Introduction
Chronic kidney disease (CKD) has been increasingly
recognized as a major public health problem in the world (1). Therefore, improvement in the current
knowledge of molecular alterations associated with CKD is required
to investigate novel strategies of diagnostics and treatment of
this disease. The progression of CKD is characterized by glomerular
hypertrophy, mesangial cell (MC) proliferation, extracellular
matrix (ECM) accumulation, glomerulosclerosis and ultimately
end-stage kidney disease (2). The
ECM produced by damaged MCs is a major factor in mesangial
proliferation and this is observed in light-chain-related
glomerular disease which is associated with an increased synthesis
of tenascin by MCs (3). MC
proliferation is reported in humans and in experimental animal
models with chronic nephron loss, and precedes the development of
secondary focal glomerulosclerosis (4–6). In
addition, progressive glomerulosclerosis is a common route for the
development of end-stage renal failure of any etiology. Therefore
MCs serve a critical role in the maintenance of renal function,
supporting the glomerular capillaries and regulating their blood
flow (7). Various growth factors
and cytokines, produced by the infiltrating cells during the
disease process and by the local kidney cells, have been implicated
in the fibrotic process (8). Among
these, aldosterone (ALD) produced by the adrenal cortex and the
MCs, in addition to other extra-adrenal tissues, including cardiac
myocytes and vascular smooth muscle cells, serves a significant
role in the pathogenesis of mesangial matrix expansion (9–19).
Part of the intracellular mechanisms involved in the proliferative
and fibrotic effect of ALD in CKD has been reported. For example,
ALD upregulates protein synthesis, and mRNA expression of
fibronectin and transforming growth factor-β1 (TGF-β1) in cultured
rat MCs partly by enhancing extracellular signal-regulated kinase
1/2 (ERK1/2) and c-Jun N-terminal kinase (JNK) activities, and
subsequent activity of transcription factor AP-1 (AP-1) (20,21).
ALD stimulates intracellular adhesion molecule-1 and connective
tissue growth factor transcription via activation of
serine/threonine-protein kinase Sgk1 and nuclear transcription
factor p65, which may be involved in the progression of ALD-induced
mesangial fibrosis and inflammation (22). ALD stimulates the mitogen-activated
protein kinase pathway, which promotes the proliferation of MCs
(23). ALD can also increase
plasminogen activator inhibitor-1 mRNA and protein expression in
cultured MCs (24). Although these
signaling pathways have been identified to serve important roles
during the pathogenesis of CKD, there remain few effective
therapeutic treatments to cure CKD by targeting these molecules.
Therefore, it is necessary for novel molecular potential targets to
be investigated.
Long non-coding RNA (lncRNA) has received attention
in the investigation of the complex mechanisms underlying malignant
processes, including tumorigenesis, drug-resistance and metastasis
of different types of cancer (25). In the beginning, the majority of
transcriptional outputs of the mammalian genome were confirmed to
be protein noncoding genes (26)
and the lncRNAs were identified as transcriptional ‘noise’ or
cloning artifacts (27). During
the past decade, multiple lncRNAs have been demonstrated and
confirmed to be involved in the regulation of gene transcription,
chromatin methylation, post-transcriptional modification and other
biological progresses (28).
However, the systematic analysis of aberrant expression profiles of
lncRNA in MCs treated with ALD remains to be performed. In the
present study, the expression pattern of lncRNAs was investigated
by high-throughput microarray in MCs treated with ALD. Furthermore,
to couple the observed differential expression of lncRNA with the
expression of mRNAs, an mRNA transcriptome analysis by microarray
was conducted. The aim of the present study was to clarify the
roles of differentially expressed lncRNAs in MCs treated with ALD
and provide a novel insight into CKD pathogenesis, and to identify
potential biomarkers and therapeutic targets for CKD.
Materials and methods
Cell culture
Cultured rat MCs were purchased from the China
Center for Type Culture Collection (Wuhan, China). MCs from
passages 7–9 were used in the experiments. The cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal calf serum (FCS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a humid atmosphere containing 5%
CO2 at 37°C. MCs were grown to 75% confluence in 100
cm2 flasks (BD Biosciences, Franklin Lakes, NJ, USA)
were incubated in serum-free medium for 24 h and then treated with
ALD (R&D Systems, Inc., Minneapolis, MN, USA) at a
concentration of 10−6 M in a 96-well plate for 24 h at
37°C.
Preparation of model
Performed as previously described by Zhang et
al (29), with 10−6
M ALD-treated MCs being used as the experimental cells. An equal
amount of solvent (DMEM+10% FCS) was added to the solvent control
group. Cells were cultured in 6-well plates for 24 h. MCs were
washed with PBS three times, then 200 µl EDTA (Gibco; Thermo Fisher
Scientific, Inc.) was added to each well followed by 750 µl
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The cells were stored at −80°C and sent to the LncRNA
Expression Microarray (Arraystar, Rockville, MD, USA).
lncRNA microarray analysis
The lncRNAs were constructed using public
transcriptome databases (Refseq, www.ncbi.nlm.nih.gov/refseq; University of California
Santa Cruz knowngenes, genome.ucsc.edu; and Gencode, www.gencodegenes.org), in addition to a publication
(30). The lncRNA expression
microarray used in the present study classifies its probes as the
following subtypes: i) Enhancer lncRNAs: Contains profiling data of
all lncRNAs with enhancer-like function; ii) Rinn lncRNAs: Contains
profiling data of all lncRNAs based on studies by Khalil et
al (31) and Guttman et
al (32); iii) homeobox
protein (HOX) cluster: Contains profiling data of all probes in the
four HOX loci, targeting 407 discrete transcribed regions, lncRNAs
and coding transcripts; iv) lncRNAs located near coding genes:
These contain differentially expressed lncRNAs and nearby coding
gene pairs (distance, 300 kb); and v) enhancer lncRNAs located near
coding genes: These contain differentially expressed enhancer-like
lncRNAs and their nearby coding genes (distance, 300 kb). Following
hybridization and washing, processed slides were scanned with an
Agilent DNA Microarray Scanner (part no. G2505B; Agilent
Technologies, Inc., Santa Clara, CA, USA). Agilent Feature
Extraction software version 10.7.3.1 (Agilent Technologies, Inc.)
