Introduction
Osteosarcoma (OS) is a highly malignant and
metastatic disease, characterized by developing rapidly and
difficulty in treatment, these characteristics result in its
association with high mortality in children and adolescents
(1). Currently, tumor resection
operation, chemical therapy and radiation therapy are mainly
treatments for OS. However, the high degree of limb pain after
surgery and resistance to radiation therapy are still difficult to
overcome (2). Previous studies
about OS have focused on the immune gene, tumor suppressor gene,
antisense gene, suicide gene, gene combined and gene vectors in
gene therapy of this disease (3).
Nevertheless, a better understanding of the molecular mechanisms in
OS is still needed and will be helpful in diagnosing and treating
this disease.
MicroRNAs (miRNAs) are endogenous small molecule
single-stranded RNAs (about 22–25 nucleotides long), known to play
important posttranscriptional regulatory roles in plants and
animals (4). MiRNAs are involved
in the complex life process, including organism growth and
development, organ formation, cell proliferation and cell
apoptosis. Moreover, it is quite remarkable that a single miRNA can
regulate the expression of key proteins which involved in different
signaling pathways and different processes in cell physiology
(5). MiRNA expression and
functions that are associated with the etiology of diseases were
first reported in the case of Fragile X syndrome (6). Thereafter, several studies proposed
that the abnormal expression of miRNAs was closely related with
tumor development and metastasis, and miRNAs were expected to be a
breakthrough in the treatment of cancer (7). An increasing number of studies have
reported that multiple miRNAs served as tumor suppressors and key
regulators in cancers (8).
However, the known of miRNAs target genes and its role in various
tumorigenesis stages were still limited. Previous studies have
confirmed that abnormal expression of miR-300 was related with
cancer cell proliferation and apoptosis, and a previous study in
MG63 cell line indicated the pro-proliferation and pro-invasion
roles of miR-300, but the exactly role of miRNA-300 in OS has not
been exhaustively investigated to our knowledge (9).
The main objective of the present study was to
investigate the effect of miR-300 in OS cells and the associated
mechanism. In the present study, miR-300 mimic, mimic control,
miR-300 inhibitor, or inhibitor control were transfected into MG63
cells (a human OS cells line), respectively. The effect of miR-300
on MG63 cells viability, migration and apoptosis were investigated
in vitro. Furthermore, the transcription factor Twist1 has
been confirmed as a target gene of miR-106b, miR-720 and miR-33b
(10), and the correlations
between Twist1 and miRNAs have been proved to be involved in cancer
cells modulations (11).
Therefore, we also assessed Twist1 as the target of miR-300 in MG63
cells and the protein level changes of main factors in NF-κB
signaling pathway to detect the possible molecular mechanisms of
miR-300 in OS. The present study might provide supplementing
certification for basic understanding of miR-300 in OS and
possibilities of it usage in OS treatment.
Materials and methods
Cell culture and transfection
Human OS cell line MG63 were obtained from the
American Type Culture Collection (Rockville, MD, USA), and cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island,
NY, USA) containing 10% heat-inactivated feral bovine serum (FBS;
HyClone, Logan, UT, USA), 100 U/ml penicillin and 100 U/ml
streptomycin (Gibco) in the incubator at 37°C with 5%
CO2 (12).
MG63 cells were transfected with miR-300 mimic,
miR-300 inhibitor, or their scramble controls (Shanghai GenePharma
Co., Ltd., Shanghai, China), respectively. Twist1 coding sequence
was amplified by PCR and cloned into pcDNA™ 3.1 vector (Thermo
Fisher Scientific, Inc., Beijing, China) to construct
pcDNA3-Twist1, then the recombinant vector and empty vector as
control were transfected into MG63 cells, respectively. The
transfection was performed by using Lipofectamine 3000 (Invitrogen
Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. After 48 h of transfection, cells were
collected for the further analysis.
Cell viability assay
After transfection, cell viability was assessed
using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) colorimetric assay according to the standard methods.
In brief, cells were seeded into 96-well plates at a density of
2×103 cells/well. After 1–4 days of incubation, 20 µl of
MTT (Sigma, St. Louis, MO, USA) was added into each well and
incubated for another 4 h at 37°C. Then, 150 µl dimethylsulfoxide
(DMSO; Sigma) was added and the plates were shaken for 10 min. Each
experiment was performed 3 times. Absorbance was measured at 570 nm
(OD570) with a 680 microplate enzym-linked immunosorbent assay
(ELISA) reader (Bio-Rad Laboratories, Inc., Hemel Hempstead,
UK).
Cell migration assay
Cell migration was determined by using a modified
two-chamber with 8.0 µm pore membranes (Greiner 662638). The
transfected MG63 cells were suspended in 200 µl of serum-free
culture medium, and were added in the uper compartment of 24-well
transwell culture chamber. Then 600 µl complete culture medium was
added to the lower comparment, cells were incubated at 37°C for 12
h. After that, cells were fixed with 4% methanol (NIST, USA) for 30
mim, non-transferred cells were removed from the upper surface of
the filter carefully with a cotton swab. Traversed cells in the
lower were stained with 0.1% crystal violet for 20 min and counted
under an optical microscope (Leica Microsystems GmbH, Wetzlar,
Germany).
