Introduction
Lung cancer is the most significant health problem
worldwide in males and females, accounting for >1.6 million new
cases (~13% of total cancer diagnoses). Lung cancer was the leading
cause of cancer mortality in males, and the second leading cause of
cancer mortality in females, in 2008 (1). Histologically, lung cancer may be
categorized as small cell lung cancer or non-small cell lung cancer
(NSCLC), the latter accounting for ~85% of all lung cancer cases
(1,2). The majority of lung cancer cases are
diagnosed at the advanced stages of the disease, rendering curative
surgery unavailable. However, current targeted therapies appear
promising (2). Therefore, further
research on the molecular mechanisms and pathogenesis of lung
cancer may lead to novel strategies for the effective control of
NSCLC.
In the present study, the effects of tumor
suppressor genes, cellular tumor antigen p53 (p53) and G
protein-coupled receptor class C group 5 member A (GPCR5A), on cell
survival in NSCLC were investigated. Previous studies have
demonstrated that p53 protein functions to maintain genomic
stability and regulate cell apoptosis (3,4), and
GPCR5A acts as a lung cancer suppressor gene whose expression is
induced by retinoids (5–8). In addition, p53 mutation occurs in
the majority of NSCLC tissues, and is associated with NSCLC
tumorigenesis and poor survival in patients with NSCLC (9). Therefore, detailed investigation of
p53 in NSCLC may aid the understanding of the association between
genetic mutations and human carcinogenesis. A previous study
demonstrated that GPRC5A may be a target gene of p53 in breast and
ovarian cancers (10). GPRC5A is a
retinoid-induced gene and is associated with human carcinogenesis.
In NSCLC, GPRC5A was demonstrated to be a putative tumor suppressor
gene (5,6,11–13).
GPRC5A was demonstrated to be expressed in fetal and adult lung
tissues, although the expression was decreased or absent in NSCLC
tissue specimens (14). A previous
study in GPRC5A−/− mice demonstrated that 76% exhibited
lung adenomas and 17% exhibited lung adenocarcinoma at 1–2 years of
age, whereas only 11% of heterozygous mice and 10% of wild-type
mice developed lung adenoma with no mice developing lung cancer
(6). GPRC5A-knockout mice
exhibited an increased lung cancer incidence following treatment
with nicotine-derived nitrosamine ketone (12). Therefore, GPRC5A was considered to
be a putative lung cancer suppressor gene. However, other studies
have demonstrated that GPRC5A may act as an oncogene (10,15).
In breast and ovarian cancers, GPRC5A expression was observed to be
upregulated leading to promotion of tumor cell proliferation
(10).
The present study investigated the role of p53 in
NSCLC cell lines by manipulating p53 expression and measuring
GPRC5A regulation, in addition to NSCLC cell behaviors, in
vitro. The results of the present study provide an insight into
the role of GPRC5A in NSCLC and the mediation of p53 antitumor
activity, which may be useful for future treatments of NSCLC.
Materials and methods
Cell lines and culture
Human NSCLC H1299 (p53-null) and A549 [wild-type
(WT)-p53] cell lines were obtained from the American Type Culture
Collection (Manassas, VA, USA) and grown in RPMI-1640 medium
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), penicillin (100 U/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and streptomycin
(100 g/ml; Sigma-Aldrich; Merck KGaA) in a humidified incubator
with 5% CO2 at 37°C. H1299 cells were reported to be
p53-null, whereas A549 cells express wild-type p53 (p53.free.fr/Database/Cancer_cell_lines/HB_cell_lines.html).
For the serum-free culture experiment, cells were plated with
complete medium containing 10% FBS overnight, and were then washed
with serum-free medium twice and maintained in serum-free medium
for the indicated time periods.
p53 plasmid and gene transfection
pcDNA-p53 plasmid carrying p53 cDNA was provided by
Dr Xufeng Chen (University of California Los Angeles, CA, USA)
(16). In order to overexpress p53
protein, H1299 cells were cultured and stably transfected with
pcDNA-p53 plasmid or vector-only control pcDNA3.1 using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Stable population
transfection was obtained following selection with 1 µg/ml G418
(Amresco, LLC, Solon, OH, USA) for 2 weeks.
