Introduction
Glioma is the most common type of malignant tumors
in the brain; it originates from neural stromal cells and accounts
for 81% of all malignant brain tumors in adults (1,2). The
formation and progression of glioma is extremely complex and
involves aberrant activation of proto-oncogenes and inactivation of
tumor suppressor genes (3–5). Glioma could be graded from grade I to
grade IV, based on the degree of malignancy according to the World
Health Organization (WHO) classification (6). Glioblastoma (GBM), the highest-grade
glioma, is characterized by a heterogeneous population of cells
that are genetically unstable, highly infiltrative, angiogenic, and
resistant to chemotherapy (7).
Despite recent advances in multimodal therapies, the prognosis of
GBM patients remains poor, with an average five-year survival rate
of only 4 to 5% (8,9). The average survival time of GBM
remains only 14.6 months following surgical treatment combined with
radiation and chemotherapy (10).
The high mortality of GBM is partially due to rapid growth,
angiogenesis and metastasis of GBM cells (11,12).
Therefore, improving understanding regarding the underlying
mechanisms involved in GBM occurrence and progression, which would
be essential to identify novel therapeutic methods for patients
with this disease, is clinically significant.
MicroRNAs (miRNAs) are a group of endogenous,
single-stranded and non-protein-coding short RNAs with
approximately 19–24 nucleotides in length (13). miRNAs negatively regulate the gene
expression through imperfect base pairing with the 3-untranslated
regions (3′-UTRs) of target messenger RNAs (mRNAs), resulting in
translation repression or mRNA degradation (14). Each miRNA could directly modulate
hundreds of target genes, and more than one-third of genes may be
regulated by miRNAs (15). miRNAs
play important roles in diverse biological processes, such as cell
proliferation, cycle, death, differentiation, metastasis and drug
resistance (16–18). The deregulation of miRNAs has been
detected in a wide variety of human cancers, such as glioma
(19), prostate cancer (20), gastric cancer (21), breast cancer (22) and lung cancer (23). Increasing evidence suggested that
the abnormal expression of some specific miRNAs are closely related
to tumorigenesis and tumor development, and some miRNAs also act as
tumor suppressors or oncogenes depending on the characteristic of
their target genes (24–26). Therefore, further investigation of
the expression and roles of miRNAs in GBM is important to develop
novel and efficient therapeutic targets for patients with GBM.
The expression and roles of miR-543 have been
reported in numerous types of human cancers (27–29).
However, miR-543 had not been well studied in GBM. In our present
study, we investigated the miR-543 expression pattern in GBM, the
effects of miR-543 on GBM cells and the involved molecular
mechanism.
Materials and methods
Tissue sample and cell lines
This study was approved by the Ethical Committee of
Shenzhen Second People's Hospital. All patients enrolled in this
study gave written informed consent. A total of 19 cases of GBM
tissues (age range, 31–69 years; median age, 45; twelve males and
seven females) and 8 cases of normal brain tissues were obtained
from Department of Neurosurgery, Shenzhen Second People's Hospital
between February 2013 and May 2015. Normal brain tissues were
collected from internal decompression patients treated with
surgical operation. None of these patients had any radiotherapy or
chemotherapy before surgical resection. All tissue samples were
immediately snap-frozen and stored at −80°C until further use.
Cell lines and transfection
Four GBM cell lines U87, U251, LN229, and T98 were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China), and maintained in Dulbecco's modified
Eagles medium (DMEM) containing 10% fetal bovine serum (FBS) (both
from Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100
mg/ml streptomycin (Invitrogen). Normal human astrocytes (NHAs)
purchased from ScienCell Research Laboratories (Carlsbad, CA, USA)
were cultured in astrocyte medium (ScienCell Research
Laboratories). All cells were grown at 37°C in a fully humidified
atmosphere containing 5% CO2 in air.
MiR-543 mimics and miRNA mimics negative control
(NC) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai,
China). A disintegrin and metalloproteinase 9 (ADAM9) overexpressed
plasmid (pcDNA3.1-ADAM9) and blank plasmid (pcDNA3.1) were
synthesized by Chinese Academy of Sciences (Changchun, China). Cell
transfeciton was performed by using Lipofectamine 2000
(Invitrogen), according to the manufacturer's instructions.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using
TRIzol (Invitrogen) according to the manufacturer's protocol. To
examine miR-543 expression levels, total RNA was transcribed by
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Carlsbad, CA, USA). qPCR was performed using TaqMan MicroRNA PCR
kit on an ABI Prism 7500 Sequence Detection system (both from
Applied Biosystems). To quantify mRNA expression of ADAM9, cDNA was
synthesized using a First-Strand cDNA Synthesis kit (Invitrogen)
and followed by qPCR with SYBR Premix Ex Taq™ (Takara Biotechnology
Co., Ltd., Dalian, China). U6 and GAPDH was used as internal
standard to normalize the expression of miR-543 and ADAM9 mRNA,
respectively. The primers were designed as follows: miR-543
forward, 5′-CCAGCTACACTGGGCAGCAGCAATTCATGTTT-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACATATACT-3′
and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′; ADAM9 forward,
5′-GCTAGTTGGACTGGAGATTTGG-3′ and reverse,
5′-TTATTACCACAGGAGGGAGCAC-3′; GAP DH forward,
5′-CATCACCATCTTCCAGGAGCG-3′ and reverse,
5′-TGACCTTGCCCACAGCCTTG-3′. Relative expression was calculated
using the 2−ΔΔCt method (30).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assays (Dojindo Molecular Technologies,
Kumamoto, Japan) were carried out to assess the cell proliferation.
Briefly, transfected cells were trypsinized, collected and seeded
into 96-well plate at a density of 3,000 cells/well. Cells were
then incubated at 37°C with 5% CO2, and CCK-8 assay was
performed on days 0, 1, 2, and 3. Ten microliters of CCK-8 were
added to each well. The plates were incubated at 37°C for another 4
h. The absorbance was determined with a microplate reader (Bio-Rad,
Richmond, CA, USA) at the wavelength of 450 nm. Each sample was
performed in triplicate and repeated three times.
Transwell invasion assay
The invasion assay was performed using Transwell
chambers (8 mm pores; Costar, Corning, NY, USA) pre-coated with
Matrigel (BD Biosciences, San Jose, CA, USA).
