Introduction
Acute pancreatitis (AP) is a common critical illness
in the general surgery department and intensive care unit (ICU). AP
can develop rapidly and cause multiple organ dysfunction syndrome
and even endanger lives (1). AP is
associated with long-term drinking, increased triglyceride levels,
and mechanical pancreatic duct obstruction. The development of AP
is usually attributed to stimulation of the pancreas by excessive
production, release, or early activation of proteolytic enzymes
(2). Injured acinar cells release
inflammatory mediators to activate immune cells and induce
inflammatory cell infiltration, thus promoting the development and
aggravation of AP (3,4).
Severe AP usually causes functional injury of
distant organs, especially lung injury. Multiple studies have
indicated that the mechanism underlying AP-induced lung injury may
be associated with excessive release of cytokines and inflammatory
mediators, cell apoptosis, and excessive accumulation of
neutrophils and macrophages (5).
During the induction of distant organ dysfunction by AP through
intercellular and interorgan signaling, reactive oxygen species
might be a key factor (6,7). However, the specific mechanism and
the processes in the inflammatory reactions and injury of distant
organs induced by AP remain unclear.
The present study evaluated the extracellular
microRNA (miRNA) expression profiles of the culture medium from
activated pancreatic acinar cells and of mesenteric lymph samples
from an AP animal model, using microarray analysis. The
differential miRNA expression was then validated in plasma samples
from patients with AP, in order to provide evidence for potentially
novel therapeutic targets in the treatment of AP.
Materials and methods
Measurement of miRNAs in the culture
medium of activated AR42J cells
Cell experiments and grouping
Rat pancreatic acinar AR42J cells were purchased
from the China Center for Type Culture Collection (Wuhan, China).
The AR42J cell line was cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin in a 37°C and 5% CO2
incubator (8).
AR42J cells were divided into two groups: The
control group did not receive any treatment, while the experimental
group was treated with 200 µM TLC-S for 40 min. Following
replacement with fresh medium, the cells were then cultured for an
additional 2 h prior to collection of the culture medium.
Microarray detection and data analysis
Total RNA from culture medium was extracted using a
Plasma/Serum Circulating and Exosomal RNA Purification kit (cat.
no. 42800; Norgen Biotek Corp., Thorold, ON, Canada) according to
the manufacturer's instructions. Quality control was performed
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) on has-miR-16 and has-miR-192 (9). According to the recommended Ct value
range of indicators in a literature report, the extracted total RNA
was determined to have sufficient quality for use in the subsequent
microarray experiments (9). The
microarray platform used was a rat µParaflo microRNA microarray
assay (LC Sciences, Houston, TX, USA). The negative control
demonstrated a lower hybridization signal value than the background
signal value. It was concluded that the microarray assay met the
quality standards. Labeling and hybridization experiments were
performed according to the microarray manufacturer's instructions.
The hybridization images were collected using a laser
scanning-based system (GenePix 4000B; Molecular Devices LLC,
Sunnyvale, CA, USA), and digital conversion of images was performed
using the Array-Pro Analyzer image analysis software (Media
Cybernetics, Inc., Rockville, MD, USA). For data analysis, the
background value was first subtracted and then data were normalized
using LOWESS (Media Cybernetics) (2), and the locally weighted regression.
miRNAs with a fold change <0.5 or >2 were considered
differentially expressed miRNAs. P<0.01 was used as the
threshold value for differentially expressed miRNAs (10).
Detection of miRNAs in the mesenteric
lymph of AP rats
Source of gene microarray data
The gene expression profile data were downloaded
from the Gene Expression Omnibus database (National Center for
Biotechnology Information, http://www.ncbi.nlm.nih.gov/geo) with the series
number GSE42455. In the original publication by Blenkiron et
al, intraductal infusion of sodium taurocholate in male Wistar
rats was performed with continuous infusion to prepare the mild AP
model (11). The individual
microarray results from the mild AP samples are available with the
accession numbers GSM1040489, GSM1040490, GSM1040491, GSM1040492,
and GSM1040493. Intraductal infusion of sodium taurocholate
combined with intermittent infusion and maintaining blood pressure
at 60–65 mmHg was performed to prepare the relatively severe AP
model. The individual microarray results for the severe AP samples
are available with the accession numbers GSM1040484, GSM1040485,
GSM1040486, GSM1040487, and GSM1040488. The results of the
sham-operated group (control) are available with the accession
numbers GSM1040479, GSM1040480, GSM1040481, GSM1040482, and
GSM1040483.