Cell apoptosis assay
Flow cytomety analysis was performed to identify and
quantify the apoptotic cells by using Annexin V-FITC/PI apoptosis
detection kit (Beijing Biosea Biotechnology Co., Ltd, Beijing,
China) according to manufacturer's recommendations. MiR-transfected
cells were collected and suspended in 200 µl of binding buffer
containing 10 µl Annexin V-FITC and 5 µl of PI, then incubated at
room temperature for 30 min in the dark. Apoptotic cells were
measured with flow cytometer (Beckman Coulter, Miami, FL, USA)
(13).
Dual-Luciferase activity assay
The 3′-untranslated regions (3′-UTR) of Twist1 was
amplified by PCR and placed into the pMiR-report vector (Ambion,
Carlsbad, CA, USA). MG63 cells were cotransfected with 0.5 µg of
the recombin Twist1 report vector and miR-300 mimics using
Lipofectamine 3000 (Invitrogen Life Technologies). The recombinant
repot vector carrying the Twist1 mutation sequence or the empty
report vector were also cotransfected with miR-300 into MG63 cells
and activted as controls, respectively. Forty eight hours after
transfection, cells were collected and Dual-Luciferase activities
were measured using the Dual-Luciferase reporter assay kit (Promega
Corp., Madison, WI, USA) following to the manufacture's information
(14).
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was extracted with Trizol (Invitrogen Life
Technologies) and reversely transcribed by MultiScribe RT kit
(Applied Biosystems Life Technologies, Foster City, CA, USA)
according to the manufacturer's instructions. All reverse
transcriptase reactions, including no template controls and
real-time minus controls, were run in a master cycler gradient
(Eppendorf AG, Hamburg, Germany). For the qPCR analysis, FastStart
Universal SYBR-Green Master (15)
(Roche Diagnostics, Indianapolis, IN, USA) was used according to
the manufacturer's instructions. The qPCR was performed in
triplicate, including no template controls. MiR-300 and Twist1
expressions were respectively normalized to U6 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expressions using
2−∆∆CT method (16).
All primers for qPCR were synthesized by GenePharma Co. The data
were analyzed with Real-Time StatMiner (Integromics, Granada,
Spain).
Western blot analysis
The protein expression changes in miR-300
transfected cells were detected by western blot analysis. The
simples were extracted by lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) supplemented with protease
inhibitors (Roche Diagnostics Ltd., Guangzhou, China). Proteins
were quantified using the BCA™ Protein Assay kit (Pierce, Appleton,
WI, USA). Equal amounts of protein were separated by 10% sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride (PVDF) membranes. Membranes
were incubated with specific primary antibodies (all 1:1,000):
Twist1 (sc15393), inhibitor of NF-κB alpha (IκBα, sc847),
phosphorylated IκBα (p-IκBα, sc101713), p65 (sc109), phosphorylated
p65 (p-p65, sc101749), B cell lymphoma/lewkmia-3 (Bcl-3, sc185)
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or GAPDH (ab8245;
Abcam, Cambridge, MA, USA), overnight at 4°C. Then the membranes
were incubated with horseradish peroxidase-conjugated secondary
antibodies (1:5,000, sc2004; Santa Cruz Biotechnology, Inc.) for 1
h at room temperature. Protein bands were developed by enhanced
chemiluminescence and were analyzed by Image Lab™ Software (Bio-Rad
Laboratories Co., Shanghai, China).