Following transfection for 48 h, 5×104
cells were added into the top of the Transwell chambers in FBS-free
DMEM, and DMEM containing 20% FBS was added to the lower chambers.
The cells were incubated at 37°C with 5% CO2 for 24 h.
Cells that did not invade through the pores were removed from the
top chambers with a cotton swab while the invasive cells were fixed
with 100% methanol and stained with 0.1% crystal violet. The number
of invasive cells was counted in five random visual fields under an
inverted microscope (Olympus Corporation, Tokyo, Japan) and images
were captured under ×200 magnification.
Flow cytometry analysis
Transfected cells were collected at 48 h
post-transfection, washed with ice-cold PBS and then fixed in 80%
ice-cold ethanol. Subsequently, cells were re-suspended in cold PBS
to a concentration of 1×104 cells. Annexin V-FITC
apoptosis detection kit (Invitrogen) was used to examine cell
apoptosis, as described by the manufacturer. The cell apoptotic
rates of the transfected cells were determined using a flow
cytometry (Beckman Coulter Inc., Brea, CA, USA).
Bioinformatics analysis
The candidate targets of miR-543 predicted by
computer-aided algorithms were obtained from TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org).
Luciferase reporter assay
Luciferase reporter plasmids,
psiCHECK2-ADAM9-3′-UTR-wild type (Wt) and
psiCHECK2-ADAM9-3′-UTR-mutant (Mut), were synthesized and purchased
from GenePharma. For luciferase reporter assay, cells were
co-transfected with the psiCHECK2-ADAM9-3′-UTR-Wt or
psiCHECK2-ADAM9-3′-UTR-Mut, and miR-543 mimics or NC using
Lipofectamine 2000, following to the manufacturer's instructions.
After co-transfection for 48 h, the Dual-Luciferase Reporter Assay
system (Promega, Manheim, Germany) was used to measure the
activities of Renilla luciferase and firefly luciferase,
according to the manufacturer's protocol. Renilla luciferase
activity was normalized to firefly luciferase activity.
Western blot analysis
Total protein was extracted with RIPA lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China) supplemented
with protease inhibitor cocktail (Roche Applied Science,
Indianapolis, IN, USA), and the protein concentration was measured
using the bicinchoninic acid protocol (Pierce, Rockford, IL, USA).
Equal amounts of protein was loaded onto 10% sodium dodecyl
sulphate-polyacrylamide gel and then transferred to polyvinylidene
difluoride membranes (Millipore, Billerica, MA, USA). Subsequently,
the membrane was blocked with 5% skimmed milk in TBS/0.1% Tween
(TBST) at room temperature for 1 h, and incubated with primary
antibodies overnight at 4°C. The primary antibodies used in this
study were mouse anti-human monoclonal ADAM9 antibody (1:1,000
dilution; sc-377233; Santa Cruz Biotechnology, Santa Cruz CA, USA)
and mouse anti-human monoclonal GAPDH antibody (1:1,000 dilution;
ab125247; Abcam, Cambridge, UK), at 4°C overnight. Following a 2 h
incubation with a goat-anti-mouse horseradish peroxidase
(HRP)-conjugated secondary antibody (1:5,000 dilution; sc-2005;
Santa Cruz Biotechnology), the protein bands were visualized using
the enhanced chemiluminescence kit (Millipore, Billerica, MA, USA).
GAPDH was used as a loading control.
Statistical analysis
Data were expressed as mean ± SD. Statistical
analyses were performed using Mann-Whitney's U test or one-way
ANOVA with SPSS 16.0 software (SPSS, Chicago, IL, USA). P<0.05
was considered to be statistically significant.
Results
Reduced miR-543 expression levels in
GBM tissues and cell lines
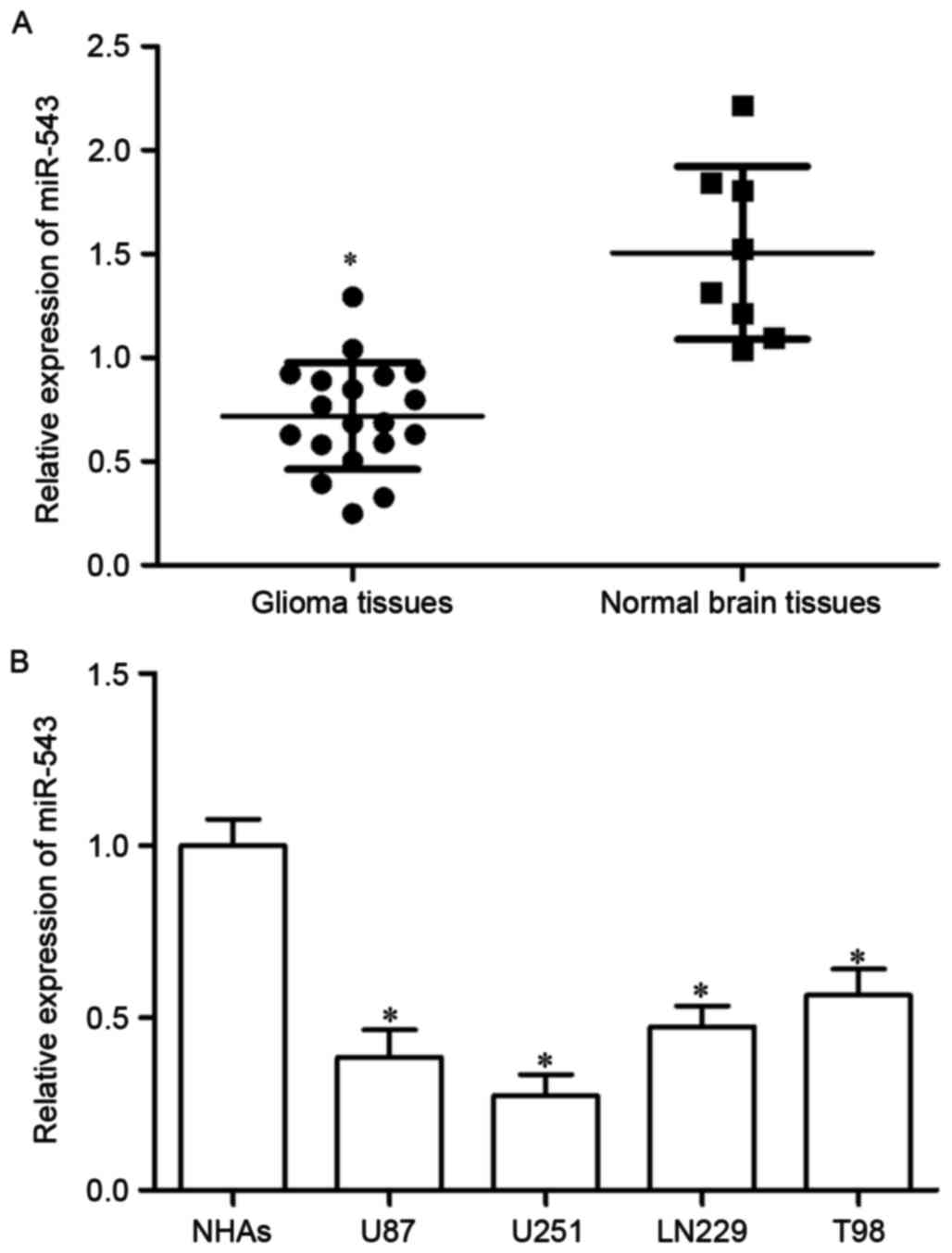
The expression of miR-543 in 19 cases of GBM tissues
and 8 cases of normal brain tissues was investigated using RT-qPCR
to explore the potential roles of miR-543 in GBM. As shown in
Fig. 1A, miR-543 expression level
was significantly decreased in GBM tissue compared with that in
normal brain tissues (P<0.05). The expression of miR-543 in four
GBM cell lines (U87, U251, LN229, T98) and NHAs was determined to
confirm the reduced miR-543 expression level in GBM tissues.