was used to analyze all acquired array images. Quantile
normalization and subsequent data processing were performed using
the GeneSpring GX version 11.5.1 software package (Agilent
Technologies, Inc.) (33). The
profile of microarray data of the 8,459 lncRNAs was detected by
third-generation lncRNA microarray. The general characteristics of
the differentially-expressed lncRNAs were summarized, including
chromosomal, source, relationship and fold-change distribution
using the most widely-used public transcriptome databases (Ensembl,
www.ensembl.org/index.html; UCSC,
genome.ucsc.edu/index.html; NONCODE,
www.noncode.org; NCBI, www.ncbi.nlm.nih.gov). The source of the lncRNA was
collected from RefSeq_NR (RefSeq validated non-coding RNA),
RefSeq_XR (RefSeq un-validated non-coding RNA), mouse_ortholog (rat
lncRNAs which are obtained by sequence comparison with mouse
lncRNAs), ‘ultra-conserved regions’ among human, mouse and rat
(users.soe.ucsc.edu/~jill/ultra.html), and misc_lncRNA
(other sources).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was used to verify the differential
expression of seven lncRNAs and six associated genes that were
detected by the lncRNA and mRNA expression microarray. The cDNA was
synthesized using reverse transcriptase (Takara Bio, Inc., Otsu,
Japan) and oligo (dTs) primers with 1 µl RNA from the same samples
as those used in the microarray. The reaction consisted of 2 µl 5X
PrimeScript buffer, 0.5 µl PrimeScript RT Enzyme Mix I, 0.5 µl
Oligo dT Primer, 0.5 µl Random 6 mers, 500 ng total RNA and RNase
free dH2O up to 10 µl, and was performed for 15 min at
37°C. Primers for each lncRNA and mRNA are reported in Table I. qPCR was performed on an Applied
Biosystems ViiA™ 7 Dx (Thermo Fisher Scientific, Inc.) using the
SYBR-Green method, according to the manufacturer's protocol. Each
RT-qPCR reaction (in 10 µl) contained 5 µl SYBR−Green
real-time PCR Master mix (Thermo Fisher Scientific, Inc.), 1.0 mM
primer and 1 µl template cDNA. The cycling conditions consisted of
an initial single cycle of 2 min at 50°C; 2 min at 95°C; followed
by 40 cycles of 15 sec at 95°C, 15 sec at 56°C and 60 sec at 72°C.
PCR amplifications were performed in triplicate for each sample.
Gene expression levels were quantified relative to the expression
of GAPDH (primers: Sense, 5′-CAAGTTCAACGGCACAGTCAA-3′; antisense,
5′-TGGTGAAGACGCCAGTAGACTC-3′) using an optimized comparative
2−ΔΔCq method (34).
RT-qPCR was performed in triplicate on the diluted cDNA and the
experiments were performed twice in the control and ALD-treated MC
groups.
 | Table I.Primers for quantitative polymerase
chain reaction of long non-coding RNA and mRNAs. |
Table I.
Primers for quantitative polymerase
chain reaction of long non-coding RNA and mRNAs.
|
| Primers
(5′-3′) |
|---|
|
|
|
|---|
| Genes | Forward | Reverse |
|---|
| BC168211 |
CACCTGGCCACTGTTTCCTAA |
TGATACTCGGCTAGGGAAGCA |
| BC088254 |
CCCAGAAAGCTCTCAGGTCCTA |
TGCTGGGTGCTTTATTTACACAA |
| AF336872 |
TGGCCAGGAGTGCCATTC |
CCCCCCAATGCCATGA |
| AY325162 |
CCCATGTCCCTCATTCATTACC |
GGTGACACGAAGCATCCAAGT |
| BC168687 |
CATTGCTCCTGTCTTAGGTCGTT |
GGTGGCGATAGGGTTAATTTCC |
| AF230638 |
TCCTTTTGCAAGAATCCATACTCA |
CGGTGCTAACGGTGAATCAGA |
| BC167085 |
TGGAGGCCGCCAAGTGT |
GAATCCCACCGGGTCACA |
| NM_001108598 |
CACCTGGCCACTGTTTCCTAA |
TGATACTCGGCTAGGGAAGCA |
| NM_001109190 |
CCAGGCTATGAACGGTTTCC |
AGTAGGGTCTGTTTGCATCCTTAGG |
| NM_001101018 |
TCACCAAGACCCAGTTCAGTTAGA |
GAAGGCCGTGCCAATGAG |
| NM_019347 |
ACACACCTGTTGGCACTTGTCT |
CGGTGGCACACCAACCA |
| NM_177962 |
CCTTGCACCTGTTTCAAATCAA |
GGGCAGAGGGAACGAATCA |
| NM_001108823 |
AACCCTCAGGAGCCATGCT |
TGGGCACTGCAGGTGAGA |
Gene ontology (GO) analysis and
pathway analysis
Previous studies have demonstrated that lncRNAs are
preferentially located next to genes with developmental functions
(26). For each lncRNA locus the
nearest protein-coding neighbor within 100 kb was identified. For
antisense overlapping and intronic overlapping lncRNAs, the
overlapping gene was identified. GO (http://www.geneontology.org) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analyses (http://www.genome.jp/kegg) were applied to determine
the roles of these closest coding genes in GO terms or biological
pathways.
GO analysis was used to assess the main function of
the closest coding genes according to the GO database, which
provides the key functional classifications for the National Center
for Biotechnology Information (35). The ontology covers two domains:
Biological processes and molecular function. Fisher's exact test is
used to identify if there is more overlap between the
differentially expressed list and the GO annotation list than would
be expected by chance. The P-value denotes the significance of GO
term enrichment in the differentially expressed genes. Pathway
analysis is a functional analysis mapping genes to the KEGG
pathways. The P-value (expression analysis systematic
explorer-score, Fisher-P-value or Hypergeometric-P-value) denotes
the significance of the pathway correlated with the conditions.
P<0.05 was considered to indicate a statistically significant
difference.
