followed by 40 cycles of denaturation at 95°C for 15 sec and
annealing and elongation at 60°C for 30 sec, and 1 cycle at 95°C
for 10 sec, at 65°C for 5 sec and at 95°C. GAPDH was used as an
internal control. All reactions were performed in triplicate. Fold
changes in TIMP3 mRNA expression were calculated using the
2−ΔΔCq method (16).
Primer sequences were as follows: GAPDH, forward
5′-CCTGCCGGTGACTAACCCT-3′, reverse 5′-AGGCGCCCAATACGACCAAA-3′; and
TIMP3, forward 5′-ACCGAGGCTTCACCAAGATG-3′ and reverse
5′-CCCCGTGTACATCTTGCCAT-3′.
RT-qPCR for miR-221 expression
Total RNA, including miRNAs, was extracted using a
miRNeasy mini kit (Qiagen, Inc.) according to the manufacturer's
protocol. The miRNAs were converted to cDNA, to which a universal
tag was added, using the miScript II RT kit (Qiagen, Inc.)
according to the manufacturer's protocol. Quantification of miR-221
expression levels was conducted by RT-qPCR using the miScript SYBR
Green PCR kit (Qiagen, Inc.), which contains a primer for the
detection of the universal tag on cDNAs of miRNAs. The RT-qPCR
assay was performed in triplicate with U6 small nuclear (sn)RNA
used as an internal control. The following primers were used:
miR-221, 5′-GCAGAGCTACATTGTCTGCT-3′ and U6,
5′-ACGCAAATTCGTGAAGCGTT-3′. The reaction was performed in a final
volume of 25 µl, containing 2 µl cDNA, 2.5 µl specific primer, 2.5
µl universal primer, 12.5 µl 2X QuantiTect SYBR-Green PCR Master
Mix and 5.5 µl H2O. The PCR cycling conditions were as
follows: 95°C for 15 min, followed by 40 cycles at 94°C for 15 sec,
55°C for 30 sec and 70°C for 30 sec. The expression levels of
miR-221 were normalized to U6 snRNA. The fold-change in miR-221
levels was calculated using the 2−ΔΔCq method (16).
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay
lysis buffer (1% NP-40, 0.5% Sodium deoxycholate and 0.1% SDS;
Wuhan Boster Biological Technology, Ltd.) and protein concentration
of whole cell lysates was determined using the Bicinchoninic Acid
Protein Assay kit (Beyotime Institute of Biotechnology, Haimen,
China). Briefly, 30 µg whole cell lysates were loaded and separated
by electrophoresis on 10% SDS-polyacrylamide gels and were then
transferred to polyvinylidene fluoride membranes (Beyotime
Institute of Biotechnology). After blocking with 5% non-fat milk
for 1 h at room temperature, the membranes were incubated with
rabbit anti-human TIMP3 (cat no. ab39184; 1:1,000; Abcam,
Cambridge, MA, USA) and rabbit anti-human β-actin (cat no. ab8227;
1:2,000; Abcam) overnight at 4°C. The membranes were then washed
three times with TBST containing 0.1% Tween-20 and incubated with
goat anti-rabbit secondary antibody (cat no. A0208; 1:1,000;
Beyotime Institute of Biotechnology) for 1 h at room temperature.
Subsequently, membranes were washed a further three times with TBST
and were incubated with BeyoECL Plus chemical luminescence solution
(Beyotime Institute of Biotechnology). The protein bands were
visualized using a ChemiDoc XRS imaging system and were analyzed
using Quantity One software version 4.3.0 (Bio-Rad Laboratories,
Inc.).