miR-543 expression level was lower in GBM cell lines than that in
NHAs (Fig. 1B, P<0.05). These
results suggested that miR-543 was downregulated in GBM and may act
as a tumor suppressor.
MiR-543 inhibits the proliferation and
invasion, as well as enhances the apoptosis of GBM cells
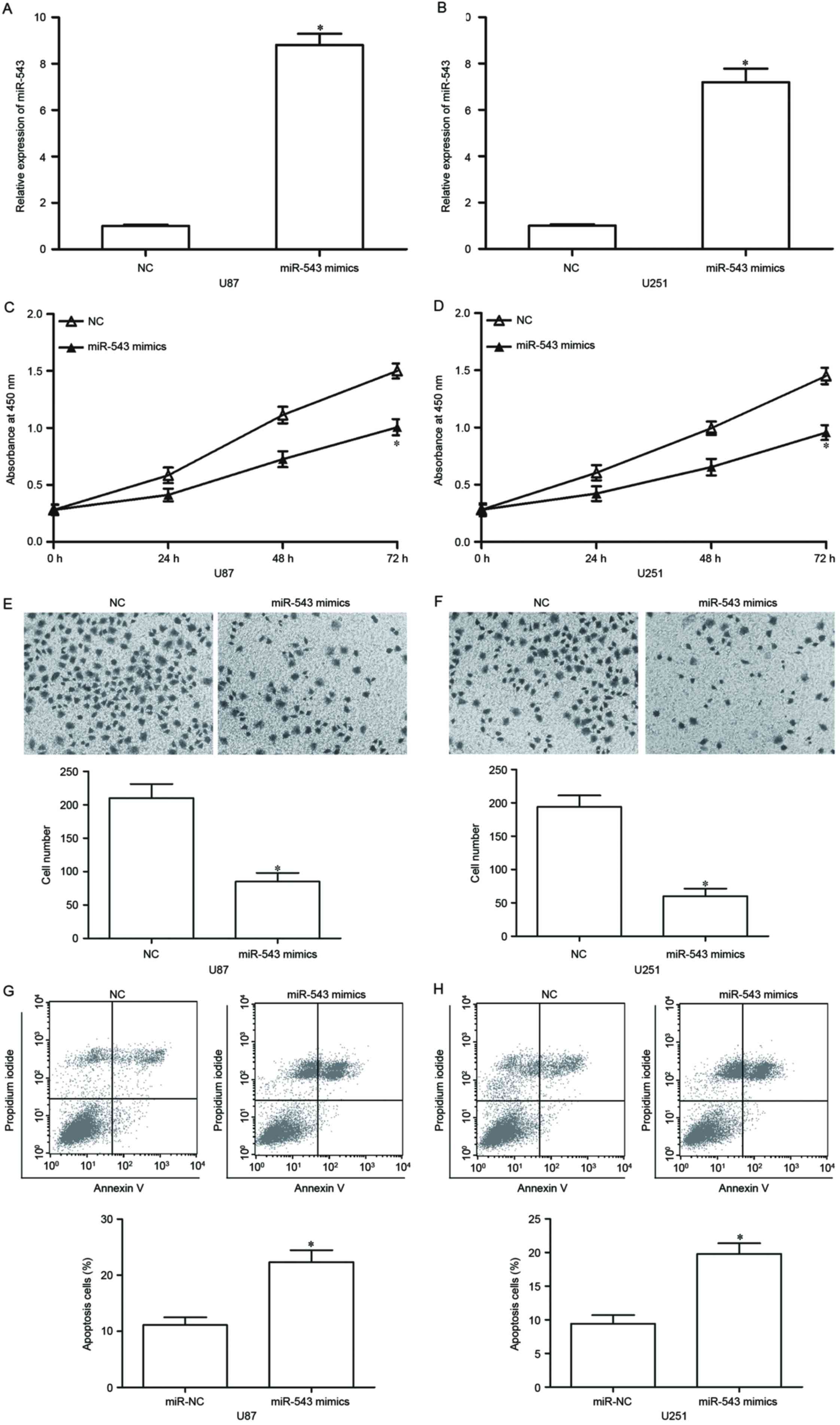
To ascertain the functional effect of miR-543 on
GBM, we transfected the two lowest miR-543 expressing cell lines,
U87 and U251, with miR-543 mimic to increase its endogenous
expression levels. After 48-h transfection, RT-qPCR confirmed that
miR-543 was markedly upregulated in U87 and U251 cells transfected
with miR-543 mimics (Fig. 2A and
B, P<0.05). CCK-8 assays were utilized to investigate the
effect of miR-543 on GBM cell proliferation. As shown in Fig. 2C and D, U87 and U251 cells with
high miR-543 expression levels exhibited a significant inhibition
of proliferation compared with the cells with NC. Then, the effect
of miR-543 on GBM cell invasion was analysed using Transwell
invasion assay. Results showed that the upregulation of miR-543
suppresses the U87 and U251 cell invasion abilities compared with
cells transfected with NC (Fig. 2E and
F, P<0.05). Flow cytometry analysis was performed in U87 and
U251 cells transfected with miR-543 mimics or NC to examine whether
miR-543 overexpression has a positive effect on GBM cell apoptosis.