Statistical analysis
Each qPCR experiment was performed at least three
times. Numerical data were presented as the mean ± standard error
of the mean. Relative expression levels of lncRNAs between the two
groups were analyzed using the Student's t-test. All statistical
analyses were performed using SPSS software (version 18; SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Profile of lncRNA microarray data
A gene chip study was performed in the normal and
ALD-treated MC group to investigate the possible lncRNA alteration
in expression using the Arraystar probe dataset, which included
8,459 lncRNAs. The lncRNAs were constructed using public
transcriptome databases (Refseq, University of California Santa
Cruz knowngenes and Gencode), in addition to a publication
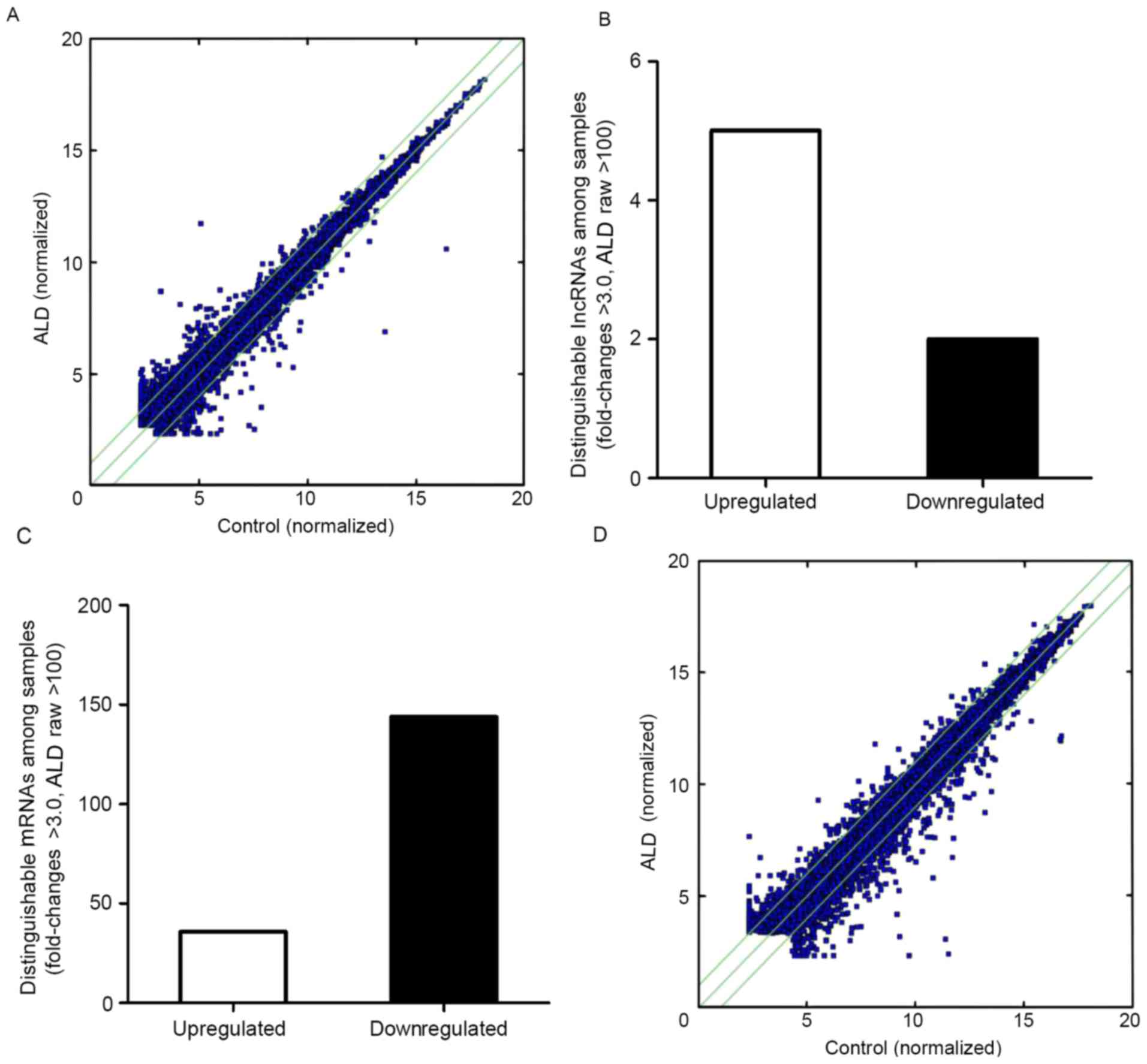
(30). The scatterplot is useful
for assessing the variation in the expression of lncRNAs and coding
transcripts between the two MCs (Fig.
1A); the dot above the green line represents a difference of
>2 times. The number of points above the top and below the
bottom green lines indicated lncRNAs that exhibit >2.0
fold-change when comparing the control and ALD-treated MC groups.
By setting a filter of fold-change >3.0, raw >100 of the
expression level between the ALD-treated group and control group, 5
upregulated and 2 downregulated lncRNAs were identified (Fig. 1B).
Differently expressed mRNAs in the
ALD-treated rat MC group
An Affymetrics gene array containing 13,214 gene
transcripts was used to perform a comprehensive analysis of mRNA
expression in control compared with ALD-treated rat MCs group.
Comparing the rat MCs treated with ALD with the control group, 180
genes were differentially expressed (fold-change >3.0; raw
>100), of which 36 genes were upregulated and 144 genes were
downregulated (Fig. 1C). A
scatter-plot illustrating the expression patterns of these
differentially expressed mRNAs between control and ALD-treated rat
MCs is exhibited in Fig. 1D.
RT-qPCR analysis of microarray
hybridization
To quantify the microarray hybridization results,
RT-qPCR was performed on five upregulated lncRNAs (BC168211,
BC088254, AF336872, AY325162 and BC168687), two downregulated
lncRNAs (AF230638 and BC167085) (Fig.
2A), three upregulated mRNAs (NM_001108598, NM_001109190 and
NM_001101018) (Fig. 2B) and three
downregulated mRNAs (NM_019347, NM_177962 and NM_001108823)
(Fig. 2C), selected on the basis
of their levels of expression on the microarray and their
biological significance. The RT-qPCR data was demonstrated to be
consistent with the microarray results, with BC168211, BC088254,
AF336872, AY325162 and BC168687 being upregulated and AF230638,
BC167085 being downregulated compared with the control. The RT-qPCR
data were again consistent with the microarray results for the
mRNAs with NM_001108598, NM_001109190 and NM_001101018 being
upregulated and NM_019347, NM_177962 and NM_001108823 being
downregulated (P<0.05 vs. the control group).