Statistical analysis
Data were presented as mean ± standard deviation
(SD), which were representative of at least three independent
experiments and analyzed using GraphPad Prism 5 software (GraphPad
Software, Inc., La Jolla, CA, USA). The P-values were calculated
using the two-tailed Student's t-test to evaluate the significance
of differences between two groups, and one-way analysis of variance
for multiple groups (17).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of miR-300 inhibited
cell viability and migration while increased apoptosis of MG63
cells
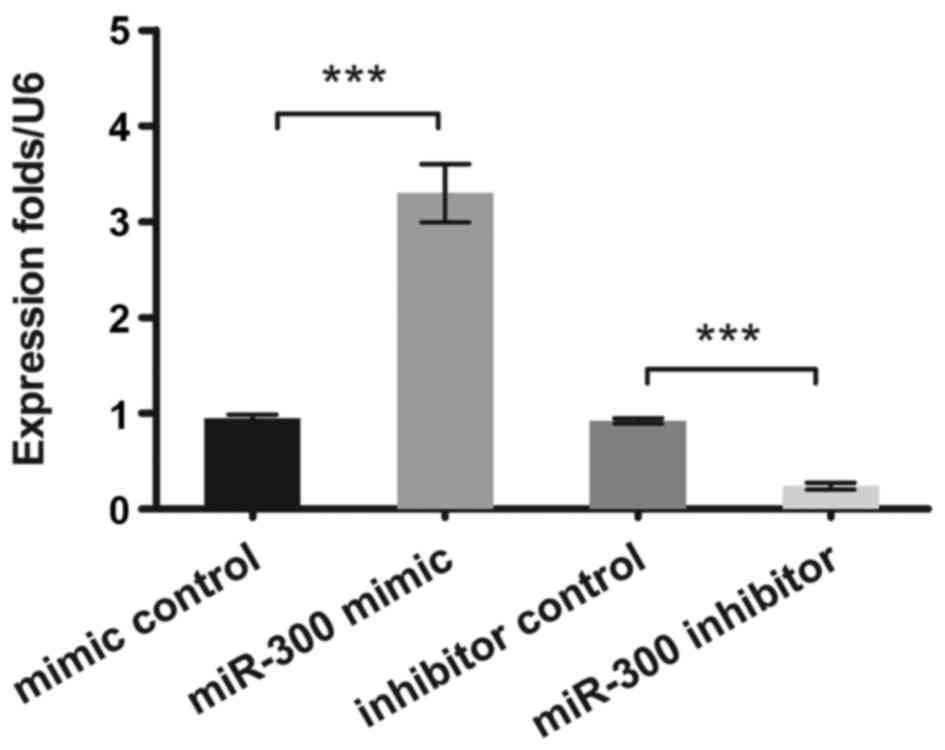
In the present study, the expression levels of
miR-300 in OS cells were measured by qPCR. It was verified that
miR-300 was effectively overexpressed or suppressed (P<0.001) in
MG63 cells after transfection (Fig.
1).
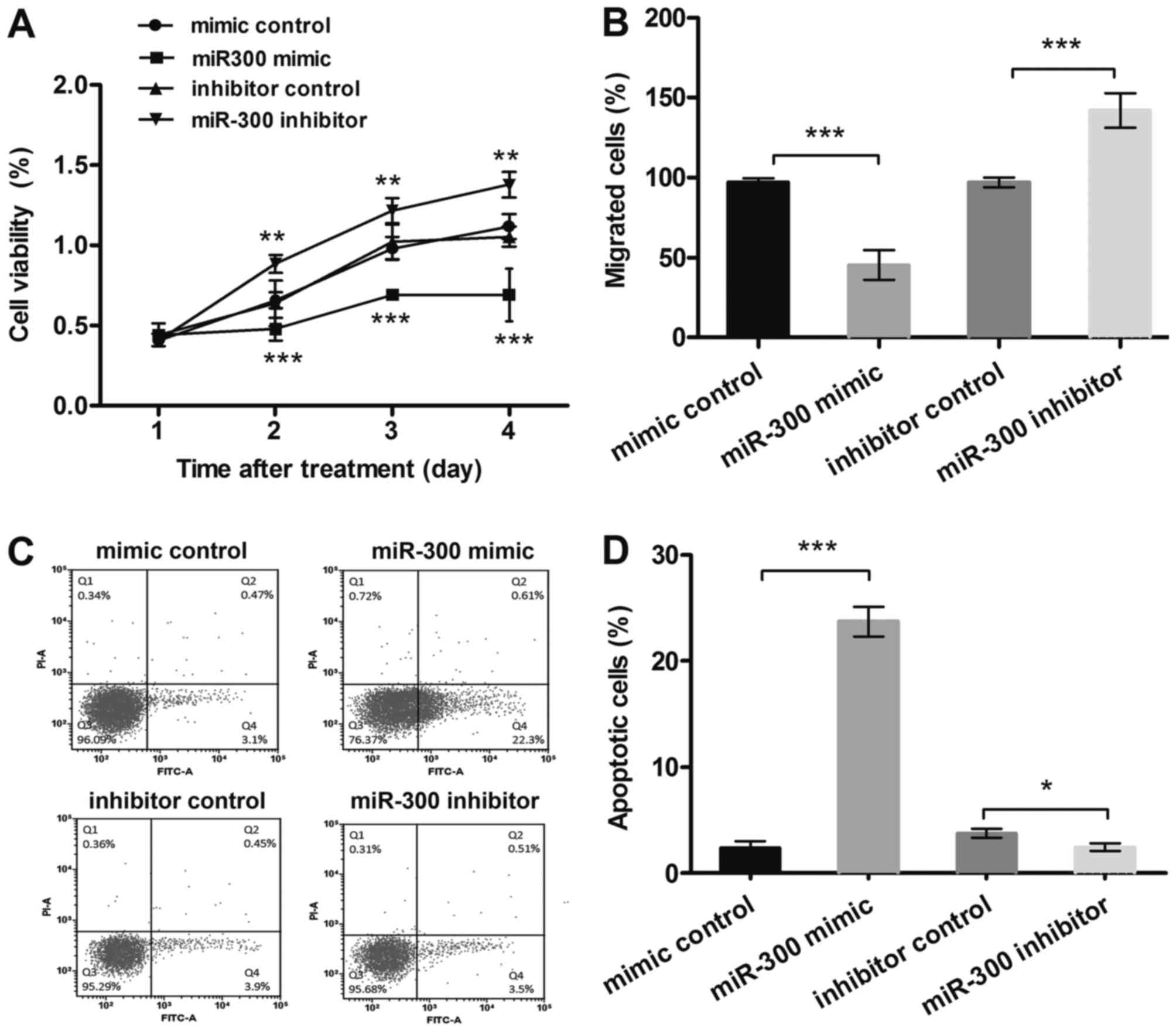
MTT results (Fig.