p53 and GPRC5A small interfering
(si)RNA and gene knockdown
The siRNA duplexes targeting p53 and GPRC5A were
obtained from Invitrogen; Thermo Fisher Scientific, Inc. In order
to knockdown p53 or GPRC5A expression in A549 cells,
sub-confluently cultured A549 cells were transfected with p53
siRNA, GPRC5A siRNA, or negative control siRNA using the RNAiMAX
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. p53 and GPRC5A expression
was assessed 3 days post-transfection using western blotting. The
p53 siRNA sequence was 5′-GACUCCAGUGGUAAUCUACTT-3′; the GPRC5A
siRNA sequences was 5′-AGGCAGCAUUUUUCGCCUGTT-3′ and the negative
control (NC) siRNA sequence was 5′-GUAGAUUACCACUGGAGUCTT-3′. All
siRNA oligos were from Invitrogen; Thermo Fisher Scientific,
Inc.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed into cDNA using the Thermoscript RT system (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocols. qPCR amplification was conducted in the ABI7000 sequence
detector (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
the SYBR-Green PCR Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Expression levels of GPRC5A and p53 mRNA were expressed as a ratio
compared with the expression level of GAPDH mRNA and measured using
the 2−ΔΔCq method (17). GPRC5A primer sequences were
5′-GCCTCACCTTCGCCTTCATC-3′ and 5′-CAACTCGTTTCGATTTCTGACAA-3′; p53
primer sequences were 5′-CCTCAGCATCTTATCCGAGTGG-3′ and
5′-TGGATGGTGGTACAGTCAGAGC-3′; and GAPDH primer sequences were
5′-GGCTGAGAACGGGAAGCTTGTCAT-3′ and 5′-CAGCCTTCTCCATGGTGGTGAAGA-3′.
PCR conditions were set to an initial denaturation at 95°C for 5
min; and 40 cycles of 95°C for 30 sec, 65°C for 60 sec and 72°C for
45 sec, using a mixture containing 2 µl cDNA, 1 µl primer, 5 µl
SYBR-Green PCR Master mix and ddH2O to make a total of
20 µl.
Protein extraction and western
blotting
Cells were washed with ice-cold PBS and lysed in
lysis buffer (150 mM NaCl; 1% Triton X-100; 0.1% SDS; 50 mM
Tris-HCl, pH 8.0; and protease inhibitor mixture), followed by
centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was
collected and assayed for protein concentration using the Bio-Rad
DC Protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA),
and 50 µg of each protein sample was subjected to 12% SDS-PAGE gel
followed by transfer onto a polyvinylidene difluoride membrane (EMD
Millipore, Billerica, MA, USA). Membranes were blocked with 5%
(w/v) milk in TBS-Tween-20 (150 mM NaCl; 10 mM Tris-HCl, pH 7.5;
and 0.1% Tween-20) for 1 h, and were then incubated overnight at
4°C with the corresponding primary antibodies as follows: anti-p53
(sc-126; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
anti-GAPDH (sc-47724; 1:5,000; Santa Cruz Biotechnology, Inc.) and
anti-GPRC5A (10309–1-AP; 1:1,000; ProteinTech Group, Inc., Chicago,
IL, USA). After three time washing with TBST, the membranes were
incubated with secondary antibodies at room temperature for 1 h.
Secondary antibodies (anti-rabbit IgG-HRP; 7074; 1:3,000 and
anti-mouse IgG-HRP; 7076; 1:3,000) were from Cell Signaling
Technology (Danvers, MA, USA). The specific bands were visualized
using enhanced chemiluminescence (MultiSciences Biotech Co., Ltd.,
Hangzhou, China), and the protein levels were quantified by using
NIH ImageJ version 1.6.0_24 (National Institutes of Health,
Bethesda, MD, USA).
Cell viability MTT assay
Cells in the log growth phase were seeded in a
96-well plate at a density of 2×103 cells/well and grown
overnight, followed by transfection with p53, GPRC5A, or NC siRNA
as described above. The cells were cultured for 2, 4 and 6 days. At
the end of each time point, 20 µl 5 mg/ml MTT reagent
(Sigma-Aldrich; Merck KGaA) was added to the culture and incubated
for an additional 4 h. Subsequently, the cell culture medium in
each well was replaced with 150 µl dimethyl sulfoxide and the plate
was analyzed using a BioTek Synergy 2 microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA) at a wavelength of 490 nm.
Cell viability was expressed as a % of the control. Experiments
were performed in triplicate and repeated ≥3 times.
Cellular apoptosis assay
The cellular apoptosis rate was measured using a
luminogenic caspase-3/7 substrate (Caspase-Glo 3/7), which detects
cleaved caspase 3/7 in apoptotic cells as a luminescent signal.