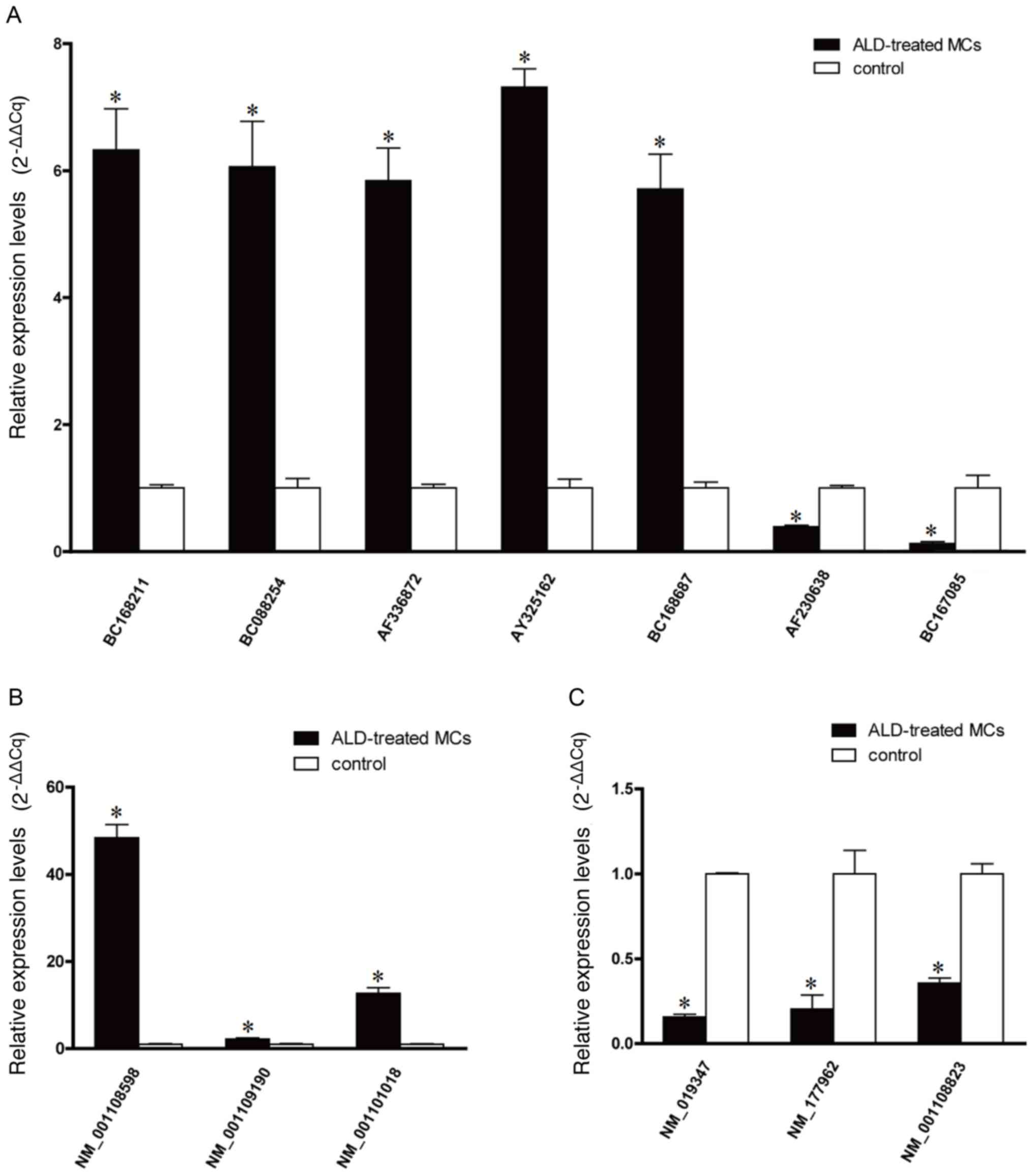 | Figure 2.Reverse transcription-quantitative
polymerase chain reaction quantification of microarray
hybridization. (A) The relative expression level of lncRNAs
BC168211, BC088254, AF336872, AY325162, BC168687, AF230638 and
BC167085. (B) The relative expression levels of upregulated mRNAs,
NM_001108598, NM_001109190 and NM_001101018, and (C) downregulated
mRNAs, NM_019347, NM_177,962 and NM_0,011,08823. *P<0.05 vs. the
control group. ALD, aldosterone; lncRNAs, long non-coding RNAs;
MCs, mesangial cells. |
Expression signatures of
differentially expressed lncRNAs
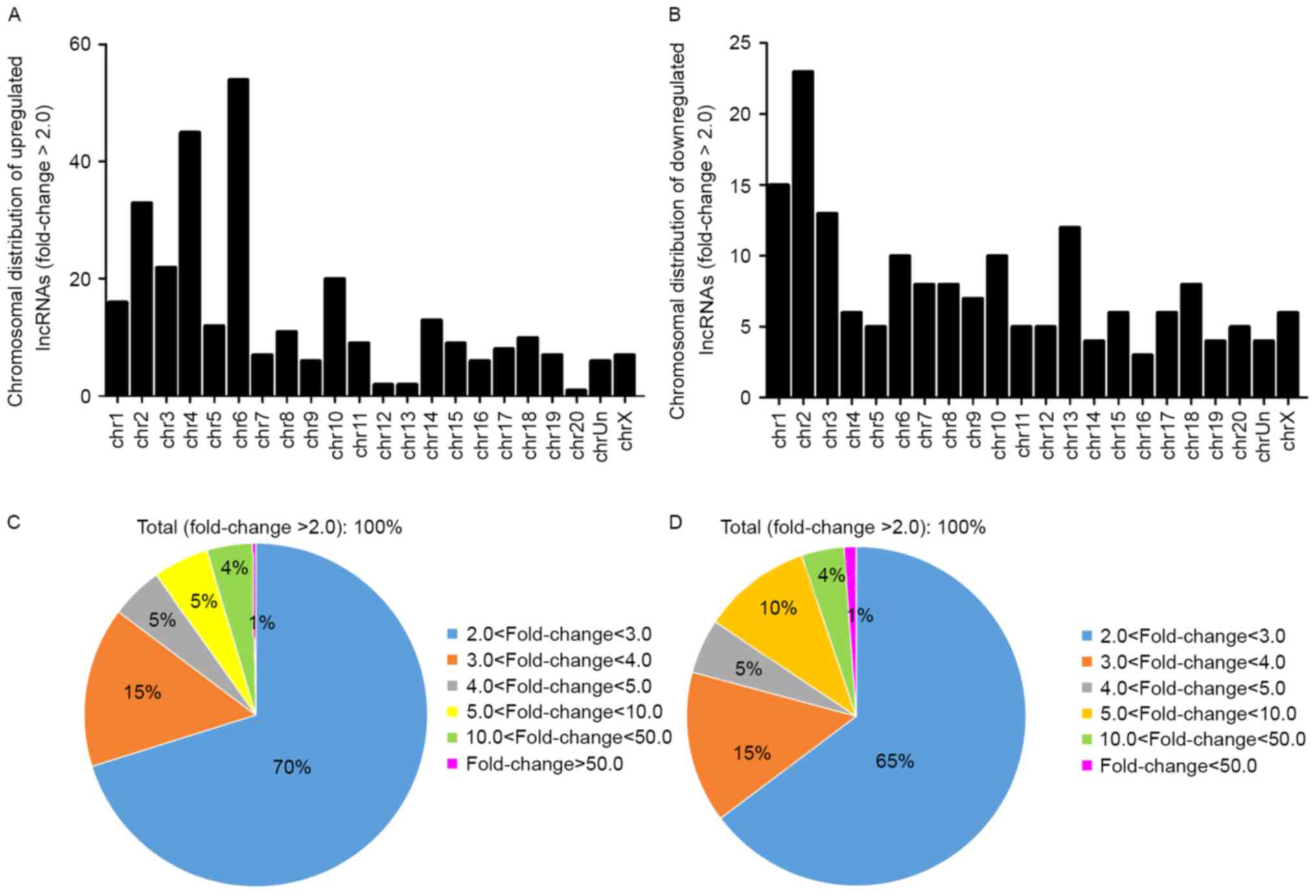
The general characteristics of the differentially
expressed lncRNAs were summarized, including chromosomal, source,
relationship and fold-change distribution. Chromosomal distribution
of the number of up- or downregulated lncRNAs located on different
chromosomes was demonstrated (Fig. 3A
and B). Fold-change distribution demonstrated the differential
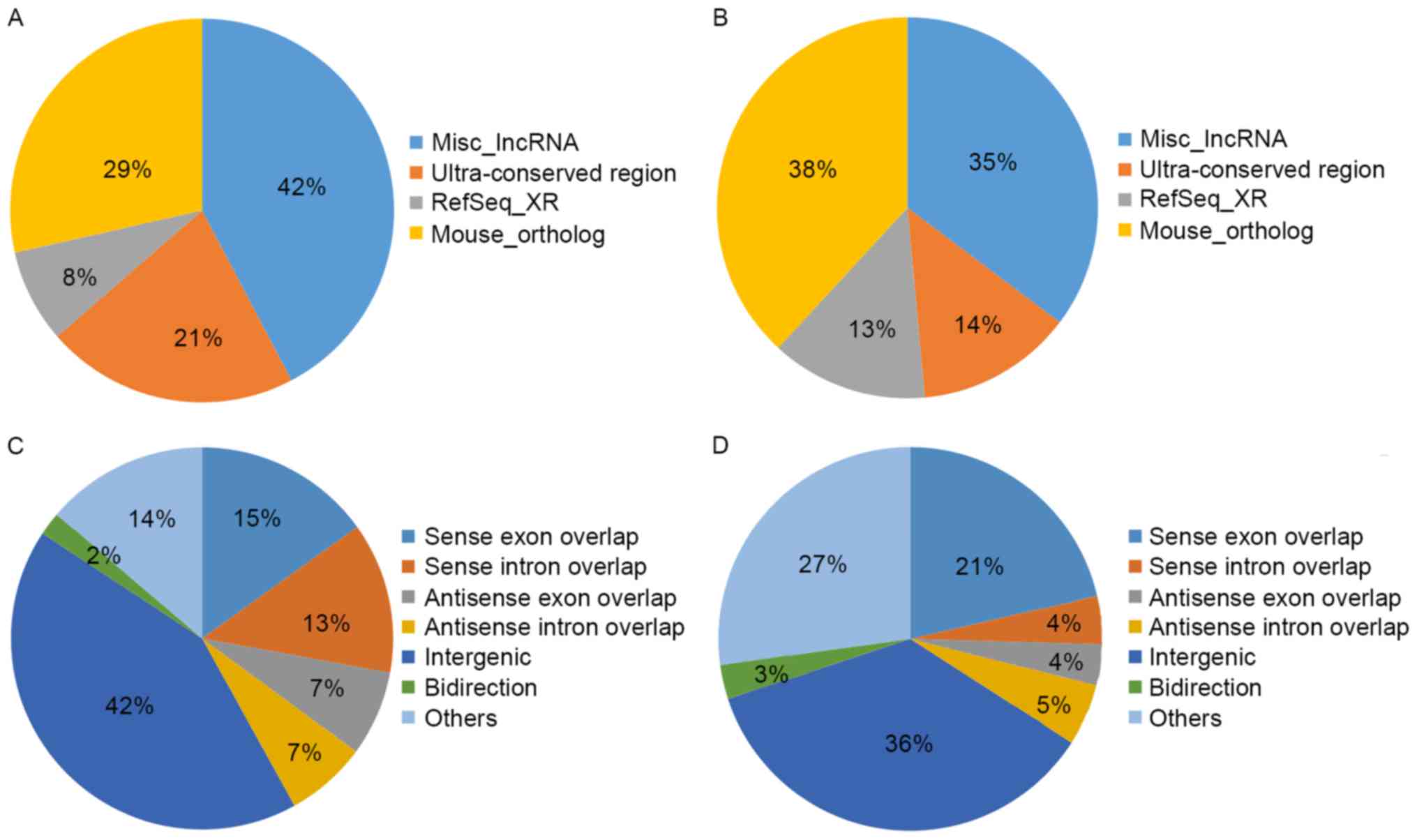
expression of up- and downregulated lncRNAs (Fig. 3C and D), respectively. Source
distribution respectively demonstrated the percentages of up- and
downregulated lncRNAs collected from different sources (Fig. 4A and B), including misc_lncRNA,
ultra-conserved region, Refseq-XR and mouse_ortholog. Relationship
distribution demonstrated the association of up- and downregulated
lncRNAs (Fig. 4C and D),
respectively.
GO and pathway analysis
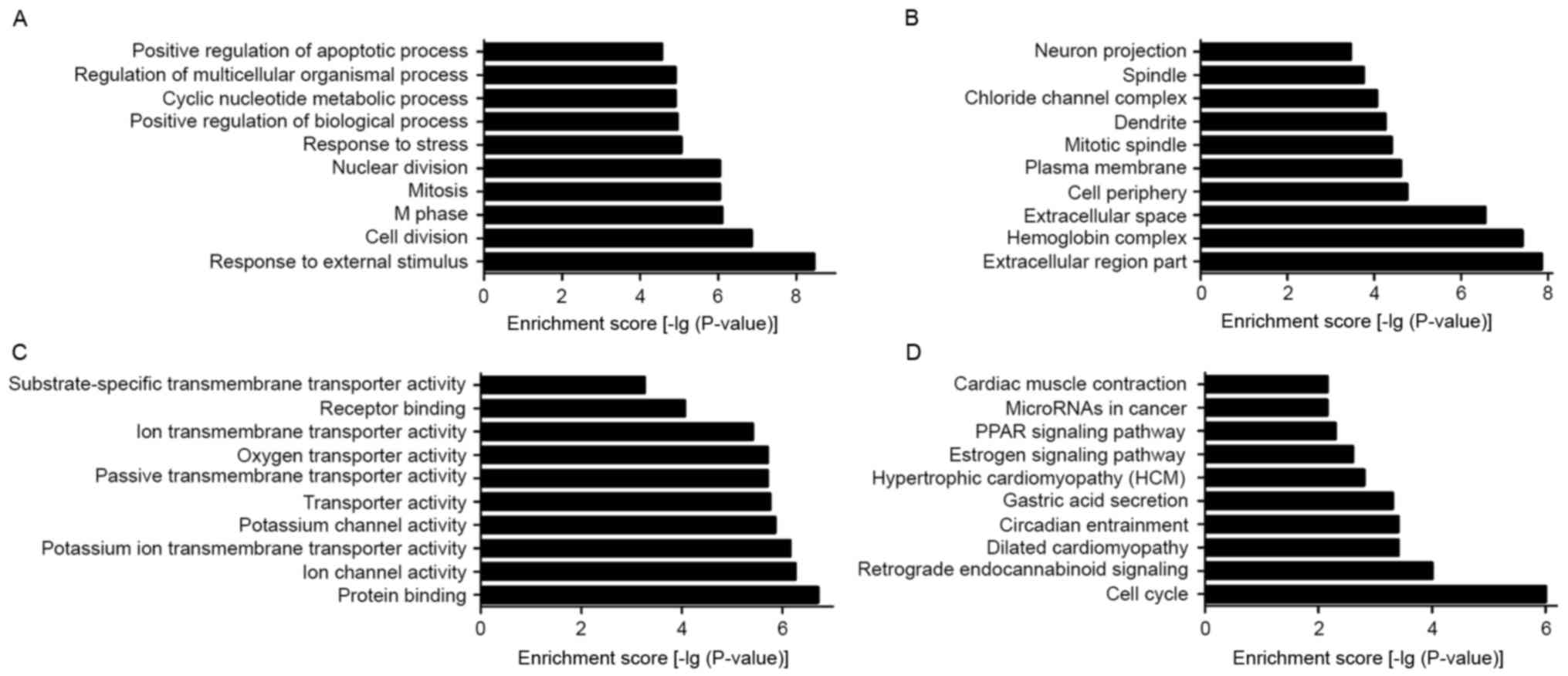
To elucidate the biological processes and functional
classification of differentially expressed lncRNAs, GO and pathway
analyses were performed. The functions of coding genes adjacent to
dysregulated lncRNAs included (in size order, most prevalent first,
the top ten): i) Response to external stimuli; ii) response to
stress; iii) positive regulation of biological processes; iv)
cyclic nucleotide metabolic process; v) regulation of multicellular
organism processes; vi) positive regulation of the apoptotic
process; vii) cell division; viii) M phase; ix) mitosis; and x)
nuclear division (Fig. 5A). The
cellular component containing dysregulated lncRNAs included the
following (in size order, most prevalent first, the top ten): i)
Cell periphery; ii) dendrite; iii) chloride channel complex; iv)
neuron projection; v) extracellular region part; vi) hemoglobin
complex; vii) extracellular space; viii) plasma membrane; ix)
mitotic spindle; and x) spindle (Fig.
5B). The molecular function of dysregulated lncRNAs mainly