Analysis of microarray data
The platform of this experiment was the GPL8786
Affymetrix Multispecies miRNA array (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The Robust Multi-array Average (RMA)
algorithm was used to calculate the expression level, and the
Microarray Suite algorithm version 5.0 Affymetrix; Thermo Fisher
Scientific, Inc.) was used to calculate the detection calls
(11). Probes with low expression
were filtered and only the groups of samples with at least 2
detection calls were retained. The linear models for microarray
data (LIMMA) differential gene screening algorithm was used to
screen upregulated and downregulated miRNAs (11). Fold changes were calculated, and
miRNAs with a fold change <0.5 or >2 were considered
differentially expressed miRNAs. The P-value cutoff was 0.01.
Target gene prediction and functional enrichment
analysis of miRNAs
The TargetScan (http://www.targetscan.org/), miRanda, (http://www.microrna.org/microrna/home.do), and PicTar
(http://pictar.mdc-berlin.de/) programs
were used to predict the target genes of the differentially
expressed miRNAs. The target genes predicted by the three types of
software were screened based on the scoring criteria of each
program. In the TargetScan algorithm, target genes with a context
percentile <50 were excluded. In the miRanda algorithm, target
genes with Max-Energy >-10 were excluded. In the PicTar
algorithm, target genes with ddG >-5 were excluded. The results
that were common to all three programs were selected as the final
target genes of differential miRNAs.
The predicted target genes of differential miRNAs
were used for Gene Ontology (GO) functional annotation (12). Using the Fisher's exact test,
enrichment analysis was performed for each GO category. The
miRNA-target gene GO network was constructed using the Cytoscape
software version 2.8.3 (13).
GO-biological process analysis was performed with
the Database for Annotation, Visualization and Integrated Discovery
(DAVID) software (david.ncifcrf.gov/), by inputting the list of genes
that were predicted to be targets of miR-24 (14).
Patient information
In total, 32 patients were selected for the present
study that were admitted into the Department of General Surgery and
the ICU ward of Harbin Medical University between July 2015 and
December 2015. The inclusion criteria were: Newly admitted patients
were assessed within 48 h and met the diagnostic criteria of the
2012 Atlanta AP guidelines (8).
Patients with the following conditions were excluded from the
study: Did not agree to participate, were released or died within
24 h of admission, were younger than 18 years of age, had AP due to
pancreatic injury resulting from trauma, had chronic pancreatitis,
or had a past history of pancreatitis. According to the severity
classification and computed tomography (CT) classification based on
the Atlanta international guidelines (15), the included patients were divided
into the mild AP group and the moderately severe AP group. Each
group included 10 patients. In addition, plasma samples from 10
healthy individuals were used as the control. Clinical information
about the patients involved in the present study is listed in
Table I. The present study was
approved by the Ethics Committee of the First Affiliated Hospital
of Harbin Medical University (Harbin, China) and conformed to the
ethics principles in the Declaration of Helsinki of the World
Medical Association (WMA), and the International Ethical Guidelines
for Biomedical Research Involving Human Subjects of the Council for
International Organizations of Medical Sciences (CIOMS) (9,16).
The patient's family and patients agreed to join the experiment and
signed an informed consent form. The approval number was HYYKY/WZLS
201560.
 | Table I.Clinical characteristics of patients
in the mild AP group, moderately severe AP group, and healthy
control group. |
Table I.
Clinical characteristics of patients
in the mild AP group, moderately severe AP group, and healthy
control group.
| Indicator | Mild AP group | Moderately severe
AP group | Control group | Statistical
method | P-value |
|---|
| Gender (n) |
|
|
| Fisher's exact
test | 0.8792 |
|
Female | 3 (30%) | 2 (20%) | 4 (40%) |
|
|
|
Male | 7 (70%) | 8 (80%) | 6 (60%) |
|
|
|
Total | 10 | 10 | 10 |
|
|
| Age (years) | 47.90±17.95 | 47.10±15.79 | 44.00±12.96 | Fisher's exact
test | 0.8428 |
| Blood amylase
(µ/l) | 491.28±471.03 | 413.78±437.51 |
| 0.11 (t-value) | 0.9165 |
| Urine amylase
(µ/l) | 2505.60
(1160–5346) | 3957.90
(305–6000) |
| 0.2656
(z-value) | 0.7906 |
| Calcium
(mmol/l) | 2.241±0.2002 | 1.74±0.262 |
| 4.8 (t-value) | 0.0001 |
| CT
classification |
|
|
|
|
|
| a | 0 | 0 |
|
|
|
| b | 1 | 0 |
|
|
|
| c | 9 | 0 |
|
|
|
| d | 0 | 0 |
|
|
|
| e | 0 | 10 |
|
|
|
Collection of patient's peripheral blood fluid
(10 ml)
After extraction of blood, the blood was sent to The
Central Laboratory of the First Hospital Affiliated to Harbin
Medical University within 2 h. Plasma samples were obtained by
using a low speed desktop centrifuge (at 3,000 × g for 15 min at
20°C). The plasma sample was stored at −80°C. Blood, urine and
calcium analysis was determined by the automatic biochemical
apparatus (Beckman AU5800 automatic biochemical analyzer).