2A) showed that after transfection with miR-300 mimic, the MG63
cells viability was inhibited, and it was much lower than in the
control group in 2–4 days (P<0.001). However, suppression of
miR-300 displayed the opposite results at the same time points
(P<0.01). The results from migration assay (Fig. 2B) showed that, cell migration was
significantly decreased by miR-300 overexpression (P<0.001),
while increased by miR-300 suppression (P<0.001). Besides,
results in Fig. 2C and D showed
that miR-300 overexpression significantly enhanced the relative
rate of apoptosis (P<0.001), while miR-300 suppression group
showed the opposite result (P<0.05). Therefore, miR-300 might
affect cell viability and migration, and might be a key regulator
for apoptosis in MG63 cells.
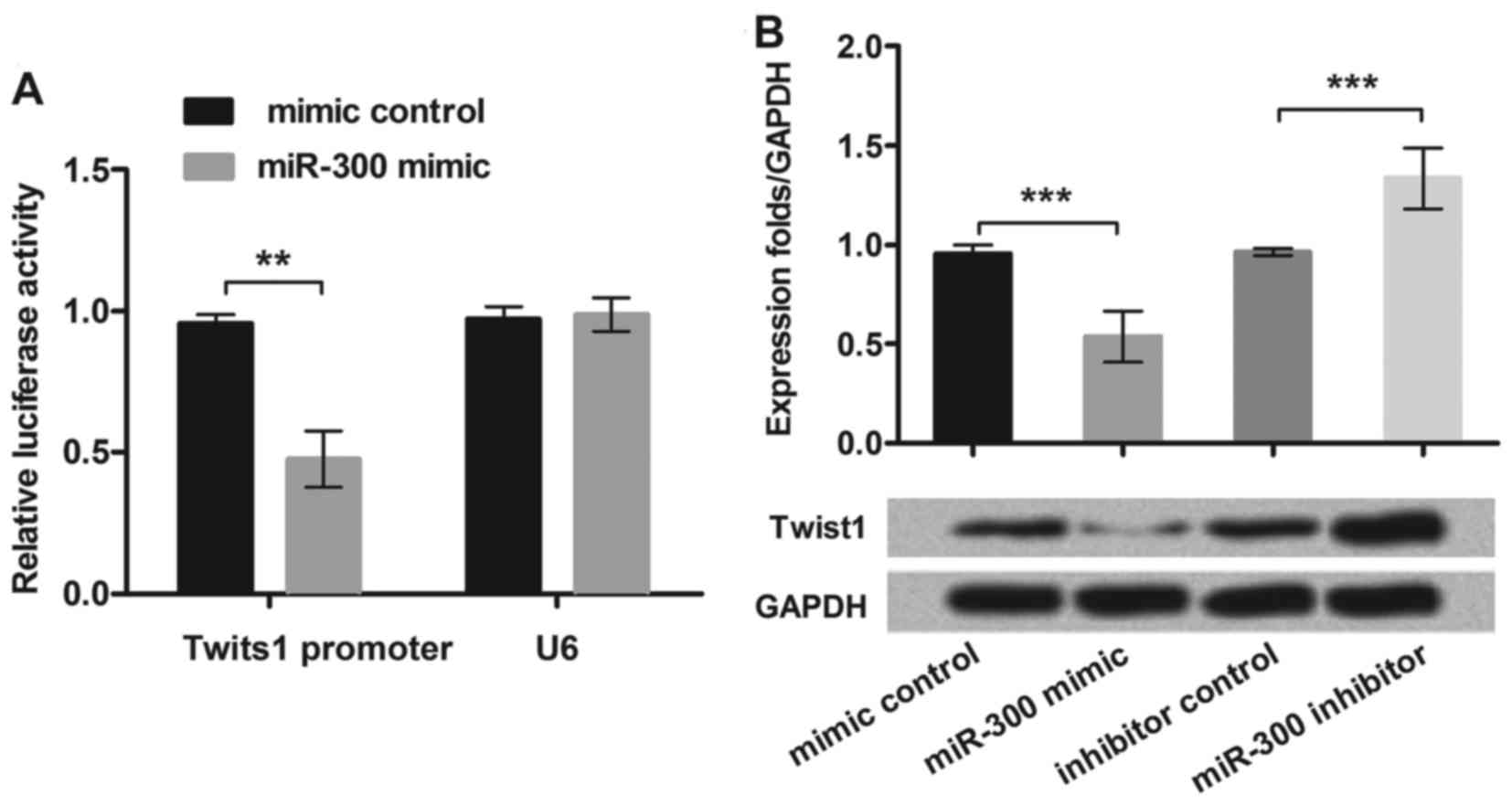
MiR-300 directly targeted Twist1 in
the MG63 cells
To test the relationship between miR-300 and Twist1,
we cloned the Twist1 3′-UTR region fragment into the luciferase
reporter system and co-transfected with miR-300 mimic into MG63
cell lines. We found that only the Twist1 reporter group displayed
obvious inhibition (P<0.01) when co-transfected with miR-300
mimic (Fig. 3A). This result
suggested that miR-300 directly targeted Twist1 3′-UTR in MG63
cells.