Luciferase reporter assay
The 3′UTR of human TIMP3 mRNA transcripts was
obtained from cDNA reverse transcribed from RNA as aforementioned,
and amplified by PCR using the following primers: TIMP3, forward
5′-CTCGAGCTGGGCAAAGAAGGGTCTTTCGC-3′ and reverse
5′-GCGGCCGCTTCCAATAGGGAGGAGGCTGGAGGA-3′. The PCR products were
cloned downstream of the Renilla luciferase stop codon in a
psi-CHECK2 vector (Promega Corporation, Madison, WI, USA) between
the XhoI (5′) and NotI (3′) sites. To perform the
luciferase assay, SCC9 cells were co-transfected with 1 µg
p3′UTR-TIMP3 and 100 nM anti-miR-221 oligonucleotides or 100 nM
non-targeting oligonucleotides using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Cells were harvested 24 h
post-transfection and were assessed using the Dual Luciferase
Reporter Assay (Promega Corporation) according to the
manufacturer's protocol, on a Synergy 2 plate reader (BioTek
Instruments, Inc., Winooski, VT, USA). The assay was performed in
triplicate. Renilla luciferase activity was normalized to
firefly luciferase activity for each well to normalize cell number
and transfection efficiency.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Student's
t-test was used to compare the means of two groups. A one-way
analysis of variance followed by Tukey post hoc test was conducted
to compare the means of more than two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-221 expression is upregulated in
SCC4 and SCC9 cells treated with doxorubicin
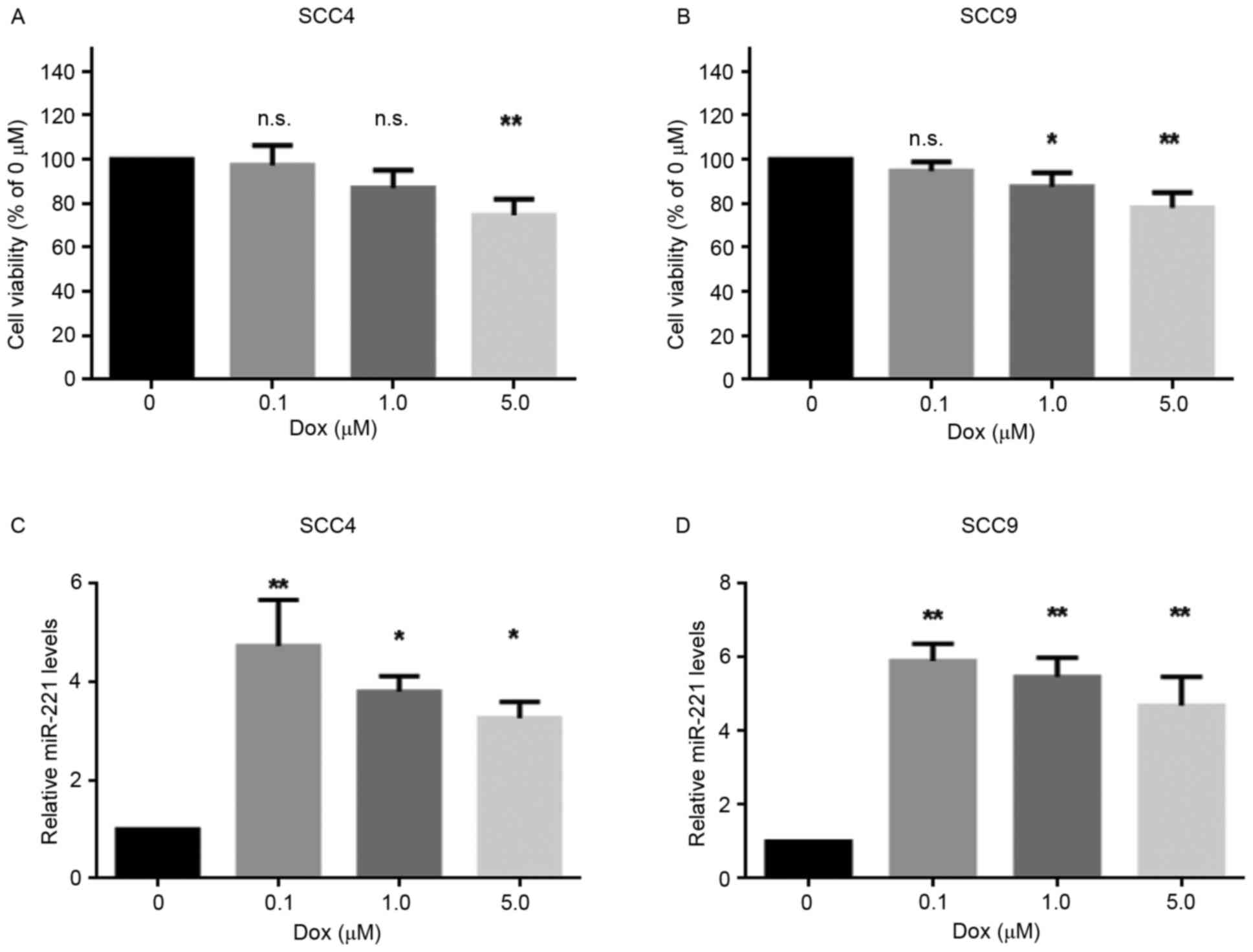
The present study aimed to investigate the effects
of doxorubicin on the viability of two OSCC cell lines, SCC4 and
SCC9, following treatment with various concentrations (0, 0.1, 1.0
and 5.0 µM) of doxorubicin for 24 h. The results revealed that 0.1
µM doxorubicin was not lethal to the cell lines, whereas 5 µM
doxorubicin significantly decreased cell viability (Fig. 1A and B). Furthermore, to determine
whether miR-221 was involved in the response of these cells to
doxorubicin, the expression levels of miR-221 were detected in SCC4
and SCC9 cells treated with various concentrations of doxorubicin.