Detection of plasma miR-24 in patients using
RT-qPCR
Total RNA was extracted from plasma samples using
the Ultrapure RNA extraction kit (cat no. CW0581; CWbio Co., Ltd.,
Beijing, China). A total of 5 µg RNA was subjected to 1% agarose
gel electrophoresis to determine the RNA integrity. The reverse
transcription of miRNAs was performed using a cDNA first-strand
synthesis reagent kit (cat. no. CW2141; CWbio Co., Ltd), according
to the manufacturer's instructions (17). The forward primer (5′-3′) of miR-24
was TGG CTC AGT TCA GCA GGA ACA G and the reverse primer was CTG
AGG TGC TGT GCG TGA C. Expression of U6 spliceosomal RNA (U6) was
also tested as the internal reference control. The U6 upstream
primer was CTC GCT TCG GCA GCA CA, and the U6 downstream primer was
AAC GCT TCA CGA ATT TGC GT. The PCR cycling conditions were as
follows: An initial pre-denaturation step at 95°C for 30 sec,
followed by denaturation at 95°C for 5 sec, annealing at 60°C for
20 sec and extension at 72°C for 30 sec for a total of 40 cycles.
Fluorescence-based quantification (SYBR Green) was achieved using
the Exicycler™ 96 Real-Time Quantitative PCR system. An ABI 7500
fluorescence quantitative PCR machine was used. Relative
quantification of the data was performed using the
2−∆∆Cq method (9).
Statistical analysis
SPSS software version 17.0 (SPSS, Inc., Chicago, IL,
USA) was used. The Fisher's exact probability test was performed to
compare the differences in gender and age among the three groups.
Blood amylase and calcium levels were analyzed using a two-way
analysis of variance followed by the Newman-Keuls post hoc test.
miR-24 expression results from the patient samples were
statistically analyzed using analysis of variance followed by least
significant difference test for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference. Each
experiment was repeated 3 times independently.
Results
Differentially expressed miRNAs in
activated AR42J cells and in mesenteric lymph samples from AP
rats
A gene microarray analysis of activated AR42J cells
compared with untreated cells (group 1) was performed in the
present study to identify differentially expressed extracellular
miRNAs released in the culture media. For a more comprehensive
analysis, results from a gene microarray database of mesenteric
lymph samples of mild AP (group 2) and severe AP (group 3) were
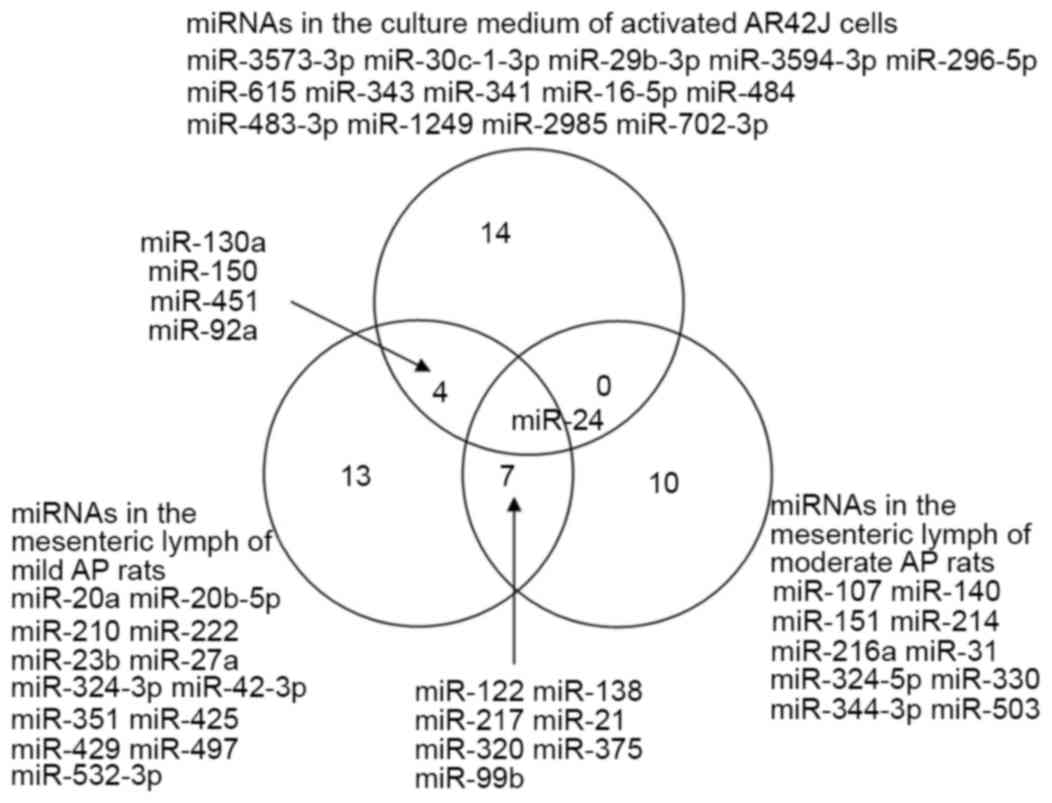
also investigated in the present study. Fig. 1 illustrates the differentially
expressed miRNAs comparison between the three groups. The results
revealed that several differentially regulated miRNAs were common
in more than one group. However, miR-24 was the only miRNA that was
significantly and differentially regulated by microarray analysis
in all three groups: The activated AR42J cells, the mesenteric