We detected a clear apoptosis activation in miR-543
mimics-transfected U87 and U251 cells (Fig. 2G and H, P<0.05). These results
indicated that miR-543 exerts tumor-suppressing roles in GBM
cells.
ADAM9 is a direct target of miR-543 in
GBM
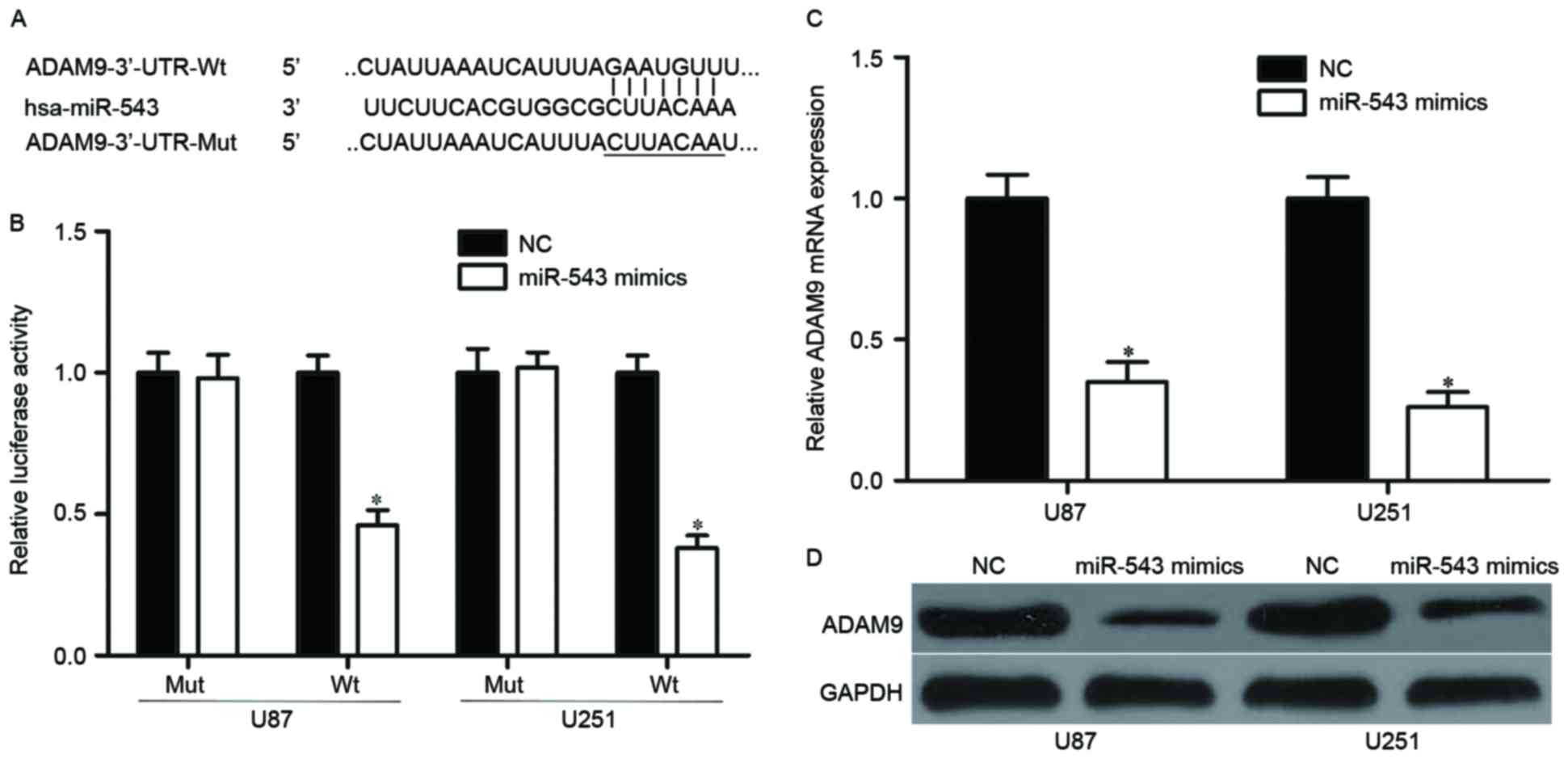
Bioinformatics analysis was conducted to identify
the candidate target genes of miR-543 and to investigate the
underlying mechanism by which miR-543 regulates GBM cell
proliferation, invasion and apoptosis. Among these predicted
targets, ADAM9, which is an important regulator for cancer
formation and progression (31–33),
attracted attention and was selected for further confirmation
(Fig. 3A). Luciferase reporter
assay was performed on U87 and U251 cells to validate interaction
between miR-543 and the predicted binding site of ADAM9. The
overexpression of miR-543 decreased wild-type (Fig. 3B, P<0.05) but not mutant ADAM9
reporter activity, suggesting that miR-543 specifically targeted
the 3′UTR of ADAM9. To confirm whether ADAM9 are regulated by
miR-543, the mRNA and protein expression of ADAM9 were examined in
U87 and U251 cells transfected with miR-543 mimics or NC. Results
showed that restoration expression of miR-543 reduced the ADAM9
expression levels in U87 and U251 cells at both mRNA and protein
levels (Fig. 3C and D, P<0.05).
These results suggested that ADAM9 is a direct target of miR-543 in
GBM.