To further demonstrate how miR-300 affected Twist1,
we overexpressed or suppressed miR-300 in MG63 cells, then assessed
the protein expression levels of Twist1. We found that
overexpression of miR-300 caused the Twist1 protein level
decrement, while the miR-300 inhibitor enhanced the Twist1
expression in MG63 cells (P<0.001; Fig. 3B). The data demonstrated that
Twist1 was negatively regulated by miR-300 in MG63 cells.
MiR-300 downregulated expression of
NF-κB regulated proteins in MG63 cell
To further explore the underlying molecular
mechanisms of miR-300 in MG63 cells, we detected the expression of
selected NF-κB pathway-related proteins (IkBα, p65 and Bcl-3). MG63
cells were transfected with miR-300 mimic, exogenous expressed
Twist1, or both of them, respectively. As shown in Fig. 4A and B, the expression of p-IkBα,
p-p65 and Bcl-3 were significantly decreased in miR-300 mimic
treated group (P<0.05). However, Twist1 significantly
upregulated the expression of p-IkBα, p-p65 and Bcl-3, and reversed
the inhibitive effects of miR-300 on these proteins expressions
(P<0.001). Therefore, we inferred miRNA-300 suppressed
activation of the NF-κB signaling pathway by inhibiting Twist1.
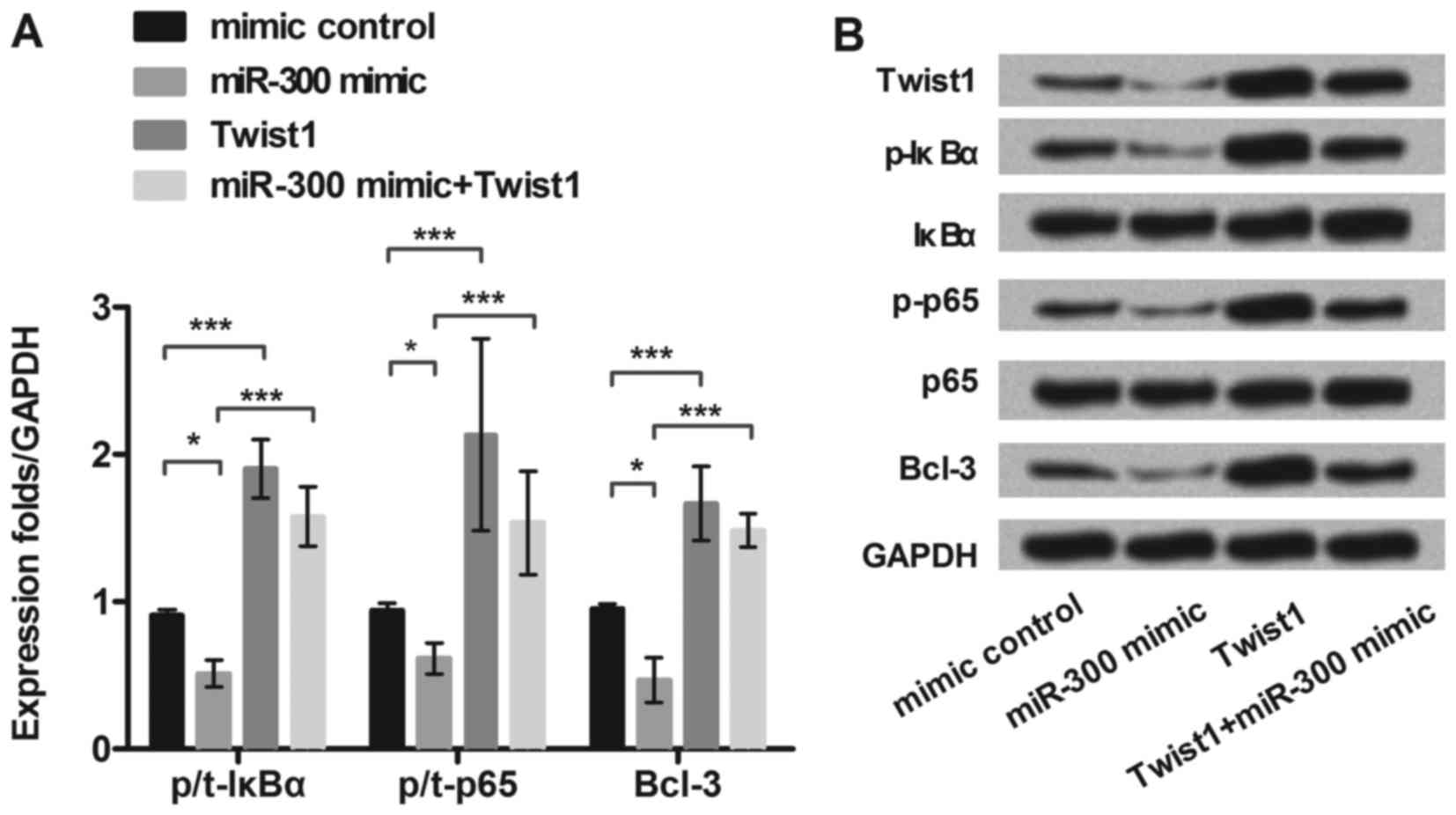 | Figure 4.MiR-300 downregulates expression of
NF-κB regulated proteins in MG63 cells. MG63 cells were transfected
with miR-300 mimic, exogenous expressed Twist1, or both of them,
respectively. (A and B) The protein level of Twist1 and NF-κB
regulated proteins (p/t-IκBα, p/t-p65, and Bcl-3) expression in
miR-300 treated MG63 cells were detected by western blot. GAPDH
acted as an internal control. MiR-300, microRNA-300; NF-κB, Nuclear
factor kappa beta; IκBα, inhibitor of NF-κB alpha; Bcl-3, B cell
lymphoma/lewkmia-3; p-IκBα, phosphorylated-IκBα; p-p65,
phosphorylated-p65 protein; **P<0.01, ***P<0.001. |
Discussion
In cancer, miRNAs serve as proto-oncogenes or tumor
suppressor genes, depending on their regulatory effect (18). Nowadays, plenty of miRNAs have been
found as oncogenes or anti-oncogenes in OS (19–21).
However, little literatures were reported the potential role of
miR-300 in the pathogenesis of OS. The present study aimed to
investigate the role of miR-300 in OS cells, and explored the
possible mechanisms. We found that miR-300 level was negatively
correlated with cell viability and migration in MG63 cells, and was
positively correlated with cell apoptosis. Twist1 was a direct
target gene of miR-300, and in vitro investigations revealed