lymph in the mild AP rats, and the mesenteric lymph of the severe
AP rats (Fig. 1).
Prediction of functions regulated by
miR-24
Three programs, TargetScan, miRanda, and PicTar,
were used for prediction of the potential target genes of miR-24.
Then the potential target genes that were identified in common by
all the three programs were used for further GO enrichment
analysis, in order to predict the cellular functions that miR-24
may be important in. According to the enrichment index, the most
significant functions identified were: ‘Positive regulation of
calcium-mediated signaling’, ‘activation of JUN kinase activity’,
‘calcium ion transport’, ‘regulation of Rho protein signal
transduction’, ‘negative regulation of protein kinase B signaling
cascade’, and ‘T cell receptor signaling pathway’ (Fig. 2). As illustrated in Fig. 3, the predicted miR-24 target genes
associated with these functions included vav guanine nucleotide
exchange factor 2 (Vav2), spleen associated tyrosine kinase (Syk),
luteinizing hormone/choriogonadotropin receptor (Lhcgr), SLC9A3
regulator 1 (Slc9a3r1), calcium channel voltage-dependent beta 1
subunit (Cacnb1), calcium channel voltage-dependent N type alpha 1B
subunit (Cacna1b), B-cell lymphoma/leukemia 10 (Bcl10) and FYVE
RhoGEF and PH domain-containing protein 3 (Fgd3).
Clinical characteristics of the
patients in the mild AP group, the moderately severe AP group, and
the healthy control group
A statistical analysis was performed for the various
clinicopathological features of the patients involved in the
present study. The results revealed that age and gender did not
significantly differ among the control group, the mild AP group,
and the moderately severe AP group (Table I). The levels of blood and urine
amylases did not significantly differ between the mild AP group and
the moderately severe AP group (Table
I). However, the calcium levels differed significantly between
the mild and severe AP groups (P=0.0001; Table I). According to the CT
classification guidelines (15),
CT in the mild AP group indicated that there was no obvious body
fluid accumulation, whereas CT in the moderately severe AP group
revealed the presence of obvious and large amounts of effusion and
even the presence of encapsulated effusion and necrotic tissues
(Table I).
miR-24 levels in patients with AP
The miR-24 levels were measured in the plasma of
patients in the healthy control group, the mild AP group, and the
moderately severe AP group. The results revealed that the mild AP
group and the moderately severe AP group had significantly higher
expression levels of miR-24 compared with the control group
(Fig. 4). However, there was no
difference between the mild AP group and the moderately severe AP
group (Fig. 4).