ADAM9 is upregulated in GBM tissues
and inversely correlates with miR-543 levels
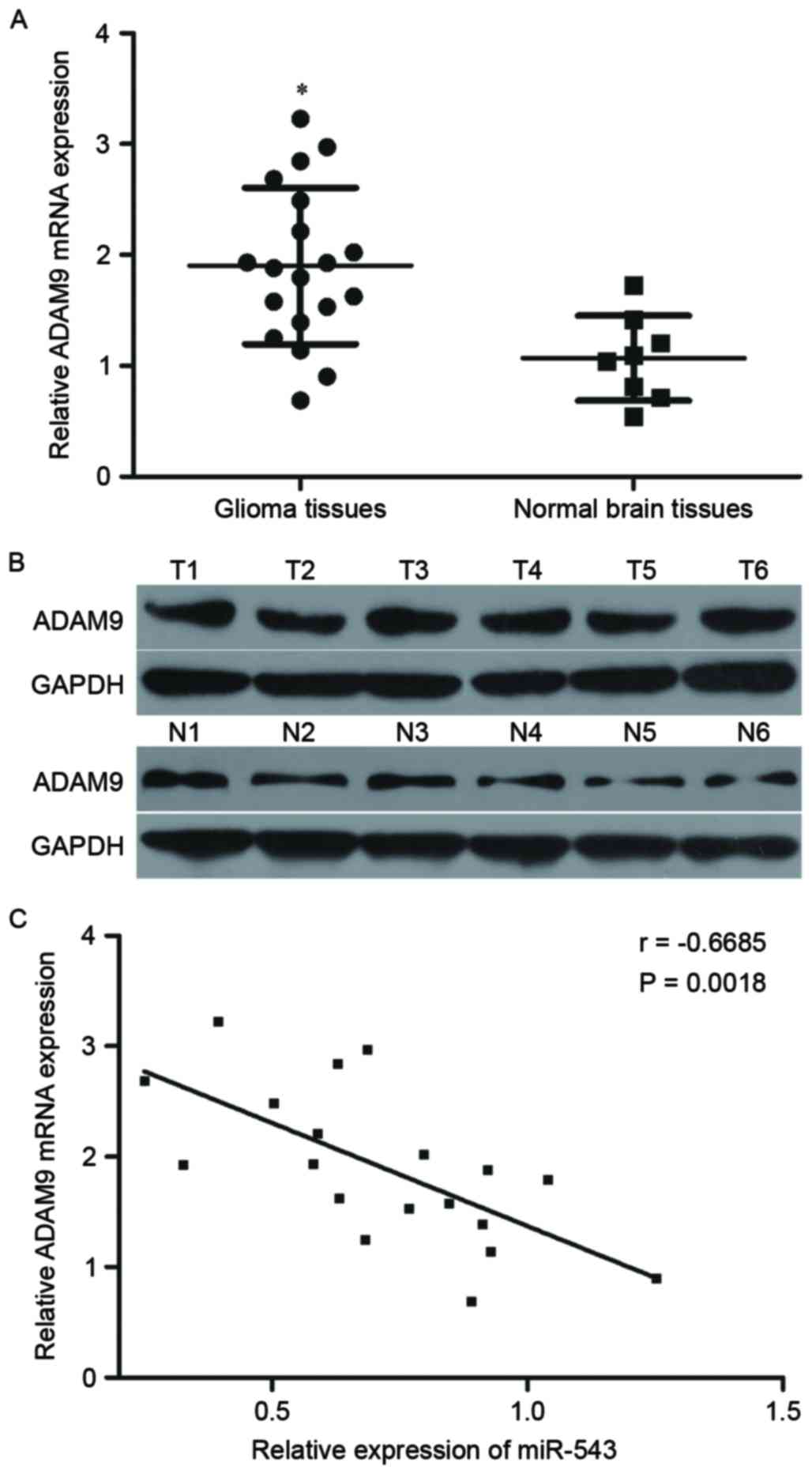
Further experiments were utilized to detect the
ADAM9 expression in 19 cases of GBM tissues and eight cases of
normal brain tissues. RT-qPCR and Western blot results revealed
that ADAM9 mRNA and protein expression levels were higher in GBM
tissues relative to that in normal brain tissues (Fig. 4A and B, P<0.05). Spearman's
correlation analysis was then performed to explore the relationship
between miR-543 and ADAM9 mRNA expression in GBM tissues. As shown
in Fig. 4C, ADAM9 mRNA was
inversely correlated with miR-543 levels in GBM tissues (Fig. 4C, r=−0.6685, P=0.0018), suggesting
that the downregulation of miR-543 may be an important cause for
the upregulation of ADAM9 in GBM.