that Twist1 was negatively regulated by miR-300. More importantly,
NF-κB pathway was implicated in the role of miR-300 in OS
cells.
Previous studies have demonstrated the versatile
roles of miRNAs in cancer. MiRNA abnormal expression were found in
a variety of tumors, and dysregulation of miRNAs has been proposed
to be a rising feature in cancer (22,23).
Deregulated expression of miR-300 has been studied in a lot of
cancers, such as urothelial carcinoma of the bladder (BUC)
(24) and colorectal cancer
(25). It also has been
demonstrated that overexpression of miR-300 inhibited cell
proliferation, cell cycle and invasion in glioblastoma cell line
(9). In terms of OS, miR-300 have
been reported to be associated with progression of OS and might act
as predictor biomarker in the prognosis of OS (26). A previous study in MG63 cell line
indicated the pro-proliferation and pro-invasion roles of miR-300
(27). In the present study, the
discrepant functions of miR-300 were found in the same OS cell
line. Our data indicated that overexpression of miR-300 notably
suppressed cell viability and migration while enhanced apoptosis in
MG63 cells. Actually, many different types of miRNAs were
abnormally expressed and affected OS cells proliferation and
apoptosis in different degree. For instance, miR-125b has been
reported as oncogenic as well as anti-oncogenic, depending on the
tumor type, and both miR-300 and miR-125b were probably involved in
pathogenesis of OS (28). The
disparity of miR-300 effect on MG63 cells might be caused by the
complex function of miR-300, also particular environment tested
would lead to the alteration of other miRNAs expressions which was
related with miR-300 expression, and it might be another reason of
triggering the contradictory results regarding miR-300 in the MG63
cells. Therefore, further studies on the exact role of miR-300 are
urgently needed.
The Twist1 transcription factor was known to promote
tumor metastasis, and induce epithelial-mesenchymal transition
(EMT) which is the important evidence of degree for malignancy
increased (11). Twist1 was also
related to the formation of cancer halting terminal
differentiation, inhibiting apoptosis, and interfering with the p53
tumor-suppressor pathway (29).
Besides, Twist1 has been confirmed as a target gene of miR-106b,
miR-720 and miR-33b, which are involved in cancer cell modulation
(30–32). However, our data suggested that
Twist1 might also be a direct target of miR-300. Additionally, we
found that Twist1 was negatively regulated by miR-300 in MG63
cells.