Cells were seeded in a 96-well plate at a density of
2×103 cells/well and transfected with siRNA specific for
p53, GPRC5A or NC siRNA as described above. A total of 6 days
post-transfection, Caspase-Glo 3/7 reagent (Promega Corporation,
Madison, WI, USA) was added into the cell culture and mixed using a
plate shaker at 500 rpm for 30 sec, and cells were incubated for a
further 1 h. The luminescence activity (relative caspase 3 and 7
activities) were measured using the BioTek Synergy 2 Microplate
Reader. Experiments were performed in triplicate and repeated ≥3
times. The cellular apoptosis rate was expressed as a % of the
control.
Statistical analysis
Experimental data are expressed as the mean ±
standard error. A two-tailed Student's t-test was performed to
compare the two different treatment groups. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using SPSS (version 13.0; SPSS,
Inc., Chicago, IL, USA), GraphPad Prism (version 5.0; GraphPad
Software, Inc., La Jolla, CA, USA) and Excel 2007 software
(Microsoft Corporation, Redmond, WA, USA).
Results
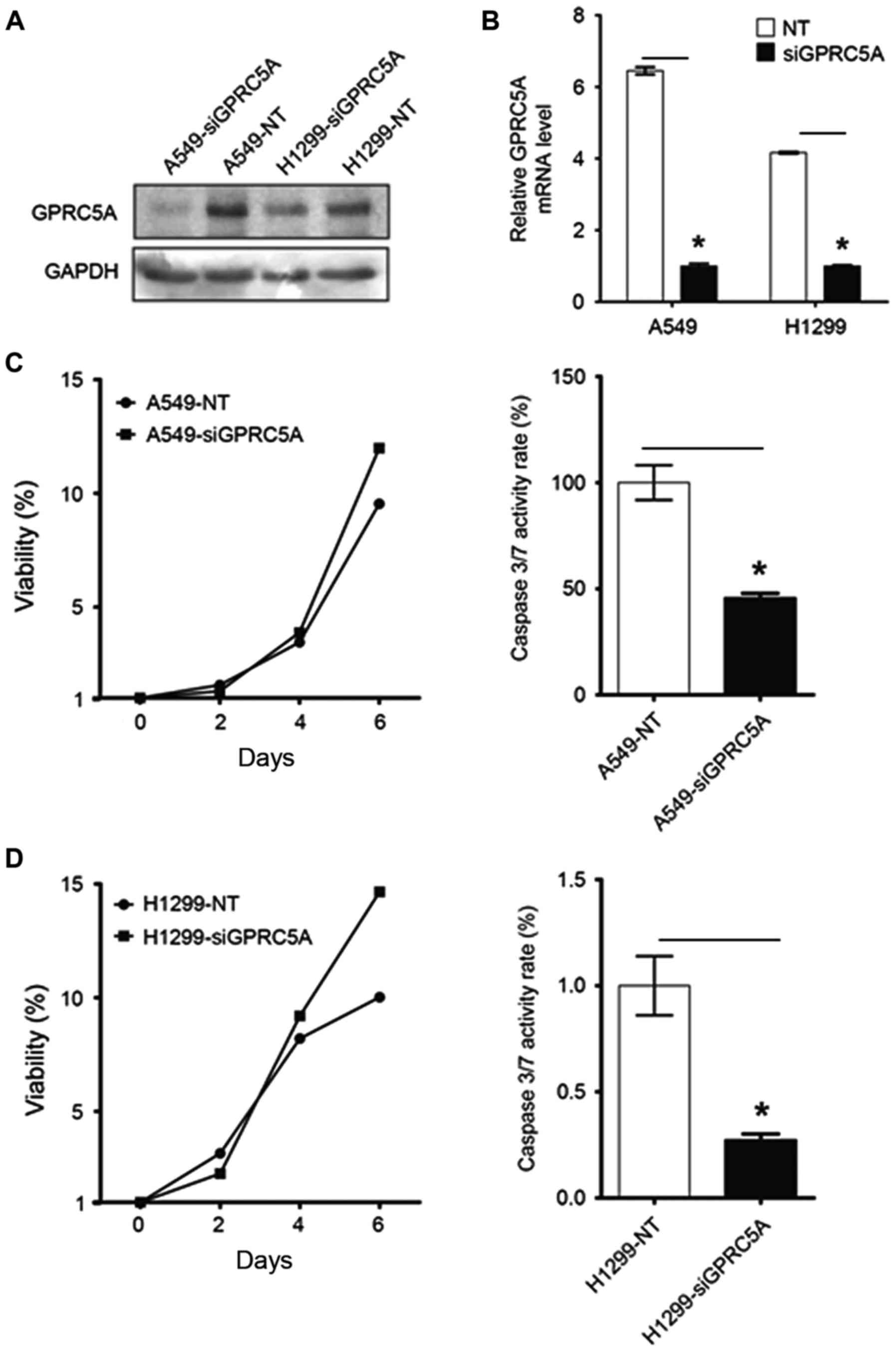
Effects of GPRC5A knockdown on NSCLC
cell viability
In the present study, GPRC5A expression in NSCLC
A549 and H1299 cell lines was knocked down using siRNA constructs,
and the alterations in the mRNA and protein levels of GPRC5A were
assessed using RT-qPCR and western blot analyses. It was observed
that downregulation of GPRC5A expression decreased apoptosis and
enhanced cell survival (Fig.