Upregulation of ADAM9 reverses the
effects of miR-543 on GBM cell proliferation, invasion and
apoptosis
Rescue experiments were performed in U87 and U251
cells to further explore the role of ADAM9 on the biological roles
induced by miR-543 in GBM. Then, western blot analysis was used to
measure the ADAM9 expression in U87 and U251 cells transfected with
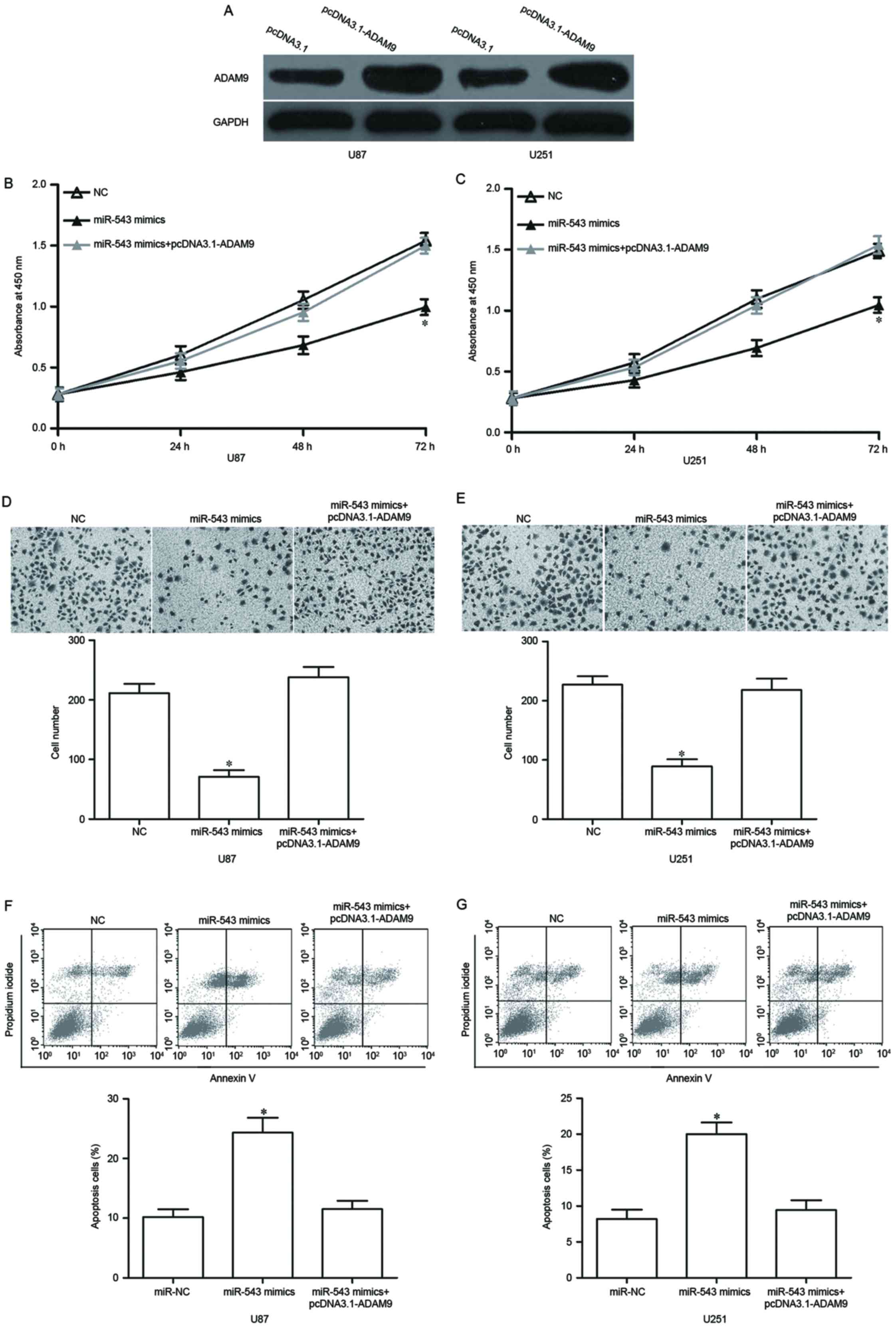
pcDNA3.1-ADAM9 or pcDNA3.1. Our data showed that ADAM9 protein was
significantly upregulated in U87 and U251 cells after transfection
with pcDNA3.1-ADAM9 (Fig. 5A,
P<0.05). Functional rescue results showed that the
reintroduction of ADAM9 reversed the effects of miR-543
overexpression on GBM cell proliferation (Fig. 5B and C, P<0.05), invasion
(Fig. 5D and E, P<0.05) and
apoptosis (Fig. 5F and G,
P<0.05). These results indicated that miR-543 inhibits GBM cell
proliferation and invasion, as well as promotes apoptosis by
targeting ADAM9.
Discussion
Recently, accumulated studies demonstrated that
abnormally expressed miRNAs may act as oncogenes or tumor
suppressor genes and is important on the formation and progression
of various human cancers, including GBM (34–36).
Therefore, investigation on the expression and roles of miRNAs may
have potential tumor diagnostic and prognostic values, as well as
therapeutic values for patients with GBM (37). In our current study, miR-543 was
lowly expressed in GBM tissues and cell lines. The resumption
expression of miR-543 inhibited the proliferation and invasion, as
well as enhanced the apoptosis of GBM cells. In addition, ADAM9 was
identified as a novel target of miR-543. Therefore, miR-543 played
tumor suppressing roles through the downregulation of ADAM9 in
GBM.
miR-543 was aberrantly expressed in numerous types
of human cancers. For example, in colorectal cancer, miR-543 was
downregulated in tumor tissues and cell lines. The low miR-543
expression level was inversely correlated with the metastatic
status of patients with colorectal cancer and the metastatic
potential of colorectal cancer cell lines (27). miR-543 was lowly expressed in
endometrial cancer (28). However,
contrary to these results, miR-543 was upregulated in
gefitinib-resistant non-small cell lung cancer (29), hepatocellular carcinoma (38), gastric cancer (39) and osteosarcoma (40). These conflicting findings indicated
that the expression pattern of miR-543 in human cancer has tissue
specificity.
miR-543 has been demonstrated to be a tumor
suppressor. Fan et al reported that the enforced expression
of miR-543 suppressed the proliferation and metastasis of
colorectal cancer cells both in vitro and in vivo
(27). Bing et al revealed
that miR-543 re-expression inhibited cell monolayer proliferation,
anchorage-independent growth, migration and invasion of endometrial
cancer (28). However, in
non-small cell lung cancer, Bi et al found that miR-543 acts
as an oncogene by attenuating tumor cell proliferation and invasion
(29). Yu et al
demonstrated that the upregulation of miR-543 enhanced the cell
growth and motility of hepatocellular carcinoma (38). Li et al showed that the
ectopic expression of miR-543 promoted the cell proliferation and
cell cycle progression of gastric cancer (39). Besides, the restoration expression
of miR-543 promoted the osteosarcoma cell growth in vitro
and in vivo (40).
These studies may appear contradictory, because
miR-543 acted as an oncogene in certain types of cancer and a tumor
suppressor in others. These contradictory results may be explained
by the imperfect complementarity of the interactions between miRNAs
and their target genes (41).
Identification of cancer-specific miRNAs and their
target genes is pivotal for understanding their roles in
tumorigenesis and tumor development (42,43).
Several target genes of miR-543 have been identified, such as KRAS,
MTA1 and HMGA2 in colorectal cancer (27), FAK and TWIST1 in endometrial cancer
(28), PTEN in lung cancer
(29), PAQR3 in hepatocellular
carcinoma (38), SIRT1 in gastric
cancer (39) and PRMT9 in
osteosarcoma (40). Herein, we
focused on the mechanisms of miR-543 in regulating cell
proliferation, invasion and apoptosis of GBM cells. Bioinformatics
analysis was performed to search candidate targets of miR-543,
among which ADAM9 was predicated as a potential target of miR-543.