Discussion
This study screened the extracellular miRNA profile
in the culture medium of activated rat pancreatic acinar AR42J
cells and of the mesenteric lymph of AP rats and discovered that
miR-24 was differentially expressed in both groups with high
expression. In addition, analysis of expression by RT-qPCR in
plasma samples of clinical AP patients demonstrated that miR-24 was
significantly upregulated in both the mild AP group and the
moderately severe AP group compared with healthy individuals. These
results indicated that during the development of AP, miR-24 might
be released in excess into circulating blood. Combined with the
findings of a previous study on AP and distant organ injury
(18), the present results
suggested that miR-24 might have an important role in the process
of AP-induced distant organ dysfunction.
Extracellular miRNAs have gradually become a focus
of intense study in the medical field (19,20).
In 2008, Jeyasee et al first discovered the presence of
miRNAs in circulating blood, and since then miRNAs have been
demonstrated to be stably present in various extracellular body
fluids, including plasma, serum, urine, tears, and lymph (11,21,22).
Extracellular miRNAs could integrate into recipient cells and
interfere with target mRNAs to regulate the function of recipient
cells (23–25). The miRNAs in circulating plasma or
serum are packaged into microvesicles and exosomes. Exosomes are
small endocytic vesicles, 30–100 nm in size. They are released into
the extracellular environment and can fuse with the plasma membrane
of recipient cells to induce signal transduction (26,27).
Valadi et al (28) prepared
total RNA from exosomes and discovered small RNAs, including
miRNAs. Hunter et al (29)
reported that miRNAs were also expressed in microvesicles in
circulating plasma and peripheral blood mononuclear cells in normal
individuals (29). Extracellular
miRNAs may be useful as novel biomarkers for diagnostic and
prognostic purposes and as potential novel therapeutic targets
(30.31).
Previous studies on AP and miRNAs rarely evaluated
the mechanism and treatment of distant organ injury. Anatomically,
mesenteric lymph nodes reach the subclavian vein through the
thoracic duct and bypass the portal vein and liver. It is generally
considered that potential toxic factors influence other internal
organs through mesenteric lymph nodes and bypass the portal vein
and liver (32). A previous study
has indicated that the intestinal ischemia/reperfusion injury
caused by the release of unknown factors into mesenteric lymph
nodes could aggravate AP (33).
Several studies have suggested that the significantly and
specifically upregulated miRNAs in pancreatic tissues, including
miR-216a, miR-216b, and miR-375, had pancreatic tissue specificity.
The miRNA results from the mesenteric lymph samples of AP mice
revealed miR-216a, miR-375 and miR-217 to be significantly
upregulated (11,34). These results indicated that
mesenteric lymph is likely to contain miRNAs derived from pancreas.
Therefore, miRNAs that were present in both pancreatic acini and
mesenteric lymph had the greatest possibility of serving a role in
the process of the distant organ injury caused by AP or in
potential treatments through regulation of signaling pathways.
Furthermore, systemic inflammatory response syndrome
(SIRS) occurs in combination with compensatory anti-inflammatory
response syndrome (CARS) in AP. In the process of inflammatory
reactions, anti-inflammatory therapy with the combined inhibition
of multiple pro-inflammatory factors has been proven to be
effective. For example, treatment using an interleukin (IL)-10
receptor agonist and an anti-IL-8 antibody showed beneficial
effects, and antagonizing intercellular adhesion molecule-1 could
attenuate lung injury in mice (30). Although the association between
pancreatic nuclear factor (NF)-κB and the activation of trypsinogen
in AP is still controversial, a recent study indicated that these
processes could both induce inflammation (35); therefore, a combined therapy to
inhibit pro-inflammatory responses might be more useful (35). The predominant pathological state
of patients with severe AP might be altered from SIRS to CARS by
successful treatment (36). The
endogenous anti-inflammatory lipid mediator resolvin D1 in AP could
improve pancreatic injury and lung injury through the inhibition of
NF-κB activation (37).
Circulating miRNAs serving as mediators of intercellular signal
transduction have also been confirmed to serve an anti-inflammatory
role. Hyaluronic acid-poly (ethyleneimine) nanoparticles that
encapsulated miR-223 could regulate the repolarization of
pro-inflammatory M1 macrophages to anti-inflammatory M2 macrophages
(38). The IL-10-dependent
miR-146a exerted anti-inflammatory effects through the Toll-like
receptor-4 pathway in monocytes (39). Therefore, it is foreseeable that
miRNAs with anti-inflammatory activity have potential significance
in the treatment of AP and the control of distant organ injury.