As presented in Fig. 1C and D,
miR-221 expression was upregulated in the cell lines following
doxorubicin treatment. In addition, a sublethal dose of doxorubicin
(0.1 µM) resulted in the highest upregulation of miR-221 levels.
These data indicated that miR-221 expression was induced in
response to doxorubicin treatment in SCC4 and SCC9 cells, thus
suggesting a potential role for miR-221 in the protection of OSCC
cells from doxorubicin.
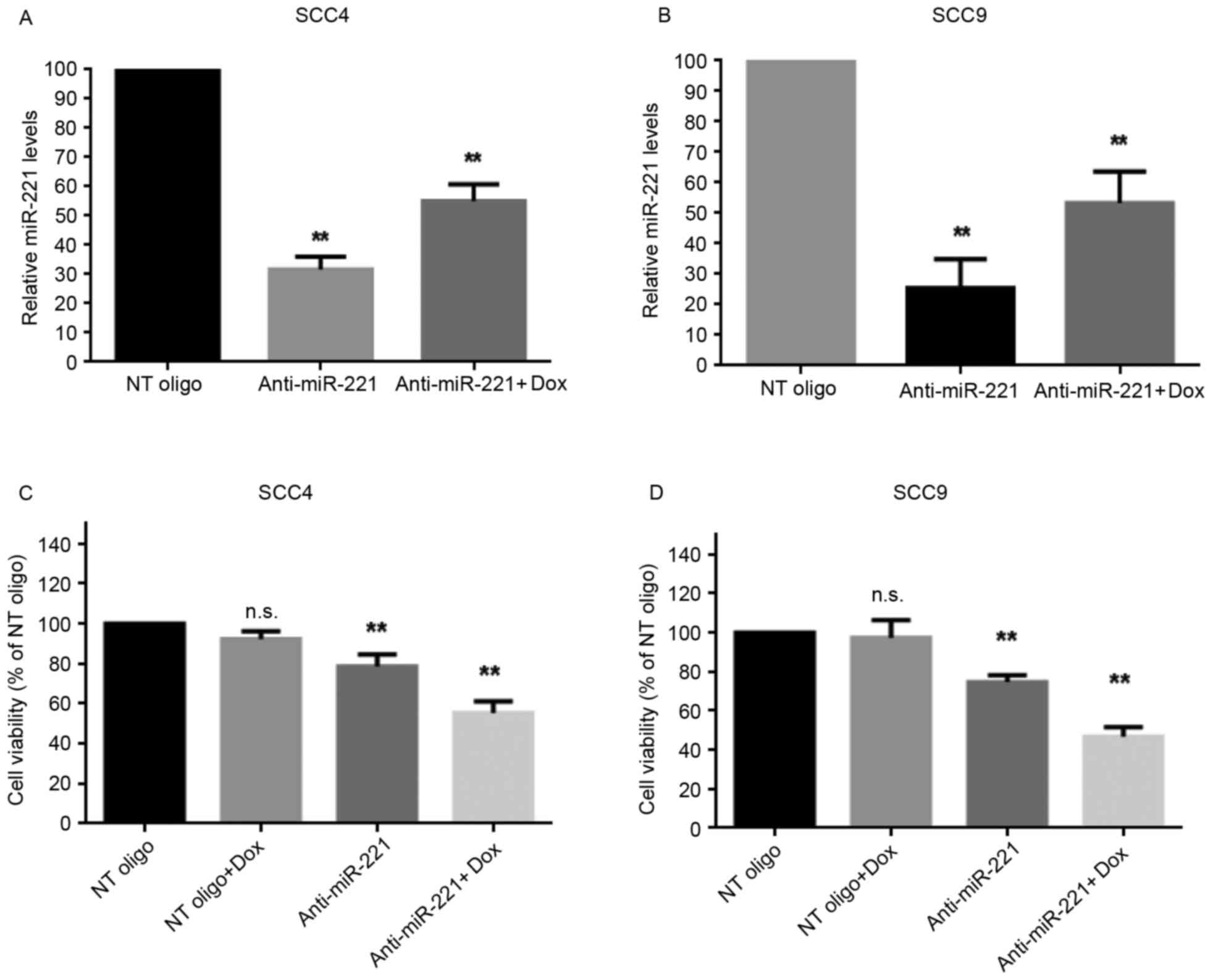
Inhibition of miR-221 sensitizes SCC4
and SCC9 cells to doxorubicin
miR-221 has been reported to protect cancer cells
from apoptosis in hepatocellular carcinoma (17), astrocytoma (18) and breast cancer (12). Therefore, the present study
investigated the roles of upregulated miR-221 in cellular
sensitivity to doxorubicin. miR-221 was knocked down in SCC4 and
SCC9 cells by transient transfection with anti-miR-221
oligonucleotides; miR-221 knockdown was confirmed using RT-qPCR.