Previous studies reported that Twist1 could decrease
OS cell survival against cisplatin by inhibiting multiple signaling
pathways, such as EMT, phosphatidylinositol-3 kinase/protein kinase
B (PI3K/AKT), NF-κB and signal transducer and activator of
transcription (STAT3) pathways, all of them formed a complex and
multichannel system (33–35). In addition, NF-κB is a major
signaling pathway nearly ubiquitous responsible for mediating DNA
transcription and cell function (36,37).
It has been detected that Twist1 regulated EMT via the NF-κB in
many cell types (38,39). Activation of NF-κB also plays a
major role in inhibiting apoptosis by causing up-regulation of key
anti-apoptotic proteins such as Bcl-3, B cell lymphoma/lewkmia-xl
(Bcl-xl), X-linked inhibitor of apoptosis protein (XIAP) and
survivin (40,41). To further explore the deeply
associated mechanism of miR-300 in MG63 cells, expression of
regulated proteins in NF-κB pathway were measured. We found that
the expression level of p-IκBα, p-p65 and Bcl-3 were decreased
significantly in miR-300 mimic transfected cells, whereas Twist1
reversed these decreases. Among these proteins, p65 is one part of
the NF-κB dimer, IκBα acts as inhibitory factor of NF-κB and Bcl-3
contributes to the regulation of transcriptional activation of
NF-κB target genes. Taken together, these data clearly indicated
that miRNA-300 could suppress activation of the NF-κB signaling
pathway by inhibiting Twist1.
Our results demonstrated that overexpression miR-300
suppressed cell viability and migration, while promoted apoptosis
in MG63 cells. These processes might be via downregulation of
Twist1, and ultimately resulting in inactivation of the NF-κB
pathway. MiRNA-300 might be a potential target for the specific
gene treatment of OS. Based on the results as outlined above,
further investigation is warranted to investigate the mechanisms of
miR-300′s effect on OS in more types of signal pathways and OS cell
lines, and more in vivo studies are also needed in the
future.
References
|
1
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng L: Progress in the treatment of
osteosarcoma. Int J Orthop. 29:91–93. 2008.
|
|
3
|
Qiu Z and Liao Q: Progress of osteosarcoma
therapy. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 24:1469–1475.
2010.(In Chinese). PubMed/NCBI
|
|
4
|
Jones-Rhoades MW and Bartel DP:
Computational identification of plant micrornas and their targets,
including a stress-induced miRNA. Mol Cell. 14:787–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zalfa F, Giorgi M, Primerano B, Moro A, Di
Penta A, Reis S, Oostra B and Bagni C: The fragile X syndrome
protein FMRP associates with BC1 RNA and Regulates the translation
of specific mRNAat synapses. Cell. 112:317–327. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Liu L, Xu Q, Wu P, Zuo X and Ji A:
MicroRNA as a novel drug target for cancer therapy. Expert Opin
Biol Ther. 12:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou F, Li Y, Hao Z, Liu X, Chen L, Cao Y,
Liang Z, Yuan F, Liu J, Wang J, et al: MicroRNA-300 inhibited
glioblastoma progression through ROCK1. Oncotarget. 7:36529–36538.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nairismägi ML, Füchtbauer A, Labouriau R,
Bramsen JB and Füchtbauer EM: The proto-oncogene TWIST1 is
regulated by microRNAs. PLos One. 8:e660702013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sánchez-Tilló E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cho WH, Lee HJ, Choi YJ, Oh JH, Kim HS and
Cho HS: Capsaicin induces apoptosis in MG63 human osteosarcoma
cells via the caspase cascade and the antioxidant enzyme system.
Mol Med Rep. 8:1655–1662. 2013.PubMed/NCBI
|
|
13
|
Zhang N, Su Y and Xu L: Targeting PKCε by
miR-143 regulates cell apoptosis in lung cancer. FEBS Lett.
587:3661–3667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi W, Bruce J, Lee M, Yue S, Rowe M,
Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW and Liu FF: MiR-449a
promotes breast cancer progression by targeting CRIP2. Oncotarget.
7:18906–18918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rojas RE, Balaji KN, Subramanian A and
Boom WH: Regulation of human CD4(+) alphabeta
T-cell-receptor-positive (TCR(+)) and gammadelta TCR(+) T-cell
responses to Mycobacterium tuberculosis by interleukin-10 and
transforming growth factor beta. Infect Immun. 67:6461–6472.
1999.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shin SS, Keshavjee S, Gelmanova IY, Atwood
S, Franke MF, Mishustin SP, Strelis AK, Andreev YG, Pasechnikov AD,
Barnashov A, et al: Development of extensively drug-resistant
tuberculosis during multidrug-resistant tuberculosis treatment. Am
J Respir Crit Care Med. 182:426–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang GC, He QY, Tong DK, Wang CF, Liu K,
Ding C, Ji F and Zhang H: MiR-367 negatively regulates apoptosis
induced by adriamycin in osteosarcoma cells by targeting KLF4. J
Bone Oncol. 5:51–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin BC, Huang D, Yu CQ, Mou Y, Liu YH,
Zhang DW and Shi FJ: MicroRNA-184 modulates doxorubicin resistance
in osteosarcoma cells by targeting BCL2L1. Med Sci Monit.