1).
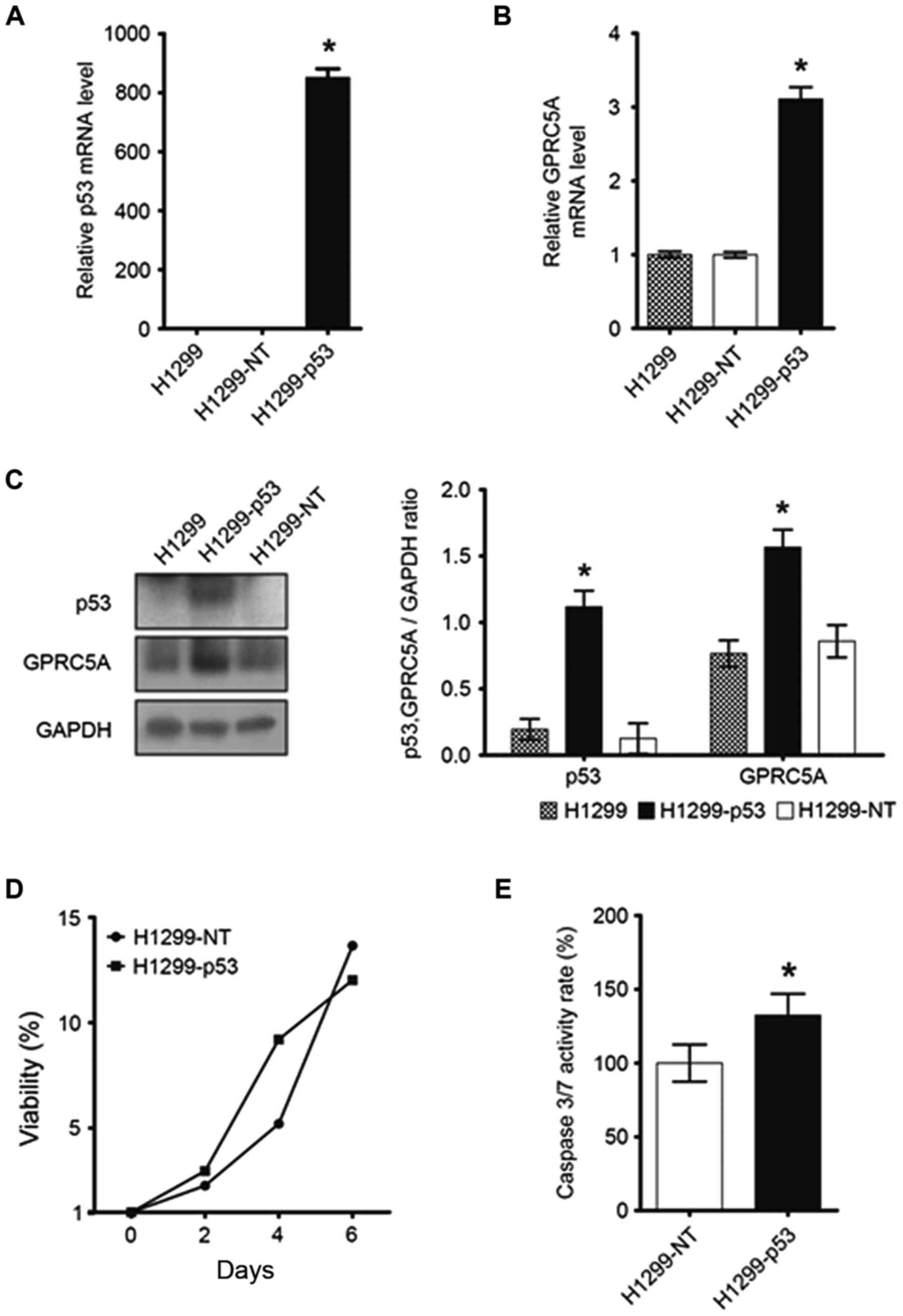
Overexpression of WT p53 protein
increases GPRC5A expression in NSCLC cells
p53-null NSCLC H1299 cells were stably transfected
with WT-p53 cDNA, and it was observed that p53 overexpression in
H1299 cells significantly increased the mRNA and protein expression
of GPRC5A (Fig. 2A-C) compared
with the vector-only controls (P<0.01). It was additionally
observed that p53 overexpression markedly reduced tumor cell
viability (Fig. 2D) and increased
the activity of caspase 3/7, an apoptosis marker, in H1299 cells
(Fig. 2E).