Luciferase reporter assay indicated that miR-543 directly targeted
the 3′-UTR of ADAM9. Following, we confirmed that ADAM9 mRNA and
protein levels were negatively regulated by miR-543 in GBM cells.
Additionally, ADAM9 was upregulated in GBM tissues and inversely
correlated with miR-543 expression level. Besides, rescue
experiments demonstrated that upregulation of ADAM9 rescued the
effects induced by miR-543 overexpression on GBM cells. These
results suggest that miR-543 can directly and negatively regulate
ADAM9 expression by binding to the 3′-UTR of ADAM9.
ADAMs are members of the metzincin superfamily of
matrix metalloproteinases (MMP) (44). To date, 21 functional ADAMs have
been described in humans and 40 in different organisms (45). ADAMs are involved in many different
biological functions, including fertilization, adhesion, migration
and proteolysis (44,46). ADAM9 is a membrane-anchored
metalloproteinase and is one of the first ADAM proteins to be
identified and characterized. It consists of an N-terminal
prodomain followed by a metalloprotease domain, a disintegrin
domain and cysteine-rich region, an epidermal growth factor repeat,
a transmembrane domain and a cytoplasmic tail with potential SH3
ligand domains (47,48). Previous studies reported that ADAM9
was upregulated in several types of human cancers, such as renal
cell cancer (31), non-small cell
lung cancer (49), prostate cancer
(50), hepatocellular carcinoma
(32), colon cancer (33) and pancreatic cancer (51). In glioma, ADAM9 was highly
expressed in tumor tissues. In addition, high expression levels of
ADAM9 were significantly correlated with poor prognosis in
lower-grade glioma. Furthermore, multivariate analysis indicated
ADAM9 expression as an independent marker of poor survival
(52). Functional assays revealed
that ADAM9 plays important roles in glioma growth and metastasis
(19,53). These studies all indicated that
ADAM9 may be a novel and promising target for therapeutic
intervention in patients with glioma.
In conclusion, miR-543 was downregulated in GBM and
played an important role in regulating GBM cell proliferation,
invasion and apoptosis. Moreover, ADAM9 is the target gene of
miR-543. These results suggested that miR-543/ADAM9 pathway may be
investigated as an important strategy for the treatment of patients
with GBM.
References
|
1
|
Jungk C, Chatziaslanidou D, Ahmadi R,
Capper D, Bermejo JL, Exner J, von Deimling A, Herold-Mende C and
Unterberg A: Chemotherapy with BCNU in recurrent glioma: Analysis
of clinical outcome and side effects in chemotherapy-naive
patients. BMC Cancer. 16:812016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Chen L,
Liu SY, Tang WL, Qiu XX and Zhou CF: Peritumoral edema on magnetic
resonance imaging predicts a poor clinical outcome in malignant
glioma. Oncol Lett. 10:2769–2776. 2015.PubMed/NCBI
|
|
3
|
Hatanpaa KJ, Burma S, Zhao D and Habib AA:
Epidermal growth factor receptor in glioma: Signal transduction,
neuropathology, imaging, and radioresistance. Neoplasia.
12:675–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen AL, Holmen SL and Colman H: IDH1 and
IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 13:3452013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Endersby R and Baker SJ: PTEN signaling in
brain: Neuropathology and tumorigenesis. Oncogene. 27:5416–5430.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Komori T, Sasaki H and Yoshida K: Revised
WHO classification of tumours of the central nervous system:
Summary of the revision and perspective. No Shinkei Geka.
44:625–635. 2016.(In Japanese). PubMed/NCBI
|
|
7
|
Minniti G, Muni R, Lanzetta G, Marchetti P
and Enrici RM: Chemotherapy for glioblastoma: Current treatment and
future perspectives for cytotoxic and targeted agents. Anticancer
Res. 29:5171–5184. 2009.PubMed/NCBI
|
|
8
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan Y, Peng Y, Ou Y and Jiang Y:
MicroRNA-610 is downregulated in glioma cells, and inhibits
proliferation and motility by directly targeting MDM2. Mol Med Rep.
14:2657–2664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pollack IF: Neuro-oncology: Therapeutic
benefits of reirradiation for recurrent brain tumors. Nat Rev
Neurol. 6:533–535. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osaki M, Okada F and Ochiya T: miRNA
therapy targeting cancer stem cells: A new paradigm for cancer
treatment and prevention of tumor recurrence. Ther Deliv.
6:323–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bushati N and Cohen SM: MicroRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Wang S, Yuan A, Yuan X and Liu B:
MicroRNA-140 represses glioma growth and metastasis by directly
targeting ADAM9. Oncol Rep. 36:2329–2338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guan H, Liu C, Fang F, Huang Y, Tao T,
Ling Z, You Z, Han X, Chen S, Xu B and Chen M: MicroRNA-744
promotes prostate cancer progression through aberrantly activating
Wnt/β-catenin signaling. Oncotarget. 8:14693–14707. 2017.PubMed/NCBI
|
|
21
|
Qi M, Liu D and Zhang S: MicroRNA-21
contributes to the discrimination of chemoresistance in metastatic
gastric cancer. Cancer Biomark. 18:451–458. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
An N, Luo X, Zhang M and Yu R:
MicroRNA-376b promotes breast cancer metastasis by targeting Hoxd10
directly. Exp Ther Med. 13:79–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, An J, Lv W, Lou T, Liu Y and Kang
W: miRNA-129-5p suppresses cell proliferation and invasion in lung
cancer by targeting microspherule protein 1, E-cadherin and
vimentin. Oncol Lett. 12:5163–5169. 2016.PubMed/NCBI
|
|
24
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng Z, Wu T, Li Y, Xu Z, Zhang S, Liu B,
Chen Q and Tian D: MicroRNA-370-3p inhibits human glioma cell
proliferation and induces cell cycle arrest by directly targeting
β-catenin. Brain Res. 1644:53–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hao Y, Zhang S, Sun S, Zhu J and Xiao Y:
miR-595 targeting regulation of SOX7 expression promoted cell
proliferation of human glioblastoma. Biomed Pharmacother.