consisted of the following (in size order, most prevalent first,
the top ten): i) Ion channel activity; ii) potassium ion
transmembrane transporter activity; iii) potassium channel
activity; iv) transporter activity; v) passive transmembrane
transporter activity; vi) ion transmembrane transporter activity;
vii) protein binding; viii) oxygen transporter activity; ix)
receptor binding; and x) substrate-specific transmembrane
transporter activity (Fig. 5C).
Pathway analysis is a functional analysis process that maps genes
to KEGG pathways. In the present study, the top 10 pathways that
were associated with coding genes of dysregulated lncRNAs involved:
i) Estrogen signaling pathway; ii) peroxisome
proliferator-activated receptor (PPAR) signaling pathway; iii) cell
cycle; iv) retrograde endocannabinoid signaling; v) circadian
entrainment; vi) hypertrophic cardiomyopathy; vii) gastric acid
secretion; viii) microRNAs in cancer; ix) cardiac muscle
contraction; and x) dilated cardiomyopathy-all Rattus
norvegicus (Fig. 5D).
Bioinformatic analysis
As the transcription of non-coding genes can affect
the expression of their flanking coding genes, relatively lowly and
highly expressed lncRNAs were selected with a < and
>3-fold-change, respectively, as well as an raw >100 in the
MCs treated with ALD and control group, and an associated coding
gene with a function in a developmental processes, including cell
cycle and PPAR signaling. It was demonstrated that dual specificity
phosphatase 15 (Dusp15) and acyl-coenzyme A thioesterase Them4
(Them4) were associated with the lncRNAs BC168211 and BC168687,
respectively (data not shown).
Discussion
Currently available therapies are not efficacious in
the treatment of CKD, suggesting that further understanding of the
molecular mechanisms underlying the pathogenesis of CKD is required
for the identification of more effective diagnostic markers and
therapeutic targets. MCs have been demonstrated to be a target of
local ALD action, which may serve an important role in glomerular
damage in CKD (36). In addition,
ALD has been proved to serve a significant role in modulating MC
function (11,23,37).
The present study, to the best of the authors'
knowledge, is the first to report the differential lncRNA
expression in MCs treated with ALD compared with normal MCs. A
threshold of >3.0 fold-change and raw >100 was set and it was
demonstrated that 5 lncRNAs were upregulated and 2 were
downregulated in MCs treated with ALD compared with the non-ALD