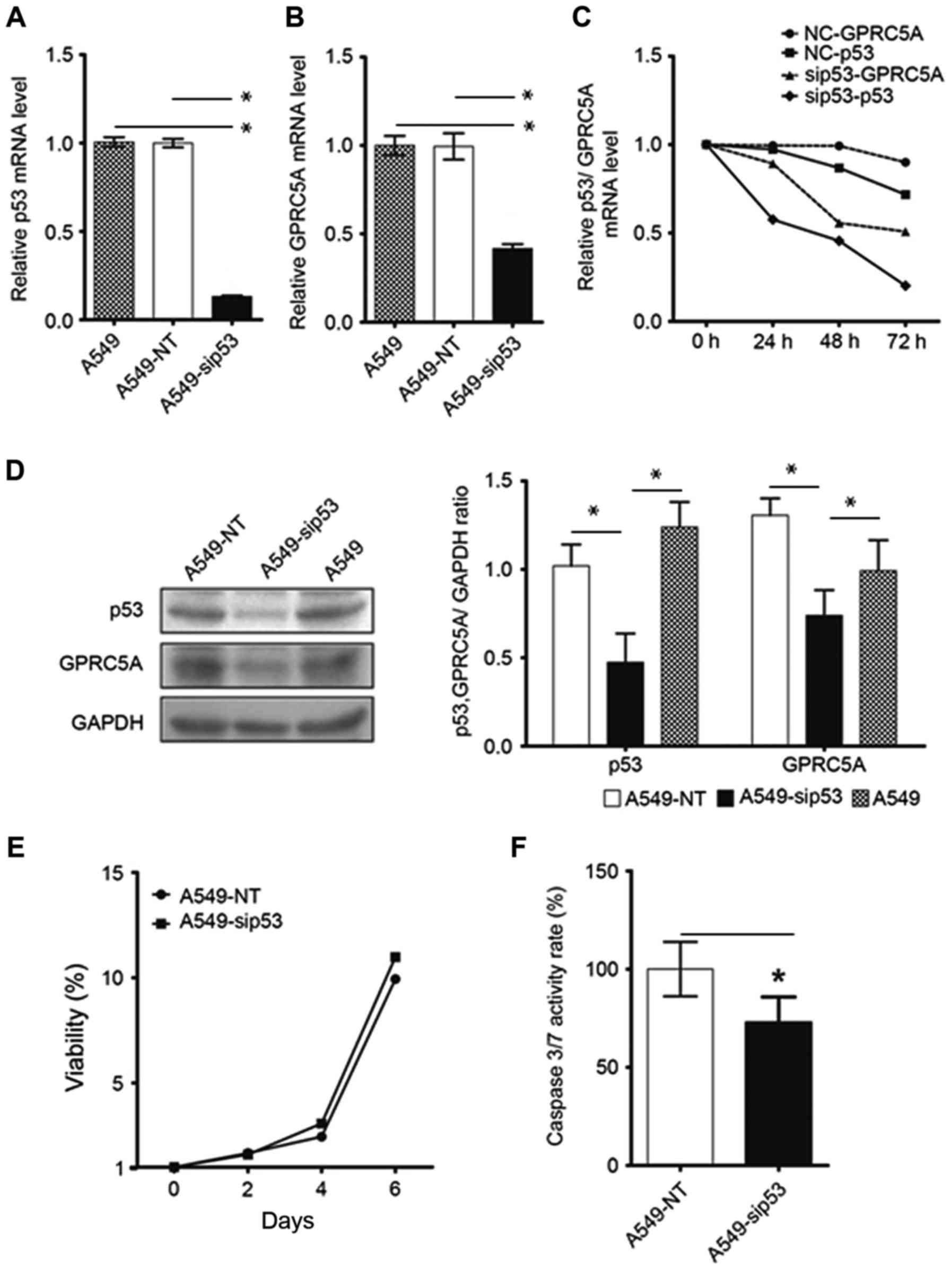
Knockdown of p53 reduces GPRC5A
expression in A549 cells
In NSCLC A549 cells expressing WT-p53, it was
demonstrated that knocking down p53 expression using p53 siRNA
transfection led to downregulated expression of GPRC5A mRNA and
protein (Fig. 3A-D). In addition,
knocking down p53 expression enhanced cell survival (Fig. 3E) and decreased tumor cell
apoptosis (Fig. 3F).