The anti-miR-221 oligonucleotides significantly decreased the
levels of miR-221 compared with in cells transfected with
non-targeting oligonucleotides, with or without doxorubicin
treatment (Fig. 2A and B).
Subsequently, the effects of silencing miR-221 on cell viability,
with or without treatment of doxorubicin, were determined. The
results revealed that inhibition of miR-221 alone moderately
reduced the viability of SCC4 and SCC9 cells (Fig. 2C and D). However, transfection with
miR-221 inhibitory oligonucleotides alongside treatment with
doxorubicin significantly inhibited the viability of both cell
lines (Fig. 2C and D). The effects
of miR-221 inhibition on cell apoptosis, with or without
doxorubicin treatment, were then investigated. Without doxorubicin
treatment, inhibition of miR-221 in SCC4 and SCC9 cells led to a
moderately increased percentage of apoptotic cells compared with
the control cells (P<0.05; Fig. 2E
and F). However, alongside doxorubicin treatment, transfection
of SCC4 and SCC9 cells with anti-miR-221 oligonucleotides further
increased the population of apoptotic cells compared with the cells
transfected with control oligonucleotides (P<0.01; Fig. 2E and F). These results indicated
that knockdown of miR-221 sensitized SCC4 and SCC9 cells to
doxorubicin, thus suggesting a protective role for upregulated
miR-221 in doxorubicin-mediated cell death.
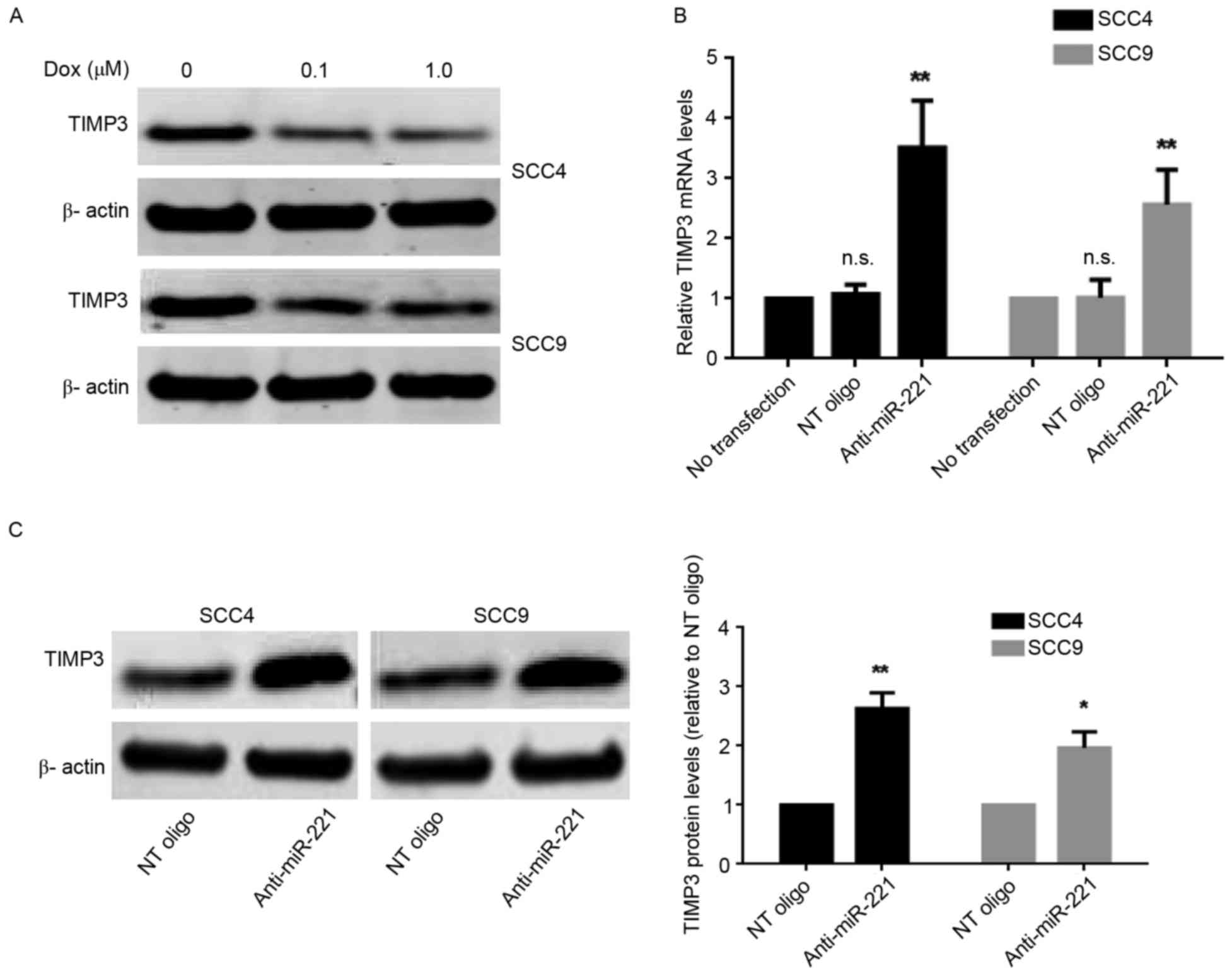
TIMP3 is downregulated in SCC4 and
SCC9 cells following doxorubicin treatment
TIMP3 has been identified as a direct target of
miR-221 in numerous types of cancer, including breast, lung and
liver cancers (12,13). To investigate whether TIMP3 is
involved in miR-221-mediated resistance to doxorubicin in SCC4 and
SCC9 cells, the present study analyzed the protein expression
levels of TIMP3 following doxorubicin treatment. The results
revealed that the protein expression levels of TIMP3 were decreased
following doxorubicin treatment (Fig.
3A) in both cell lines. However, the decrease in TIMP3 protein
levels did not occur in a dose-dependent manner, since an increase
in the concentration of doxorubicin from 0.1 to 1.0 µM did not
further decrease the protein expression levels of TIMP3 (Fig. 3A). To determine whether miR-221
regulated TIMP3 expression in SCC4 and SCC9 cells the mRNA and
protein expression levels of TIMP3 in cells transiently transfected
with anti-miR-221 or control oligonucleotides were determined.
TIMP3 mRNA (Fig. 3B) and protein
levels (Fig. 3C) were
significantly increased when miR-221 was inhibited in SCC4 and SCC9