treated MCs. The RT-qPCR results revealed that BC168211, BC088254,
BC168687, AF336872 and AY325162 were significantly upregulated in
MCs treated with ALD, and AF230638 and BC167085 were downregulated.
Furthermore, it was demonstrated that lncRNAs may act through
distinct transcription factors to modulate their target genes'
transcription, thereby being involved in the potential mechanism of
CKD. Then, mRNA microarray technology was used to evaluate
differences in the mRNA expression profiles of control and MCs
treated with ALD. GO and pathway analyses revealed that these
lncRNAs were associated with changes in key pathogenic processes of
CKD.
The collected data can be used to analyze the role
of lncRNA transcripts in ALD-induced CKD. The GO project provides a
controlled vocabulary that can be used to describe genes and gene
product attributes (38). The
biological processes involving dysregulated lncRNAs was
demonstrated to be associated with cell proliferation stimulated by
ALD included the following: Cell division; positive regulation of
biological processes; response to external stimuli; regulation of
multicellular organism processes; and cyclic nucleotide metabolic
processes. The biological processes associated with lncRNAs
included cell division, immune system processes, immune responses
and cell-cell signaling. Pathway analysis provides a method for
gaining insight into the underlying biology of differentially
expressed genes and proteins (16). Pathway analysis demonstrated that
the associated genes of dysregulated lncRNAs between the control
and MCs treated with ALD included a variety of pathways for example
cell cycle-associated, PPAR signaling and estrogen signaling
pathways. It has been reported that ALD serves a major part in the
glomerular ECM accumulation and proliferation of MCs in several
glomerular diseases, and produces renal fibrosis in rats (18–21,23,24).
The lncRNAs examined in the present study were demonstrated to be
involved in the progression of CKD, which is induced by cell
division, cell proliferation and immune deposits, in which ALD was
involved.
Due to the complexity of the transcriptome, lncRNAs
are frequently overlapping or are interspersed between multiple
coding and non-coding transcripts (39,40).
In the present study, two genes were identified to be associated
with lncRNAs through GO and pathway analysis. These results were
used to investigate the association between lncRNAs and genes,
further. Dusp15 and Them4 were demonstrated to be associated with
the lncRNAs BC168211 and BC168687, respectively.