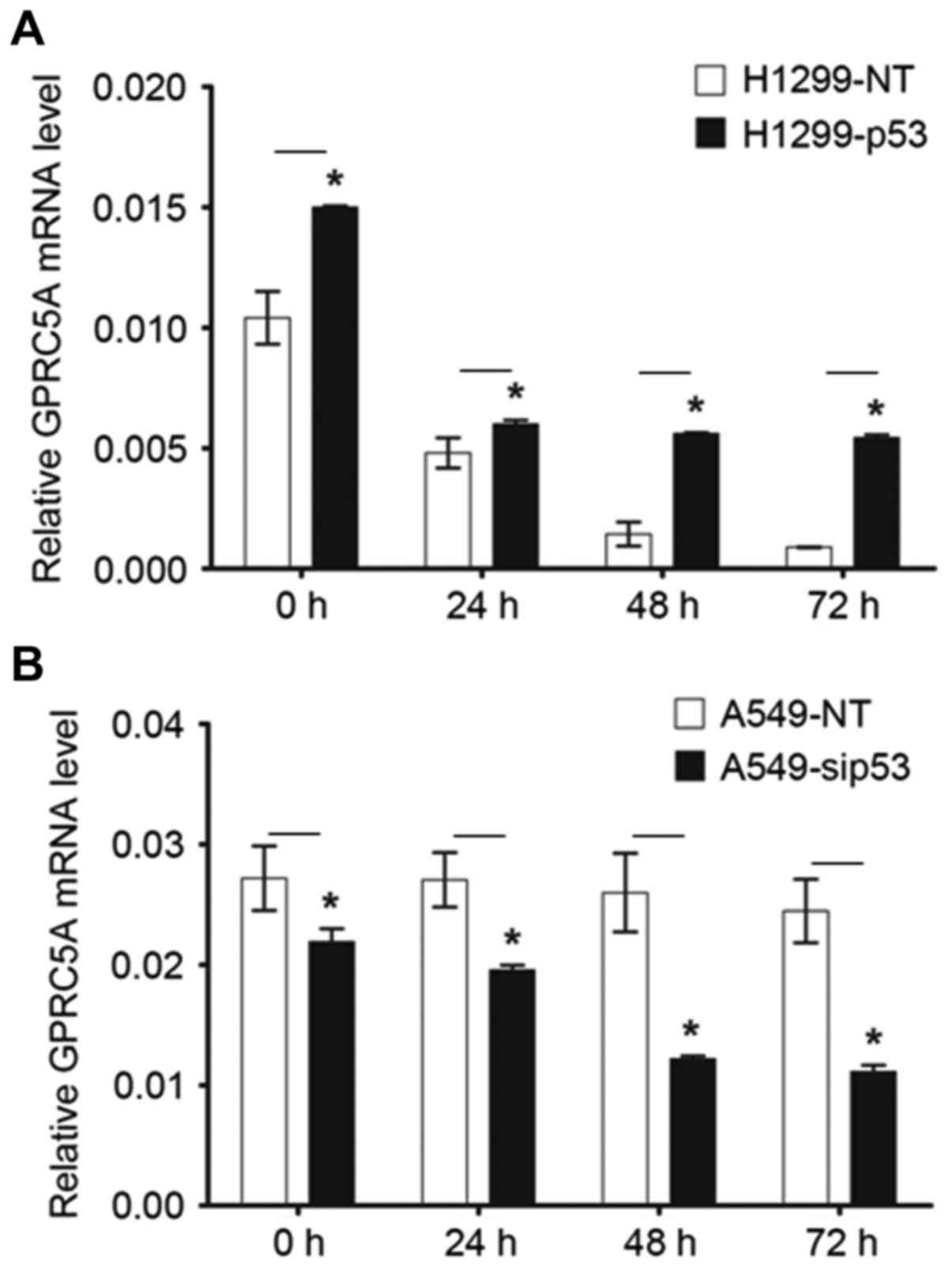
Serum-free culture altered p53 and
GPRC5A expression and cell survival in NSCLC cells
In H1299 cells transfected with control empty vector
plasmid, culturing cells in serum-free medium markedly reduced mRNA
expression of GRPC5A in a time-dependent manner. When cells were
engineered with p53 expression, a decreased level of GPRC5A mRNA
was also identified in serum-free medium cultured cells. However,
GPRC5A mRNA expression level in these cells were significantly
higher than that in cells expressing no p53 (P<0.05). It is
notable that culturing in serum-free condition medium did not
affect the level of GPRC5A mRNA in A549 cells which expressed
wild-type p53. In A549 cells that were transfected with p53-siRNA,
GPRC5A mRNA level decreased in cells cultured with serum-free
medium with statistical significance at indicated each time point
(P<0.05; Fig. 4).