cells. Subsequently, the present study confirmed that TIMP3 was a
direct target of miR-221 in OSCC using a 3′UTR luciferase reporter
assay (Fig. 3D). The reporter
assay revealed that transfection of cells with anti-miR-221
oligonucleotides significantly increased the luciferase activity of
the TIMP3 3′UTR reporter plasmid in SCC9 cells (Fig. 3E). These data suggested that TIMP3
expression is downregulated through direct regulation by miR-221 in
response to doxorubicin treatment in SCC4 and SCC9 cells.
miR-221 protects OSCC cells through
downregulating TIMP3 levels
The present study investigated whether miR-221 may
protect OSCC cells from doxorubicin-mediated cell death through
downregulating TIMP3 levels. TIMP3 was knocked down in SCC4 and
SCC9 cells following transfection with TIMP3-specific siRNA; the
knockdown was confirmed by RT-qPCR (Fig. 4A) and western blotting (Fig. 4B). Suppression of TIMP3 alone did
not lead to significant alterations in cell viability (Fig. 4C). The present study then detected
the viability and apoptosis of SCC4 and SCC9 cells transfected with
anti-miR-221 oligonucleotides and TIMP3 siRNA, and compared the
results to cells transfected with the anti-miR-221 oligonucleotides
and non-targeting siRNA under doxorubicin treatment. The cell
viability results revealed that co-transfection of cells with
anti-miR-221 and TIMP3 siRNA significantly improved the survival
rate compared with cells transfected with anti-miR-221
oligonucleotides and control siRNA (P<0.05) under doxorubicin
treatment in SCC4 and SCC9 cells (Fig.
4D). Furthermore, the percentage of early apoptotic cells was
reduced in SCC4 (P<0.05, Fig.
4E) and SCC9 (P<0.05, Fig.
4F) cells transfected with anti-miR-221 oligonucleotides and
TIMP3 siRNA compared with cells transfected with anti-miR-221 and
control siRNA under doxorubicin treatment. These results indicated
that suppression of TIMP3 partially reversed the
anti-miR-221-induced inhibition of cell viability and promotion of
apoptosis in OSCC cells, thus suggesting that miR-221 may protect
OSCC cells from doxorubicin treatment partially by downregulating
TIMP3.
 | Figure 4.miR-221 protects OSCC cells through
downregulating TIMP3 levels. (A) mRNA expression levels of TIMP3 in
cells transfected with TIMP3 siRNA or control oligonucleotides, as
detected by reverse transcription-quantitative polymerase chain
reaction. Data are presented as the mean ± standard deviation of
three independent experiments. **P<0.01 vs. siNT group. (B)
Protein expression levels of TIMP3 in cells transfected with TIMP3
siRNA or control oligonucleotides, as detected by western blotting.