80:121–126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma
H, Yu D, Maitikabili A, Xiao H, Zhang C, et al: MicroRNA-543
suppresses colorectal cancer growth and metastasis by targeting
KRAS, MTA1 and HMGA2. Oncotarget. 7:21825–21839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bing L, Hong C, Li-Xin S and Wei G:
MicroRNA-543 suppresses endometrial cancer oncogenicity via
targeting FAK and TWIST1 expression. Arch Gynecol Obstet.
290:533–541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bi M, Chen W, Yu H, Wang J, Ding F, Tang
DJ and Tang C: miR-543 is up-regulated in gefitinib-resistant
non-small cell lung cancer and promotes cell proliferation and
invasion via phosphatase and tensin homolog. Biochem Biophys Res
Commun. 480:369–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fritzsche FR, Wassermann K, Jung M, Tölle
A, Kristiansen I, Lein M, Johannsen M, Dietel M, Jung K and
Kristiansen G: ADAM9 is highly expressed in renal cell cancer and
is associated with tumour progression. BMC Cancer. 8:1792008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao K, Qian N, Tang Y, Ti Z, Song W, Cao D
and Dou K: Increased expression of a disintegrin and
metalloprotease-9 in hepatocellular carcinoma: Implications for
tumor progression and prognosis. Jpn J Clin Oncol. 40:645–651.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Ji Z, Qiao C, Qi Y and Shi W:
Overexpression of ADAM9 Promotes Colon Cancer Cells Invasion. J
Invest Surg. 26:127–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu N, Zhang L, Wang Z, Cheng Y, Zhang P,
Wang X, Wen W, Yang H, Liu H, Jin W, et al: MicroRNA-101 inhibits
proliferation, migration and invasion of human glioblastoma by
targeting SOX9. Oncotarget. 8:19244–19254. 2017.PubMed/NCBI
|
|
35
|
Li D, Wang Z, Chen Z, Lin L, Wang Y,
Sailike D, Luo K, Du G, Xiang X and Jiafu GD: MicroRNA-106a-5p
facilitates human glioblastoma cell proliferation and invasion by
targeting adenomatosis polyposis coli protein. Biochem Biophys Res
Commun. 481:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Liu H, Tian L, Wang F, Han L,
Zhang W and Bai YA: miR-15b Inhibits the progression of
glioblastoma cells through targeting insulin-like growth factor
receptor 1. Horm Cancer. 8:49–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karsy M, Arslan E and Moy F: Current
progress on understanding MicroRNAs in glioblastoma multiforme.
Genes Cancer. 3:3–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang
J, Guo M, Liu N and Zhu L: MicroRNA-543 acts as an oncogene by
targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res.
4:897–906. 2014.PubMed/NCBI
|
|
39
|
Li J, Dong G, Wang B, Gao W and Yang Q:
miR-543 promotes gastric cancer cell proliferation by targeting
SIRT1. Biochem Biophys Res Commun. 469:15–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang H, Guo X, Feng X, Wang T, Hu Z, Que
X, Tian Q, Zhu T, Guo G, Huang W and Li X: miRNA-543 promotes
osteosarcoma cell proliferation and glycolysis by partially
suppressing PRMT9 and stabilizing HIF-1α protein. Oncotarget.
8:2342–2355. 2017.PubMed/NCBI
|
|
41
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song L, Yang J, Duan P, Xu J, Luo X, Luo
F, Zhang Z, Hou T, Liu B and Zhou Q: MicroRNA-24 inhibits
osteosarcoma cell proliferation both in vitro and in vivo by
targeting LPAATβ. Arch Biochem Biophys. 535:128–135. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu X, Zhong D, Gao Q, Zhai W, Ding Z and
Wu J: MicroRNA-34a inhibits human osteosarcoma proliferation by
downregulating ether à go-go 1 expression. Int J Med Sci.
10:676–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Blobel CP: ADAMs: Key components in EGFR
signalling and development. Nat Rev Mol Cell Biol. 6:32–43. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nath D, Slocombe PM, Stephens PE, Warn A,
Hutchinson GR, Yamada KM, Docherty AJ and Murphy G: Interaction of
metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on
different haemopoietic cells. J Cell Sci. 112:579–587.
1999.PubMed/NCBI
|
|
46
|
Edwards DR, Handsley MM and Pennington CJ:
The ADAM metalloproteinases. Mol Aspects Med. 29:258–289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guaiquil V, Swendeman S, Yoshida T,
Chavala S, Campochiaro PA and Blobel CP: ADAM9 is involved in
pathological retinal neovascularization. Mol Cell Biol.
29:2694–2703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Duffy MJ, McKiernan E, O'Donovan N and
McGowan PM: Role of ADAMs in cancer formation and progression. Clin
Cancer Res. 15:1140–1144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang J, Qi J, Chen N, Fu W, Zhou B and He
A: High expression of a disintegrin and metalloproteinase-9
predicts a shortened survival time in completely resected stage I
non-small cell lung cancer. Oncol Lett. 5:1461–1466.
2013.PubMed/NCBI
|
|
50
|
Fritzsche FR, Jung M, Tölle A, Wild P,
Hartmann A, Wassermann K, Rabien A, Lein M, Dietel M, Pilarsky C,
et al: ADAM9 expression is a significant and independent prognostic
marker of PSA relapse in prostate cancer. Eur Urol. 54:1097–1106.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Grutzmann R, Lüttges J, Sipos B, Ammerpohl
O, Dobrowolski F, Alldinger I, Kersting S, Ockert D, Koch R,
Kalthoff H, et al: ADAM9 expression in pancreatic cancer is
associated with tumour type and is a prognostic factor in ductal
adenocarcinoma. Br J Cancer. 90:1053–1058. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fan X, Wang Y, Zhang C, Liu L, Yang S,
Wang Y, Liu X, Qian Z, Fang S, Qiao H and Jiang T: ADAM9 expression
is associate with glioma tumor grade and histological type and acts
as a prognostic factor in lower-grade gliomas. Int J Mol Sci.
17(pii): E12762016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen CM, Hsieh YH, Hwang JM, Jan HJ, Hsieh
SC, Lin SH and Lai CY: Fisetin suppresses ADAM9 expression and
inhibits invasion of glioma cancer cells through increased
phosphorylation of ERK1/2. Tumour Biol. 36:3407–3415. 2015.
View Article : Google Scholar : PubMed/NCBI
|