Discussion
p53 is a well-studied protein in the literature. p53
has been described as the guardian of the human genome, as it
conserves genome stability and integrity (4). p53 is able to activate DNA repair
proteins following genomic DNA damage, and arrest cells at the G1/S
phase of the cell cycle to fix DNA damage prior to entering cell
cycle progression. When DNA damage is irreparable, p53 is able to
induce cell apoptosis (18,19).
Previous studies have demonstrated that numerous genes interact
with p53 and are involved in p53 signaling pathways (19); however, there are multiple unknown
factors which regulate the functions of p53 in human cells.
Therefore, the present study investigated the interaction of p53
and GPRC5A as a mediator of p53 function in NSCLC cells in
vitro. It was observed that p53 overexpression in NSCLC cells
markedly increased p53 and GPRC5A levels, while p53 overexpression
and GPRC5A induction reduced tumor cell viability and induced
apoptosis. However, the knockdown of p53 expression additionally
downregulated GPRC5A expression, reduced tumor cell apoptosis and
increased tumor cell viability. The results of the present study
indicate that p53 antitumor activity may be mediated by GPRC5A in
NSCLC cells.
The precise mechanism of action of GPRC5A as a
retinoid-induced gene remains unclear. However, it has been
demonstrated that GPRC5A is able to mediate retinoid antitumor
activity or cellular processes. Previous studies have demonstrated
that the GPRC5A gene contains retinoic acid binding elements
(RAREs), which facilitate the binding of all-trans retinoic acid
and thereby induce GPRC5A expression (5–7).
Cyclic adenosine 5′-phosphate has been reported to upregulate
GPRC5A expression in human aortic smooth muscle cells (20). An additional study demonstrated
that p53 was able to reduce GPRC5A expression in breast and ovarian
cancer cell lines. The oncogenic function of GPRC5A in breast and
ovarian cancer cells (10) is
contrary to the role of GPRC5A in NSCLC demonstrated in the present
study; therefore, GPRC5A may exhibit diverse functions in different
types of cancer, or alterations in the GPRC5A signaling pathway may
serve an important role in carcinogenesis. The present study
investigated the role of p53 in regulating GPRC5A expression in
NSCLC cell lines and demonstrated the opposing functions of p53,
suggesting that p53 and GPRC5A exhibit anti-tumor functions in
NSCLC. In addition, GPRC5A is a retinoid-induced gene (6), and culturing cells in serum-free
conditions inhibited GPRC5A expression. The results of the present
study demonstrated that parental H1299 cells and p53
cDNA-transfected H1299 cells grown in serum-free medium exhibited
decreased levels of GPRC5A expression. Increased expression of
GPRC5A was observed in p53 cDNA-transfected H1299 cells compared
with parental H1299 parental cells, and decreased expression of
GPRC5A was observed in A549 cells when p53 was silenced. The
results of the present study indicate that GPRC5A is able to
mediate p53 antitumor activity in NSCLC.
GPRC5A acts as a tumor suppressor gene in NSCLC and
is markedly downregulated in NSCLC tissue and cell lines (5,6).
Although the mechanism underlying GPRC5A downregulation remains
unknown, studies have demonstrated that a chromosome deletion at
12p12.3 was rare in NSCLC and that epigenetic alteration may
contribute to GPRC5A downregulation (8). In the present study, it was
demonstrated that p53 upregulated GPRC5A expression in NSCLC cells,
and that downregulation of p53 decreased GPRC5A expression in NSCLC
cells. The results of the present study indicate that the loss of
p53 expression in NSCLC may be one of the mechanisms leading to the
decreased in GPRC5A expression in NSCLC.
The results of the present study provide a
proof-of-concept, and additional studies are required to further
elucidate the role of GPRC5A regulation and the association between
GPRC5A and p53, in NSCLC.
Acknowledgements
The present study was supported in part by the
Zhejiang Provincial Health-related Research Projects (grant no.