(C) Effects of silencing TIMP3 on cell viability, measured using an
MTT assay. (D) Viability of SCC4 and SCC9 cells transfected with
anti-miR-221 oligonucleotides and TIMP3 siRNA or control
oligonucleotides under doxorubicin treatment, measured using an MTT
assay Data are presented as the mean ± standard deviation of three
independent experiments. *P<0.05 vs. anti-miR-221 + Dox + siNT
group. Effects of anti-miR-221 and TIMP3 siRNA on (E) SCC4 and (F)
SCC9 cell apoptosis under doxorubicin. Representative dot plots and
quantification of flow cytometry results are presented. Data are
presented as the mean ± standard deviation of three independent
experiments. *P<0.05 vs. anti-miR-221 + Dox + siNT group. Dox,
doxorubicin; miR, microRNA; TIMP3, tissue inhibitor of
metalloproteinase-3; NT, non-targeting; siRNA, small interfering
RNA; FITC, fluorescein isothiocyanate; n.s., not significant. |
Discussion
OSCC represents the most common type of oral cancer,
which is the sixth most common malignancy in humans (1). Despite advances in therapy, only
40–50% of patients with OSCC are likely to survive for 5 years,
which is primarily due to metastasis to distal organs (19,20).
Locally advanced tumors and metastatic disease require a
comprehensive therapeutic strategy involving chemotherapy; however,
the efficacy of chemotherapy is compromised due to acquired and
inherent tumor cell resistance to chemotherapeutic reagents.
Doxorubicin, which is a drug widely used in cancer treatment, is
less often used in OSCC due to the resistance of OSCC cancer cells.
Therefore, to improve its use in OSCC a strategy is required to
increase the sensitivity of OSCC cells to this reagent.
miR-221 has been reported to be aberrantly expressed
in OSCC and promotes the development of OSCC by improving
anchorage-independent growth, migration and invasion of cancer
cells (5,10). In addition, miR-221 is involved in
the resistance of cancer cells to chemotherapeutic reagents
(14,15,17).
In breast cancer cells, downregulation of miR-221/222 enhances the
sensitivity of MCF-7 and MDA-MB-231 to tamoxifen via the
upregulation of TIMP3 (12). In
NSCLC and hepatocarcinoma cells, miR-221/222 induces TRAIL
resistance by targeting PTEN and TIMP3 (13). Furthermore, inhibition of miR-221
in SNU449 liver cancer cells increased doxorubicin-induced cell
apoptosis (9). Therefore, the
present study aimed to determine whether miR-221 is involved in
regulating the sensitivity of OSCC cells to doxorubicin.
The results of the present study demonstrated that
miR-221 levels were upregulated in OSCC cells treated with
doxorubicin, thus suggesting that miR-221 may serve a protective
role in response to doxorubicin. In support of this hypothesis,
knockdown of miR-221 in doxorubicin-treated OSCC cells reduced cell
viability and induced cell apoptosis, indicating that inhibition of
miR-221 may increase the sensitivity of OSCC cells to doxorubicin.
miR-221 has also been reported to serve an oncogenic role in OSCC.
Gombos et al (21) reported
that miR-221 was overexpressed in OSCC samples. Yang et al
(10) indicated that miR221
increased growth and anchorageindependent colony formation of OSCC
cell lines, and promoted the tumorigenesis of an OSCC cell line in
nude mice. Furthermore, He et al (5) demonstrated that inhibition of miR-221
suppressed migration and invasion of UM1 OSCC cells by targeting
methyl-CpG-binding domain protein 2. The findings of the present
study are consistent with these results, thus indicating that
miR-221 functions as an oncogene in OSCC development.
TIMP3 promotes apoptotic cell death in melanoma
cells (22). In addition, it has
previously been identified as a target of miR-221 in breast cancer,
NSCLC and liver cancer (12,13).