22:1761–1765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Geng S, Gu L, Ju F, Zhang H, Wang Y, Tang
H, Bi Z and Yang C: MicroRNA-224 promotes the sensitivity of
osteosarcoma cells to cisplatin by targeting Rac1. J Cell Mol Med.
20:1611–1619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu F, Gao J, Zhang Z, Ge J, Wei Z and
Cheng W: Expression of miRNA-300 in urothelial carcinoma of the
bladder. J Medical Res. 2012.
|
|
25
|
Lin W: A preliminary study of miRNA-300
regulation in invasion and proliferation of colorectal cancer
(unpublished PhD thesis). Third Military Medical University.
2015.(In Chinese).
|
|
26
|
Karbasy SH, Taheriazam A, Mirghasemi A,
Sedaghati F, Shakeri M, Yahaghi E and Bahador R: RETRACTED ARTICLE:
Upregulation of miR-300 and downregulation of miR-125b act as
potential predictor biomarkers in progression, metastasis, and poor
prognosis of osteosarcoma. Tumor Biol. 2015.(Epub ahead of
print).
|
|
27
|
Xue Z, Zhao J, Niu L, An G, Guo Y and Ni
L: Up-regulation of MiR-300 promotes proliferation and invasion of
osteosarcoma by targeting BRD7. PLoS One. 10:e01276822015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banzhaf-Strathmann J and Edbauer D: Good
guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell
Commun Signal. 12:302014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maestro R, Dei Tos AP, Hamamori Y,
Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH and
Hannon GJ: Twist is a potential oncogene that inhibits apoptosis.
Genes Dev. 13:2207–2217. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou
X, Zhang ZJ, Peng YH, Yang YZ and Yun JP: miR-720 inhibits tumor
invasion and migration in breast cancer by targeting TWIST1.
Carcinogenesis. 35:469–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nairismägi ML, Füchtbauer A, Labouriau R,
Bramsen JB and Füchtbauer EM: The protooncogene TWIST1 is regulated
by microRNAs. PLoS One. 8:e660702013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong P, Kaneuchi M, Watari H, Sudo S and
Sakuragi N: MicroRNA-106b modulates epithelial-mesenchymal
transition by targeting TWIST1 in invasive endometrial cancer cell
lines. Mol Carcinog. 53:349–359. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Y, Zang X, Huang Z and Zhang C: TWIST
interacts with endothelin-1/endothelin A receptor signaling in
osteosarcoma cell survival against cisplatin. Oncol Lett.
5:857–861. 2013.PubMed/NCBI
|
|
34
|
Wu J, Liao Q, He H, Da Z and Ke Y: TWIST
interacts with β-catenin signaling on osteosarcoma cell survival
against cisplatin. Mol Carcinog. 53:440–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu G, Shen J, Yu A, Wang H, Man TK and Lau
CC: Abstract 3402: Knockdown of TWIST1 increases chemosensitivity
of osteosarcoma cells. Cancer Res. 70:34022010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wullaert A, Verstrepen L, Van Huffel S,
Adib-Conquy M, Cornelis S, Kreike M, Haegman M, El Bakkouri K,
Sanders M, Verhelst K, et al: LIND/ABIN-3 is a novel
lipopolysaccharide-inducible inhibitor of NF-kappaB activation. J
Biol Chem. 282:81–90. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Y, Li Z, Wang W, Zhang H, Chen J, Su
P, Liu L and Li W: Naringin protects against cartilage destruction
in osteoarthritis through repression of NF-κB signaling pathway.
Inflammation. 39:385–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horikawa T, Yang J, Kondo S, Yoshizaki T,
Joab I, Furukawa M and Pagano JS: Twist and epithelial-mesenchymal
transition are induced by the EBV oncoprotein latent membrane
protein 1 and are associated with metastatic nasopharyngeal
carcinoma. Cancer Res. 67:1970–1983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lv N, Shan Z, Gao Y, Guan H, Fan C, Wang H
and Teng W: Twist1 regulates the epithelial-mesenchymal transition
via the NF-κB pathway in papillary thyroid carcinoma. Endocrine.
51:469–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sethi G, Ahn KS and Aggarwal BB: Targeting
nuclear factor-kappa B activation pathway by thymoquinone: Role in
suppression of antiapoptotic gene products and enhancement of
apoptosis. Mol Cancer Res. 6:1059–1070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brandwood CP, Hoyland JA, Hillarby MC,
Berry JL, Davies M, Selby PL and Mee AP: Apoptotic gene expression
in Paget's disease: A possible role for Bcl-2. J Pathol.
201:504–512. 2003. View Article : Google Scholar : PubMed/NCBI
|