Dusp15, a member of the protein tyrosine phosphatase
family, previously only reported to be expressed in the testes, was
suggested to be a pharmacological target for promoting
remyelination in multiple sclerosis (41). Dusp15 is expressed in the kidneys
of spontaneously hypertensive rats, which indicates that it is
associated with renal injury. Dusp15 was identified to serve a role
in the regulation of cell proliferation, positive regulation of the
JNK cascade and TGF-β receptor-signaling pathway. TGF-β has been
recognized to be an important factor in the development of CKD
(42). In rat MCs, ALD upregulates
mRNA expression of TGF-β partly by enhancing the ERK1/2, JNK and
AP-1 intracellular signaling pathways, and stimulating the
progression of renal disease (20,21,43–45).
TGF-β has also been demonstrated to induce mesangial expansion,
which is caused by MC hypertrophy, proliferation and eventually
apoptosis (46). In the present
study, an association was demonstrated between BC168211 and Dusp15.
The level of Dusp15 was revealed to be increased in stimulated MCs
compared with the control cells. This may indicate that Dusp15 is
involved in promoting the proliferation of MCs. However, the
intrinsic association between BC168211 and Dusp15 is not completely
understood. Further studies on this issue are planned.
Them4, a negative regulator of RAC-α
serine/threonine-protein kinase (Akt) and activated Akt, is known
to protect the cell from apoptosis. The amino-terminal domain of
Them4 may bind to Akt (47,48).
Human Them4 has also been linked to Akt regulation and apoptosis
(49). ALD stimulates MC
proliferation via the phosphoinositide 3-kinase (PI3K)/Akt
signaling pathway (50). The
PI3K-Akt signaling pathway regulates fundamental cellular functions
including transcription, translation, proliferation, growth and
survival. PI3K catalyzes the production of
phosphatidylinositol-3,4,5-triphosphate, which in turn serves as a
second messenger that helps to activate Akt. Once active, Akt can
control key cellular processes by phosphorylating substrates
involved in apoptosis, protein synthesis, metabolism and cell cycle
(50) (from KEGG source record:
rno04151). Thus far, it has been clear that Them4 is associated
with the cell cycle (49). In the
present study it was hypothesized that Them4 is involved in the
PI3K/Akt signaling pathway in the progression of mesangial
expansion. Them4 was demonstrated to be upregulated in ALD
stimulated MCs. However, the precise mechanism of Them4 with
BC168687 in ALD induced CKD warrants further investigation.
In conclusion, to the best of the author's
knowledge, the present study is the first to provide a profile of
lncRNAs in ALD-induced MCs in vitro. A network of
differentially expressed lncRNAs was constructed, and numerous
lncRNAs are involved in the development and mechanism of CKD.
Further investigation of the biological progresses and molecular
mechanisms of the dysregulated lncRNAs is necessary. The present
study may provide novel insights into the molecular basis of CKD,
and aid the identification of potential novel biomarkers and
development of therapeutic interventions for this disease.
Acknowledgements
The present study was sponsored by the Scientific
Research Program of Nanjing Medical University (grant no.
2015NJMUZD028) and the Natural Science Fund Project of Colleges in
Jiangsu Province, China (grant no. 15KJD320005).
References
|
1
|
Zhang L, Zhang P, Wang F, Zuo L, Zhou Y,
Shi Y, Li G, Jiao S, Liu Z, Liang W and Wang H: Prevalence and
factors associated with CKD: A population study from Beijing. Am J
Kidney Dis. 51:373–384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeisberg M, Khurana M, Rao VH, Cosgrove D,
Rougier JP, Werner MC, Shield CF III, Werb Z and Kalluri R:
Stage-specific action of matrix metalloproteinases influences
progressive hereditary kidney disease. PLoS Med. 3:e1002006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jo YI, Cheng H, Wang S, Moeckel GW and
Harris RC: Puromycin induces reversible proteinuric injury in
transgenic mice expressing cyclooxygenase-2 in podocytes. Nep Exp
Nephrol. 107:e87–e94. 2007. View Article : Google Scholar
|
|
4
|
Pesce CM, Striker LJ, Peten E, Elliot SJ
and Striker GE: Glomerulosclerosis at both early and late stages is
associated with increased cell turnover in mice transgenic for
growth hormone. Lab Invest. 65:601–605. 1991.PubMed/NCBI
|
|
5
|
Kreisberg JI and Karnovsky MJ: Glomerular
cells in culture. Kidney Int. 23:439–447. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Floege J, Burns MW, Alpers CE, Yoshimura
A, Pritzl P, Gordon K, Seifert RA, Bowen-Pope DF, Couser WG and
Johnson RJ: Glomerular cell proliferation and PDGF expression
precede glomerulosclerosis in the remnant kidney model. Kidney Int.
41:297–309. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schlöndorff D and Banas B: The mesangial
cell revisited: No cell is an island. J Am Soc Nephrol.
20:1179–1187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eddy AA and Neilson EG: Chronic kidney
disease progression. J Am Soc Nephrol. 17:2964–2966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Siragy HM and Xue C: Local renal
aldosterone production induces inflammation and matrix formation in
kidneys of diabetic rats. Exp Physiol. 93:817–824. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu P, Liang X, Dai Y, Liu H, Zang Y, Guo
Z, Zhang R, Lai W, Zhang Y and Liu Y: Aldosterone biosynthesis in
extraadrenal tissues. Chin Med J (Engl). 112:414–418.
1999.PubMed/NCBI
|
|
11
|
Nishikawa T, Suematsu S, Saito J, Soyama
A, Ito H, Kino T and Chrousos G: Human renal mesangial cells
produce aldosterone in response to low-density lipoprotein (LDL). J
Steroid Biochem Mol Biol. 96:309–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martinez D, Oestreicher E, Roubsanthisuk
W, et al: Angiotensin II, aldosterone and the caveolae. IV.
Aldosterone content of the kidney after adrenalectomyproceedings of
the Journal of Hypertension. Lippincott Williams & Wilkins;
Philadelphia, PA: pp. 19106–3621. 2002
|
|
13
|
Silvestre JS, Heymes C, Oubénaïssa A,
Robert V, Aupetit-Faisant B, Carayon A, Swynghedauw B and Delcayre
C: Activation of cardiac aldosterone production in rat myocardial
infarction: effect of angiotensin II receptor blockade and role in
cardiac fibrosis. Circulation. 99:2694–2701. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Silvestre JS, Robert V, Heymes C,
Aupetit-Faisant B, Mouas C, Moalic JM, Swynghedauw B and Delcayre
C: Myocardial production of aldosterone and corticosterone in the
rat. Physiological regulation. J Biol Chem. 273:4883–4891. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kayes-Wandover KM and White PC:
Steroidogenic enzyme gene expression in the human heart. J Clin
Endocrinol Metab. 85:2519–2525. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takeda Y, Yoneda T, Demura M, Miyamori I
and Mabuchi H: Sodium-induced cardiac aldosterone synthesis causes
cardiac hypertrophy. Endocrinology. 141:1901–1904. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kornel L: Colocalization of
11beta-hydroxysteroid dehydrogenase and mineralocorticoid receptors
in cultured vascular smooth muscle cells. Am J Hypertens.