The present study demonstrated that, unlike miR-221, the protein
expression levels of TIMP3 were downregulated in response to
doxorubicin treatment. Therefore, downregulation of TIMP3 may be
induced by miR-221 and may be involved in induction of resistance
to doxorubicin. To test these hypotheses, the present study
validated that TIMP3 is a direct target of miR-221 in
doxorubicin-treated OSCC cells using a TIMP3 3′UTR luciferase
reporter assay. To further demonstrate that TIMP3 contributes to
miR-221-induced doxorubicin chemoresistance, TIMP3 siRNA was
transfected into OSCC cells to decrease TIMP3 expression under
inhibition of miR-221. The results demonstrated that depletion of
TIMP3 improved the survival of cells transfected with anti-miR-221
oligonucleotides and treated with doxorubicin. Therefore, it may be
concluded that miR-221 induced resistance to doxorubicin in human
OSCC cell lines at least in part by downregulating TIMP3. Due to
the complexity of the gene networks that are regulated by miR-221,
other factors may also contribute to miR-221-induced doxorubicin
resistance; therefore, a better understanding of how miR-221
regulates doxorubicin sensitivity is required for designing
therapeutic strategies that target miR-221 in OSCC.
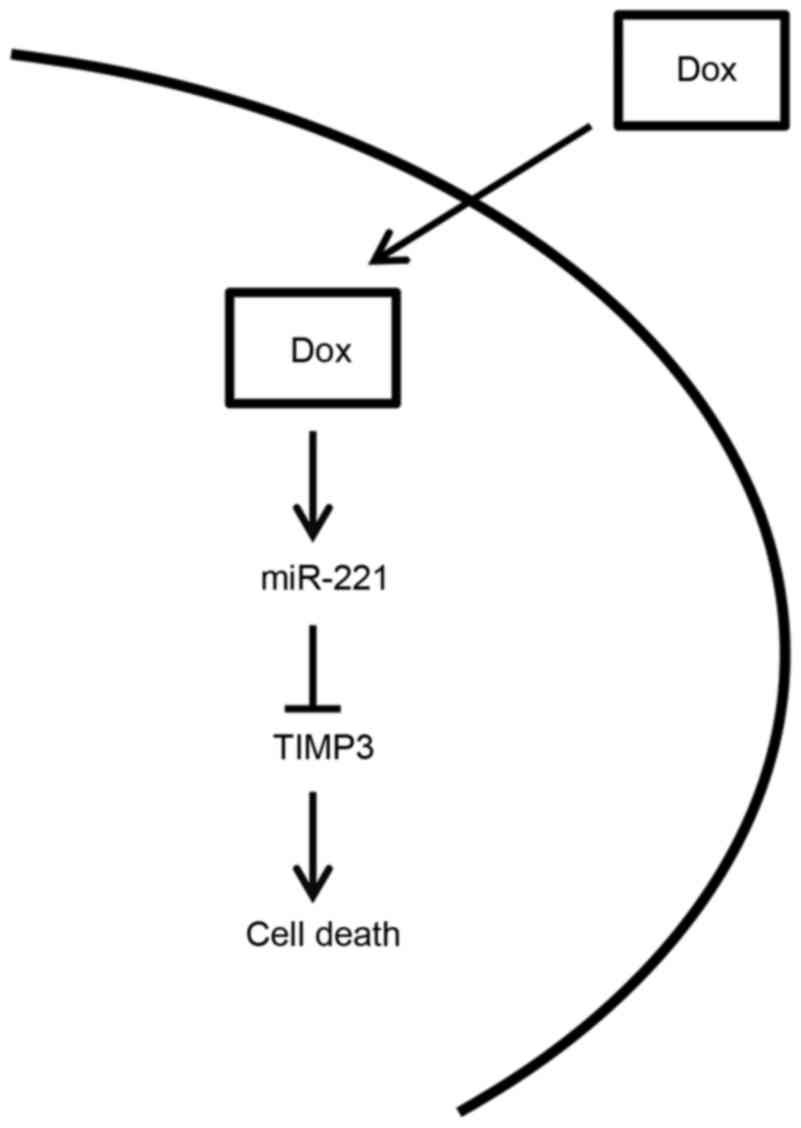
In conclusion, the present study is summarized in
the diagram presented in Fig. 5.
Briefly, miR-221 levels were upregulated in response to doxorubicin
treatment in OSCC cell lines, resulting in a decrease in the
expression levels of TIMP3. The downregulation of TIMP3 was able to
protect OSCC cells from doxorubicin-induced apoptosis, resulting in
resistance of OSCC cells to doxorubicin. In concordance with this
hypothesis, the present study indicated that inhibiting miR-221,
following transfection with inhibitory oligonucleotides,
significantly sensitized OSCC cells to doxorubicin. Therefore,
inhibition of miR-221 may represent a potential strategy to
sensitize OSCC cells to doxorubicin and improve the efficacy of
doxorubicin in OSCC.