The present study screened miRNAs in the culture
media of activated pancreatic acinar cells and the lymph of an AP
animal model and revealed that miR-24 was the miRNA that was
significantly upregulated in both. Through functional enrichment
analysis, significant functions related to miR-24 were predicted:
‘Positive regulation of calcium-mediated signaling’, ‘activation of
JUN kinase activity’, ‘calcium ion transport’, ‘regulation of Rho
protein signal transduction’, ‘negative regulation of protein
kinase B signaling cascade’, and ‘T cell receptor signaling
pathway’. Previous studies suggested that the dose-dependent
inhibition of Rho-kinase had protective effects on AP, whereas bee
venom could prevent AP and inhibit pancreatic acinar cell death
through the inhibition of c-Jun N-terminal kinase activation
(40,41). The phosphoinositide 3-kinase and
protein kinase B signal transduction pathways may participate in
the pathological process of lung injury in severe AP through the
upregulation of NF-κB, tumor necrosis factor-α, and IL-lβ (18). In addition, calcium is important in
the process of substance P (SP)-induced chemokine synthesis in
pancreatic acinar cells (42).
Drug treatment targeting the SP-calcium-mediated signaling pathway
might be beneficial for AP therapy. The upregulation of the
expression of the sarcoplasmic reticulum calcium pump in pancreatic
tissues to reduce intracellular calcium overload could also reduce
pancreatic tissue lesions (43).
In the present study, when assaying the plasma of
clinical AP patients, we demonstrated that plasma miR-24 levels
were significantly higher in patients with mild AP group and
moderately severe AP compared with the healthy volunteer control
group. These results indicated that CARS was present during the
systemic inflammatory reaction of AP, which was consistent with the
results from another study (40).
However, if the underlying mechanism of miRNA24 in the process of
inflammation is further researched, novel therapeutic targets may
be found for the treatment of AP, especially for remote organ
damage.
Previous studies on miR-24 also indicated a role in
inflammatory reactions, anti-inflammatory processes, and apoptosis.
Specifically, miR-24 inhibits atherosclerosis and increases
macrophage accumulation. Biological analysis indicated that
miRNA-24 regulated macrophage activation and polarization.
Overexpression of miR-24 inhibited cytokine secretion and exhibited
anti-inflammatory functions (24,44).
Increasing the miR-24 levels induces cell apoptosis by reducing the
Bcl-2 protein expression levels, whereas anti-miR-24 transfection
enhances the inflammatory process and cell apoptosis-related
reactions (31,45). These findings are consistent with
the miR-24 function predicted in the present study. Further studies
are required to elucidate the mechanism of miR-24 and its potential
as a treatment target.
Although experimental studies related to AP and
miRNAs were performed previously, most previous studies evaluated
plasma miRNAs only in AP patients to screen for better diagnosis
and prediction markers. The novelty of the present study was the
discovery of the uniquely expressed extracellular miR-24 in all
models tested: activated pancreatic acinar cells, mesenteric lymph
from an AP rat model, and plasma samples from patients with AP. In
addition, target gene prediction was performed to understand the
major cellular functions that miR-24 may be involved in. The
present results suggested that miR-24 might be of use as a
treatment target in AP and distant organ injury. Further studies on
the functions of miRNAs in AP-related inflammatory pathways and
mechanisms might facilitate the search for novel therapeutic
targets for treating AP and the related distant organ injury.
Acknowledgements
This work was supported by the National Natural
Science Foundation of China (grant no. 81570579). The authors thank
Fenghe Inc. (Shanghai, China) for help in bioinformatics
analysis.
References
|
1
|
An F, Zhan Q, Xia M, Jiang L, Lu G, Huang
M, Guo J and Liu S: From moderately severe to severe
hypertriglyceridemia induced acute pancreatitis: Circulating miRNAs
play role as potential biomarkers. PLoS One. 9:e1110582014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goodwin D, Rosenzweig B, Zhang J, Xu L,
Stewart S, Thompson K and Rouse R: Evaluation of miR-216a and
miR-217 as potential biomarkers of acute pancreatic injury in rats
and mice. Biomarkers. 19:517–529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qin T, Fu Q, Pan YF, Liu CJ, Wang YZ, Hu
MX, Tang Q and Zhang HW: Expressions of miR-22 and miR-135a in
acute pancreatitis. J Huazhong Univ Sci Technolog Med Sci.