7:100–103. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai LY, Gu Y, Chen J, Yu SQ, Ma J, Yang HC
and Lin SY: Production of aldosterone by rat mesangial cell and the
accumulation of extracellular matrix induced by aldosterone.
Zhonghua Yi Xue Za Zhi. 83:1900–1905. 2003.(In Chinese). PubMed/NCBI
|
|
19
|
Terada Y, Kuwana H, Kobayashi T, Okado T,
Suzuki N, Yoshimoto T, Hirata Y and Sasaki S:
Aldosterone-stimulated SGK1 activity mediates profibrotic signaling
in the mesangium. J Am Soc Nephrol. 19:298–309. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han JS, Choi BS, Yang CW and Kim YS:
Aldosterone-induced TGF-beta1 expression is regulated by
mitogen-activated protein kinases and activator protein-1 in
mesangial cells. J Korean Med Sci. 24 Suppl 1:S195–S203. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li XD, Chen XW, Tang DS, Liang D and Liu
HF: Effects of aldosterone on synthesis of fibronectin and
expression of transforming growth factor-beta1 mRNA in cultured rat
mesangial cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23:140–1422.
2007.(In Chinese). PubMed/NCBI
|
|
22
|
Terada Y, Ueda S, Hamada K, Shimamura Y,
Ogata K, Inoue K, Taniguchi Y, Kagawa T, Horino T and Takao T:
Aldosterone stimulates nuclear factor-kappa B activity and
transcription of intercellular adhesion molecule-1 and connective
tissue growth factor in rat mesangial cells via serum-and
glucocorticoid-inducible protein kinase-1. Clin Exp Nephrol.
16:81–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Terada Y, Kobayashi T, Kuwana H, Tanaka H,
Inoshita S, Kuwahara M and Sasaki S: Aldosterone stimulates
proliferation of mesangial cells by activating mitogen-activated
protein kinase 1/2, cyclin D1, and cyclin A. J Am Soc Nephrol.
16:2296–2305. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan J, Jia R and Bao Y: Aldosterone
up-regulates production of plasminogen activator inhibitor-1 by
renal mesangial cells. J Biochem Mol Biol. 40:180–188.
2007.PubMed/NCBI
|
|
25
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
61:379–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang A, Han Y, Wang B, Li S and Gan W:
Beyond gap junction channel function: The expression of Cx43
contributes to aldosterone-induced mesangial cell proliferation via
the ERK1/2 and PKC pathways. Cell Physiol Biochem. 36:1210–1222.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang
W, Chen J, Yu F, Zhou T and Wang Y: Long noncoding RNA expression
profiles of lung adenocarcinoma ascertained by microarray analysis.
PLoS One. 9:e1040442014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales D Rivea, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009;
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lai L, Chen J, Hao CM, Lin S and Gu Y:
Aldosterone promotes fibronectin production through a
Smad2-dependent TGF-beta1 pathway in mesangial cells. Biochem
Biophys Res Commun. 348:70–75. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nishiyama A, Yao L, Nagai Y, Miyata K,
Yoshizumi M, Kagami S, Kondo S, Kiyomoto H, Shokoji T, Kimura S, et
al: Possible contributions of reactive oxygen species and
mitogen-activated protein kinase to renal injury in
aldosterone/salt-induced hypertensive rats. Hypertension.
43:841–448. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Balakrishnan R, Harris MA, Huntley R, Van
Auken K and Cherry JM: A guide to best practices for Gene Ontology
(GO) manual annotation. Database (Oxford). 2013:bat0542013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kapranov P, Drenkow J, Cheng J, Long J,
Helt G, Dike S and Gingeras TR: Examples of the complex
architecture of the human transcriptome revealed by RACE and
high-density tiling arrays. Genome Res. 15:987–997. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schmidt F, van den Eijnden M, Gobert R
Pescini, Saborio GP, Carboni S, Alliod C, Pouly S, Staugaitis SM,
Dutta R, Trapp B, et al: Identification of VHY/Dusp15 as a
regulator of oligodendrocyte differentiation through a systematic
genomics approach. PLoS One. 7:e404572012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schnaper HW, Jandeska S, Runyan CE,
Hubchak SC, Basu RK, Curley JF, Smith RD and Hayashida T: TGF-beta
signal transduction in chronic kidney disease. Front Biosci
(Landmark Ed). 14:2448–2465. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chin BY, Mohsenin A, Li SX, Choi AM and
Choi ME: Stimulation of pro-alpha(1)(I) collagen by TGF-beta(1) in
mesangial cells: role of the p38 MAPK pathway. Am J Physiol Renal
Physiol. 280:F495–F504. 2001.PubMed/NCBI
|
|
44
|
Huwiler A and Pfeilschifter J:
Transforming growth factor beta 2 stimulates acute and chronic
activation of the mitogen-activated protein kinase cascade in rat
renal mesangial cells. FEBS Lett. 354:255–258. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Martínez-Salgado C, Rodríguez-Peña AB and
López-Novoa JM: Involvement of small Ras GTPases and their
effectors in chronic renal disease. Cell Mol Life Sci. 65:477–492.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
López-Hernández FJ and López-Novoa JM:
Role of TGF-β in chronic kidney disease: An integration of tubular,
glomerular and vascular effects. Cell Tissue Res. 347:141–154.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cao J, Xu H, Zhao H, Gong W and
Dunaway-Mariano D: The mechanisms of human hotdog-fold thioesterase
2 (hTHEM2) substrate recognition and catalysis illuminated by a
structure and function based analysis. Biochemistry. 48:1293–1304.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao H, Martin BM, Bisoffi M and
Dunaway-Mariano D: The Akt C-terminal modulator protein is an
acyl-CoA thioesterase of the Hotdog-Fold family. Biochemistry.
48:5507–5509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao H, Lim K, Choudry A, Latham JA,
Pathak MC, Dominguez D, Luo L, Herzberg O and Dunaway-Mariano D:
Correlation of structure and function in the human hotdog-fold
enzyme hTHEM4. Biochemistry. 51:6490–6492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nishiyama A, Yao L, Fan Y, Kyaw M, Kataoka
N, Hashimoto K, Nagai Y, Nakamura E, Yoshizumi M, Shokoji T, et al:
Involvement of aldosterone and mineralocorticoid receptors in rat
mesangial cell proliferation and deformability. Hypertension.
45:710–716. 2005. View Article : Google Scholar : PubMed/NCBI
|