Glossary
Abbreviations
Abbreviations:
|
OSCC
|
oral squamous cell carcinoma
|
|
TIMP3
|
tissue inhibitor of
metalloproteinase-3
|
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality
Worldwide: IARC Cancerbase No. 11 (Internet)International Agency
for Research on Cancer. Lyon, France: 2013 http://globocan.iarc.frAccessed. May 19–2014
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
MacFarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: Their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He S, Lai R, Chen D, Yan W, Zhang Z, Liu
Z, Ding X and Chen Y: Downregulation of miR-221 inhibits cell
migration and invasion through targeting methyl-CpG binding domain
protein 2 in human oral squamous cell carcinoma cells. Biomed Res
Int. 2015:7516722015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nassirpour R, Mehta PP, Baxi SM and Yin
MJ: miR-221 promotes tumorigenesis in human triple negative breast
cancer cells. PLoS One. 8:e621702013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y,
Li H, Zhu X, Yao L and Zhang J: Exosomal miR-221/222 enhances
tamoxifen resistance in recipient ER-positive breast cancer cells.
Breast Cancer Res Treat. 147:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Y, Zhao L, Ischenko I, Bao Q, Schwarz
B, Nieß H, Wang Y, Renner A, Mysliwietz J and Jauch KW: Antisense
inhibition of microRNA-21 and microRNA-221 in tumor-initiating
stem-like cells modulates tumorigenesis, metastasis, and
chemotherapy resistance in pancreatic cancer. Target Oncol.
10:535–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fornari F, Milazzo M, Galassi M, Callegari
E, Veronese A, Miyaaki H, Sabbioni S, Mantovani V, Marasco E,
Chieco P, et al: p53/mdm2 feedback loop sustains miR-221 expression
and dictates the response to anticancer treatments in
hepatocellular carcinoma. Mol Cancer Res. 12:203–216. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang CJ, Shen WG, Liu CJ, Chen YW, Lu HH,
Tsai MM and Lin SC: miR-221 and miR-222 expression increased the
growth and tumorigenesis of oral carcinoma cells. J Oral Pathol
Med. 40:560–566. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cruz-Munoz W and Khokha R: The role of
tissue inhibitors of metalloproteinases in tumorigenesis and
metastasis. Crit Rev Clin Lab Sci. 45:291–338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garofalo M, Di Leva G, Romano G, Nuovo G,
Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P,
et al: miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Starr TK, Scott PM, Marsh BM, Zhao L, Than
BL, O'Sullivan MG, Sarver AL, Dupuy AJ, Largaespada DA and Cormier
RT: A sleeping beauty transposon-mediated screen identifies murine
susceptibility genes for adenomatous polyposis coli (Apc)-dependent
intestinal tumorigenesis. Proc Natl Acad Sci USA. 108:5765–5770.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Than BL, Goos JA, Sarver AL, O'Sullivan
MG, Rod A, Starr TK, Fijneman RJ, Meijer GA, Zhao L, Zhang Y, et
al: The role of KCNQ1 in mouse and human gastrointestinal cancers.
Oncogene. 33:3861–3868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rong M, Chen G and Dang Y: Increased
miR-221 expression in hepatocellular carcinoma tissues and its role
in enhancing cell growth and inhibiting apoptosis in vitro. BMC
Cancer. 13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu D and Sun YC: Overexpression of
miR-221 inhibits proliferation and promotes apoptosis of human
astrocytoma cells. Int J Clin Exp Pathol. 8:4851–4856.
2015.PubMed/NCBI
|
|
19
|
Tanaka T, Tanaka M and Tanaka T: Oral
carcinogenesis and oral cancer chemoprevention: A review. Pathol
Res Int. 2011:4312462011. View Article : Google Scholar
|
|
20
|
Huang SH and O'Sullivan B: Oral cancer:
Current role of radiotherapy and chemotherapy. Med Oral Patol Oral
Cir Bucal. 18:e233–e240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gombos K, Horváth R, Szele E, Juhász K,
Gocze K, Somlai K, Pajkos G, Ember I and Olasz L: miRNA expression
profiles of oral squamous cell carcinomas. Anticancer Res.
33:1511–1517. 2013.PubMed/NCBI
|
|
22
|
Das AM, Seynhaeve AL, Rens JA, Vermeulen
CE, Koning GA, Eggermont AM and Ten Hagen TL: Differential TIMP3
expression affects tumor progression and angiogenesis in melanomas
through regulation of directionally persistent endothelial cell
migration. Angiogenesis. 17:163–177. 2014. View Article : Google Scholar : PubMed/NCBI
|