34:225–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huan C, Kim D, Ou P, Alfonso A and Stanek
A: Mechanisms of interleukin-22′s beneficial effects in acute
pancreatitis. World J Gastrointest Pathophysiol. 7:108–116. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang X, Zhang B, Chen Q, Zhang J, Lei B,
Li B, Wei Y, Zhai R, Liang Z, He S and Tang B: The mechanism
underlying alpinetin-mediated alleviation of
pancreatitis-associated lung injury through upregulating
aquaporin-1. Drug Des Devel Ther. 10:841–850. 2016.PubMed/NCBI
|
|
6
|
Shi C, Andersson R, Zhao X and Wang X:
Potential role of reactive oxygen species in
pancreatitis-associated multiple organ dysfunction. Pancreatology.
5:492–500. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Araki Y, Andoh A, Yokono T, Asano N,
Yoshikawa K, Bamba S, Ishizuka I and Fujiyama Y: The free radical
scavenger edaravone suppresses experimental closed duodenal
loop-induced acute pancreatitis in rats. Int J Mol Med. 12:121–124.
2003.PubMed/NCBI
|
|
8
|
Wang H, Jiang Y, Lu M, Sun B, Qiao X, Xue
D and Zhang W: STX12 lncRNA/miR-148a/SMAD5 participate in the
regulation of pancreatic stellate cell activation through a
mechanism involving competing endogenous RNA. Pancreatology.
17:237–246. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Y, Wang H, Lu M, Qiao X, Sun B, Zhang
W and Xue D: Pancreatic acinar cells employ miRNAs as mediators of
intercellular communication to participate in the regulation of
pancreatitis-associated macrophage activation. Mediators Inflamm.
2016:63404572016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Blenkiron C, Askelund KJ, Shanbhag ST,
Chakraborty M, Petrov MS, Delahunt B, Windsor JA and Phillips AR:
MicroRNAs in mesenteric lymph and plasma during acute pancreatitis.
Ann Surg. 260:341–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiao X, Sherman BT, da W Huang, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Balthazar EJ: Acute pancreatitis:
Assessment of severity with clinical and CT evaluation. Radiology.
223:603–613. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Council for International Organizations of
Medical Sciences, . International ethical guidelines for biomedical
research involving human subjects. Bull Med Ethics. 1–23. 2002.
|
|
17
|
Gao X, Gulari E and Zhou X: In situ
synthesis of oligonucleotide microarrays. Biopolymers. 73:579–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang X, Wang LZ, Wang YG, Liu L, Fan ZW,
Bai LZ and Lu XG: Expression and significance of
phosphatidylinositol 3-kinase/protein kinase B signal transduction
pathway in severe acute pancreatitis-associated lung injury.
Zhonghua Yi Xue Za Zhi. 90:732–737. 2010.(In Chinese). PubMed/NCBI
|
|
19
|
Usborne AL, Smith AT, Engle SK, Watson DE,
Sullivan JM and Walgren JL: Biomarkers of exocrine pancreatic
injury in 2 rat acute pancreatitis models. Toxicol Pathol.
42:195–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuśnierz-Cabala B, Nowak E, Sporek M,
Kowalik A, Kuźniewski M, Enguita FJ and Stepien E: Serum levels of
unique miR-551-5p and endothelial-specific miR-126a-5p allow
discrimination of patients in the early phase of acute
pancreatitis. Pancreatology. 15:344–351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeyaseelan K, Lim KY and Armugam A:
MicroRNA expression in the blood and brain of rats subjected to
transient focal ischemia by middle cerebral artery occlusion.
Stroke. 39:959–966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamada S, Masamune A, Kanno A and
Shimosegawa T: Comprehensive analysis of serum microRNAs in
autoimmune pancreatitis. Digestion. 91:263–271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Di Gregoli K, Jenkins N, Salter R, White
S, Newby AC and Johnson JL: MicroRNA-24 regulates macrophage
behavior and retards atherosclerosis. Arterioscler Thromb Vasc
Biol. 34:1990–2000. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Katoh M: Therapeutics targeting
angiogenesis: Genetics and epigenetics, extracellular miRNAs and
signaling networks (Review). Int J Mol Med. 32:763–767. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lakkaraju A and Rodriguez-Boulan E:
Itinerant exosomes: Emerging roles in cell and tissue polarity.