2014KYB189), the Zhejiang Provincial Chinese Medicine-related
Research Projects (grant no. 2015ZA133), the Medical and Scientific
Research Projects of Hangzhou (grant no. 2016Z02), and the Social
Development Project of Hangzhou (grant no. 20160533B10).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang P, Allen MS, Aubry MC, Wampfler JA,
Marks RS, Edell ES, Thibodeau S, Adjei AA, Jett J and Deschamps C:
Clinical features of 5,628 primary lung cancer patients: Experience
at mayo clinic from 1997 to 2003. Chest. 128:452–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fridman JS and Lowe SW: Control of
apoptosis by p53. Oncogene. 22:9030–9040. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fukasawa K, Wiener F, Woude GF Vande and
Mai S: Genomic instability and apoptosis are frequent in p53
deficient young mice. Oncogene. 15:1295–1302. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng Y and Lotan R: Molecular cloning and
characterization of a novel retinoic acid-inducible gene that
encodes a putative G protein-coupled receptor. J Biol Chem.
273:35008–35015. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tao Q, Fujimoto J, Men T, Ye X, Deng J,
Lacroix L, Clifford JL, Mao L, Van Pelt CS, Lee JJ, et al:
Identification of the retinoic acid-inducible Gprc5a as a new lung
tumor suppressor gene. J Natl Cancer Inst. 99:1668–1682. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye X, Tao Q, Wang Y, Cheng Y and Lotan R:
Mechanisms underlying the induction of the putative human tumor
suppressor GPRC5A by retinoic acid. Cancer Biol Ther. 8:951–962.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kadara H, Fujimoto J, Men T, Ye X, Lotan
D, Lee JS and Lotan R: A Gprc5a tumor suppressor loss of expression
signature is conserved, prevalent, and associated with survival in
human lung adenocarcinomas. Neoplasia. 12:499–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mogi A and Kuwano H: TP53 mutations in
nonsmall cell lung cancer. J Biomed Biotechnol. 2011:5839292011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Q, Ding W, Mirza A, Van Arsdale T, Wei
I, Bishop WR, Basso A, McClanahan T, Luo L, Kirschmeier P, et al:
Integrative genomics revealed RAI3 is a cell growth-promoting gene
and a novel P53 transcriptional target. J Biol Chem.
280:12935–12943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Huang S and Peng SB: Overexpression
of G protein-coupled receptors in cancer cells: Involvement in
tumor progression. Int J Oncol. 27:1329–1339. 2005.PubMed/NCBI
|
|
12
|
Fujimoto J, Kadara H, Men T, van Pelt C,
Lotan D and Lotan R: Comparative functional genomics analysis of
NNK tobacco-carcinogen induced lung adenocarcinoma development in
Gprc5a-knockout mice. PLoS One. 5:e118472010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barta P, Van Pelt C, Men T, Dickey BF,
Lotan R and Moghaddam SJ: Enhancement of lung tumorigenesis in a
Gprc5a Knockout mouse by chronic extrinsic airway inflammation. Mol
Cancer. 11:42012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujimoto J, Kadara H, Garcia MM, Kabbout
M, Behrens C, Liu DD, Lee JJ, Solis LM, Kim ES, Kalhor N, et al:
G-protein coupled receptor family C, group 5, member A (GPRC5A)
expression is decreased in the adjacent field and normal bronchial
epithelia of patients with chronic obstructive pulmonary disease
and non-small-cell lung cancer. J Thorac Oncol. 7:1747–1754. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagahata T, Sato T, Tomura A, Onda M,
Nishikawa K and Emi M: Identification of RAI3 as a therapeutic
target for breast cancer. Endocr Relat Cancer. 12:65–73. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Wong JY, Wong P and Radany EH:
Low-dose valproic acid enhances radiosensitivity of prostate cancer
through acetylated p53-dependent modulation of mitochondrial
membrane potential and apoptosis. Mol Cancer Res. 9:448–461. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lakin ND and Jackson SP: Regulation of p53
in response to DNA damage. Oncogene. 18:7644–7655. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nowsheen S and Yang ES: The intersection
between DNA damage response and cell death pathways. Exp Oncol.
34:243–254. 2012.PubMed/NCBI
|
|
20
|
Hirano M, Zang L, Oka T, Ito Y, Shimada Y,
Nishimura Y and Tanaka T: Novel reciprocal regulation of cAMP
signaling and apoptosis by orphan G-protein-coupled receptor GPRC5A
gene expression. Biochem Biophys Res Commun. 351:185–191. 2006.
View Article : Google Scholar : PubMed/NCBI
|