Trends Cell Biol. 18:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Wu XH, Luo CL, Zhang JM, He BC
and Chen G: Interleukin-12-anchored exosomes increase cytotoxicity
of T lymphocytes by reversing the JAK/STAT pathway impaired by
tumor-derived exosomes. Int J Mol Med. 25:695–700. 2010.PubMed/NCBI
|
|
28
|
Valadi H, Ekström K, Bossios A, Sjostrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hunter MP, Ismail N, Zhang X, Aguda BD,
Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS One. 3:e36942008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kylänpää ML, Repo H and Puolakkainen PA:
Inflammation and immunosuppression in severe acute pancreatitis.
World J Gastroenterol. 16:2867–2872. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maegdefessel L, Spin JM, Raaz U, Eken SM,
Toh R, Azuma J, Adam M, Nakagami F, Heymann HM, Chernogubova E, et
al: miR-24 limits aortic vascular inflammation and murine abdominal
aneurysm development. Nat Commun. 5:52142014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fanous MY, Phillips AJ and Windsor JA:
Mesenteric lymph: The bridge to future management of critical
illness. JOP. 8:374–399. 2007.PubMed/NCBI
|
|
33
|
Flint RS, Phillips AR, Power SE, Dunbar
PR, Brown C, Delahunt B, Cooper GJ and Windsor JA: Acute
pancreatitis severity is exacerbated by intestinal
ischemia-reperfusion conditioned mesenteric lymph. Surgery.
143:404–413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Endo K, Weng H, Kito N, Fukushima Y and
Iwai N: MiR-216a and miR-216b as markers for acute phased
pancreatic injury. Biomed Res. 34:179–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kylanpaa L, Rakonczay Z Jr and O'Reilly
DA: The clinical course of acute pancreatitis and the inflammatory
mediators that drive it. Int J Inflam. 2012:3606852012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ohmoto K and Yamamoto S: Serum
interleukin-6 and interleukin-10 in patients with acute
pancreatitis: Clinical implications. Hepatogastroenterology.
52:990–994. 2005.PubMed/NCBI
|
|
37
|
Liu Y, Zhou D, Long FW, Chen KL, Yang HW,
Lv ZY, Zhou B, Peng ZH, Sun XF, Li Y and Zhou ZG: Resolvin D1
protects against inflammation in experimental acute pancreatitis
and associated lung injury. Am J Physiol Gastrointest Liver
Physiol. 310:G303–G309. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tran TH, Krishnan S and Amiji MM:
MicroRNA-223 induced repolarization of peritoneal macrophages using
CD44 targeting hyaluronic acid nanoparticles for anti-inflammatory
effects. PLoS One. 11:e01520242016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Curtale G, Mirolo M, Renzi TA, Rossato M,
Bazzoni F and Locati M: Negative regulation of Toll-like receptor 4
signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA.
110:11499–11504. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Awla D, Hartman H, Abdulla A, Zhang S,
Rahman M, Regnèr S and Thorlacius H: Rho-kinase signalling
regulates trypsinogen activation and tissue damage in severe acute
pancreatitis. Br J Pharmacol. 162:648–658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bae GS, Heo KH, Park KC, Choi SB, Jo IJ,
Seo SH, Kim DG, Shin JY, Kang DG, Lee HS, et al: Apamin attenuated
cerulein-induced acute pancreatitis by inhibition of JNK pathway in
mice. Dig Dis Sci. 58:2908–2917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ramnath RD, Sun J and Bhatia M: Role of
calcium in substance P-induced chemokine synthesis in mouse
pancreatic acinar cells. Br J Pharmacol. 154:1339–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xue P, Deng LH, Zhang ZD, Yang XN, Xia Q,
Xiang DK, Huang L and Wan MH: Effect of Chaiqinchengqi decoction on
sarco/endoplasmic reticulum Ca2+-ATPase mRNA expression
of pancreatic tissues in acute pancreatitis rats. World J
Gastroenterol. 14:2343–2348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fordham JB, Naqvi AR and Nares S: miR-24
regulates macrophage polarization and plasticity. J Clin Cell
Immunol. 6:pii:3622015.
|
|
45
|
Zhu Y, You W, Wang H, Li Y, Qiao N, Shi Y,
Zhang C, Bleich D and Han X: MicroRNA-24/MODY gene regulatory
pathway mediates pancreatic β-cell dysfunction. Diabetes.
62:3194–3206. 2013. View Article : Google Scholar : PubMed/NCBI
|